Abstract
Objective:
To develop a visual analytic system to help medical professionals improve disease diagnosis by providing insights for understanding disease progression.
Methods:
We develop MatrixFlow, a visual analytic system that takes clinical event sequences of patients as input, constructs time-evolving networks and visualizes them as a temporal flow of matrices. MatrixFlow provides several interactive features for analysis: 1) one can sort the events based on the similarity in order to accentuate underlying cluster patterns among those events; 2) one can compare co-occurrence events over time and across cohorts through additional line graph visualization.
Results:
MatrixFlow is applied to visualize heart failure (HF) symptom events extracted from a large cohort of HF cases and controls (n=50,625), which allows medical experts to reach insights involving temporal patterns and clusters of interest, and compare cohorts in novel ways that may lead to improved disease diagnoses.
Conclusions:
MatrixFlow is an interactive visual analytic system that allows users to quickly discover patterns in clinical event sequences. By unearthing the patterns hidden within and displaying them to medical experts, users become empowered to make decisions influenced by historical patterns.
Introduction
There is a substantial challenge to diagnosing many diseases in an early state, as symptoms may rarely emerge as a solitary disease process. Comorbidities may mimic or mask the presence of a disease, and potentially can lead to false-positive or negative diagnoses. In practice, physicians often make difficult diagnoses in the moment using their clinical knowledge, and not necessarily based on a quantitative assessment of longitudinal patient data from electronic health records (EHRs). This is mainly due to the lack of available, analytical tools to help such decision makers extract meaningful patterns and insights from EHRs in a timely manner.
One motivating example is the clinical complexity and heterogeneity of heart failure (HF). HF has posed challenges to developing standardized criteria for its diagnosis. The Framingham HF criteria, originally published in 1971, were based on clinical data acquired in the 1950s and 60s13. In that study, two or more major criteria or one major and two or more minor criteria are used as the diagnosis criteria for HF. The challenges for making the correct HF diagnosis earlier are 1) how to correlate the sparse signals of a single patient across time and encounters, and 2) how to leverage historical data of other similar patients to identify the emerging pattern earlier.
To address such challenges, we propose MatrixFlow, a visual analytic tool designed to help aid medical decision makers and researchers by making the subtle trends of disease progression more obvious. The goal of our work is that by unearthing the hidden patterns in patient health records in MatrixFlow, emerging health risks may become more discoverable and earlier diagnoses of diseases can occur so clinicians and patients can proactively develop preventative strategies to reduce negative future outcomes.
Our analytics work by extracting clinical event sequences from patient EHR data and then constructing a temporal network of co-occurring events to model the relationships between events as a disease progresses over time. The patterns in the evolution of the disease are then revealed in our interactive visualization as a temporal flow of matrices, MatrixFlow. MatrixFlow provides several interactive features for analysis: 1) one can sort the events based on the similarity in order to accentuate underlying cluster patterns among those events; 2) one can compare co-occurrence events over time and across cohorts through additional line graph visualization.
We demonstrate that our approach is effective on a large population (n=50,625) by analyzing over 3.3 million clinical notes. Our system was then used in collaboration with medical experts, who provide strong evidence that our visual analytics system supports their goals of a better understanding of disease progression and potential improvement of the diagnostic capabilities of medical practitioners.
Background
There are many challenges when trying to extract meaningful patterns and insights from electronic health records, including overwhelming amounts of data, time-limited practitioners, and gaps between the type of raw data collected and the information needs of clinical decision makers9. However, visual analytics is seen as a promising solution to many of these challenges. In fact, there is already a growing use of visualization to support clinical research and patient care1,6,7,14. There have been several visual analytics to visualize clinical event sequences, notably LifeLines18, which supports the discovery of temporal categorical patterns across patient records, and LifeFlow19, which provides an overview of patient event sequences to help medical researchers understand the pattern of transfers within hospitals for quality control.
However, our work does not focus on visualizing clinical event sequences, but instead focuses on visualizing clinical event networks of a patient population based on event co-occurrence over time. There is a rich history on visualizing networks, but most employ techniques on static networks that do not change over time12. One common technique for visualizing static networks is using an adjacency matrix representation1,11. However, the recent proliferation of longitudinal network data has resulted in a need for developing visualization techniques for dynamic networks, resulting in a recent survey3. Common approaches for visualizing include rendering the network as a node-link diagram that users can advance through time or watch as a dynamic movie16. Another approach, TimeMatrix uses a matrix-based visualization, where each cell of the matrix contains a bar chart to illustrate how nodes and edges are evolving over time20. To the best of our knowledge, there have been no systems designed to support the temporal visualizations of clinical event networks to date.
Methods
We propose MatrixFlow, a visual analytics system for understanding the evolution of clinical events. We discuss how clinical events are extracted from electronic medical records, how a network of clinical events is modeled, and then how we visually represent the network over time to support insights.
Clinical Event Sequences
In this work, we aim at discovering meaningful patterns from clinical event sequences of patients. Clinical event sequences are simply a series of time-stamped events from a patient’s medical record, such as disease diagnoses, patient symptoms, lab results, and medication orders.
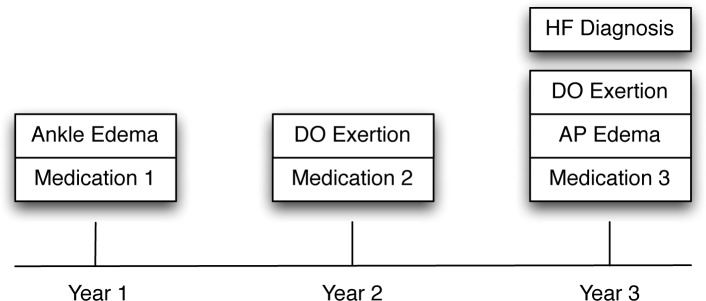
Figure 1 shows an illustrative example of a sequence of clinical events for a patient over 3 years. In the first year, the patient experienced a symptom (Ankle Edema) and was then ordered a medication (Medication 1) to resolve it. A year later, the patient once again sought medical advice due to experiencing Dyspnea On Exertion (DOExertion), and was then ordered a new medication (Medication 2) to replace Medication 1. Finally, in Year 3, the patient returned for medical care experiencing both DOExertion and Acute Pulmonary Edema (APEdema). At this point, physicians were able to diagnose this patient with heart failure (HF), and the doctor also prescribed Medication 3 to manage the symptoms instead of Medication 2.
Figure 1.
An illustrative example of a sequence of clinical events for a fictional patient.
Clinical Event Networks
Our research is interest in determining the co-occurrence of event – that is, when events simultaneously occur. Cooccurrence can be modeled by creating a network of clinical events, where events are nodes, and co-occurring events are connected by an edge.
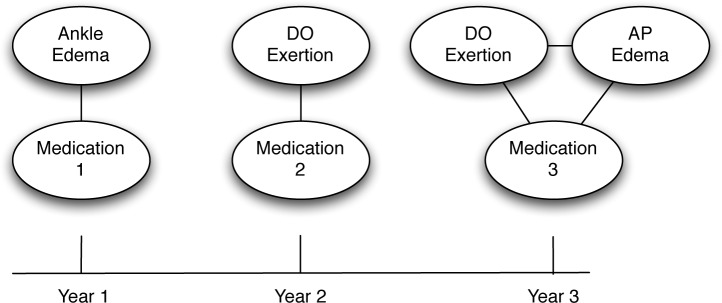
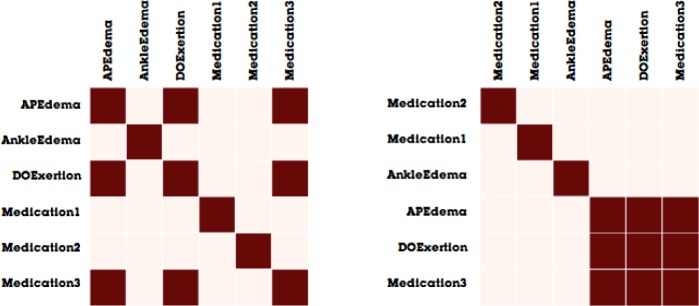
Figure 2 illustrates a clinical event network, derived from the clinical event sequence described in Figure 1. The network representation of the first year would be a simple network with two nodes (Ankle Edema and Medication1). As these two clinical events both occurred within the same time interval, the nodes are connected by an edge. Similarly, the network for the second year also contains two nodes (DOExertion and Medication2), also connected by an edge. Finally, the network representing the third year is slightly more complex network, as there are three connected nodes (DOExertion, APEdema, and Medication 3). While Figure 2 is illustrative at showing how the clinical network evolves over time, it can be difficult to interpret the differences as the graph becomes larger and denser. For this reason, we created our visual analytics system, MatrixFlow.
Figure 2.
An illustrative example of a clinical event network, derived from the sequence described in Figure 1.
Instead of using a traditional node-link diagram (such as the illustration in Figure 2), MatrixFlow relies on its namesake visualization: the adjacency matrix. In matrix visualizations, the columns and rows represent the nodes of the network, whereas each cell in a matrix represents the edge between the two nodes. Figure 3 shows a matrix visualization the same networks illustrated in Figure 2.
Figure 3.
Matrix visualizations of the clinical event networks described in Figure 2.
Matrices have several advantages for our scenario over node-link diagrams8, including: 1) a focus on showing cooccurrences, which are naturally represented by adjacency matrix, 2) a stable layout that makes comparing networks across time easier, and 3) a natural technique for representing edges that represent values. We will return to these advantages later as we describe our system.
Modeling Clinical Event Networks
While the above illustrations have focused on showing the clinical event network for only one patient, MatrixFlow was designed to show patterns of thousands of patients, mining through millions of clinical events. MatrixFlow’s input in a set of patient event sequences. MatrixFlow aligns patients on a common event (e.g., the diagnosis date of HF) and aggregates individual event graphs anchored by that alignment time point. The system then analyzes the historical clinical events before the diagnosis and extract the sequence of events that led up to the diagnosis, representing a trajectory of the disease across the patient population. Users of MatrixFlow have control over the granularity of the time intervals, so they can choose an annual interval as used in Figures 1–3, or a different time window, such as months or weeks.
Once MatrixFlow calculates the diagnosis alignment and intervals are specified, the system models the clinical networks across all patients. In practice, the system determines the nodes in the network by extracting a list of all unique clinical events, and these events become the nodes of the network. The system then efficiently computes, for each event pair, the number of distinct patients who exhibited both events during the same time interval. For instance, if 500 patients out of a population of 10,000 experienced Symptom1 and Symptom2 together in the same interval, then those two nodes would be connected by an edge with a weight of 500/10000 or 5%. However, if zero patients experienced Symptom1 and Symptom 3 in the same time interval, then there would not be an edge connected those two events. This co-occurrence network is computed using our advanced network modeling framework, Orion10. As our networks now feature edges that have varying edge weights, our matrix visualization renders the color of each cell according to a sequential color scale representing the edge weight value (such as the color scale shown at the bottom of Figure 6).
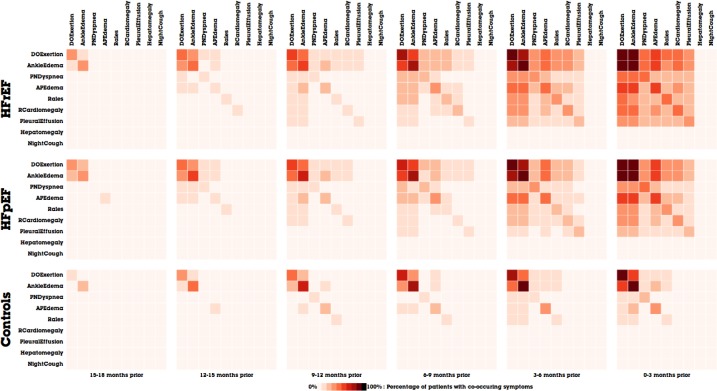
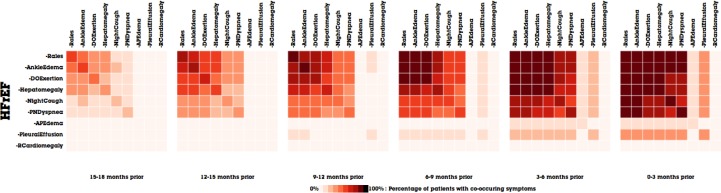
Figure 6.
The temporal evolution of the Framingham symptoms in MatrixFlow. a) The top row of matrices represents the patterns the HFrEF patient cohort. b) The middle row represents the HFpEF patient cohort. c) The bottom row represents the Controls cohort.
Visualizing Clinical Event Networks
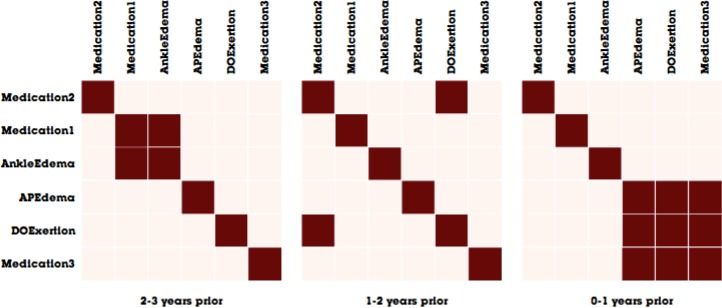
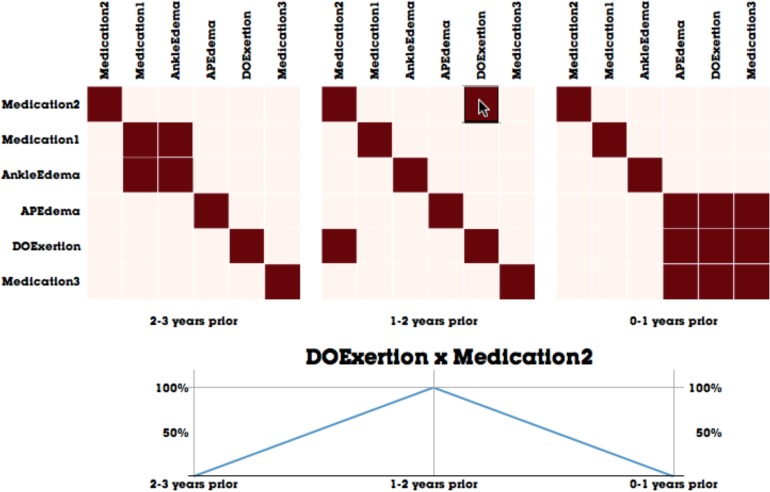
When visualizing matrices, it is important to choose an effective method to sort the order of nodes in order to reveal as many patterns as possible4. As we wish to reveal clusters of clinical events, we employ a greedy hierarchical clustering optimizing Newman’s modularity metric17. From this algorithm, we are able to obtain a sort order that minimizes the distance among connected nodes by ordering the nodes according to the cluster tree produced by our hierarchical clustering algorithm. Figure 4 shows an example of matrix visualization before and after the sorting is produced. In the latter example, well-connected nodes (APEdema, DOExertion, and Medication 3) assemble to form a large box-like structure in the visualization, which makes the cluster more apparent than in the unsorted version on the left.
Figure 4.
An illustrative example of revealing clusters by using hierarchical clustering. The matrix on the left is unsorted, whereas the matrix on the right is sorted to minimize distance among connected nodes in the clinical event network.
In addition to visualizing clinical event networks as matrices, one of MatrixFlow’s main features is the ability to compare networks as they change over time. As has been alluded in Figure 3, MatrixFlow supports this by visualizing matrices side-by-side along a horizontal axis. These small multiples provide an overview of the disease progression over time. However, in order make the temporal trends clearer, users can interact with the matrices to focus on a particular trend of interest. By moving the mouse over an edge cell, a line graph is presented that plots the evolution of the co-occuring events across all time intervals. For instance, in Figure 5, the user selected the cooccurrence of DOExertion and Medication 2 and the temporal line graph is visualized.
Figure 5.
The temporal visualization capabilities of MatrixFlow, where matrices are aligned on a horizontal axis of time, and interactive line charts clarify temporal trends of edges of interest.
Comparing the Clinical Event Networks of Cohorts
Our system also supports analyzing and comparing various cohorts of patients. For example, users may wish to compare and contrast the evolution of clinical events across different patient populations, such as patients with a disease compared to a control group. Each cohort is represented by a row of matrices, and the line graph visualization shows temporal progression for all cohorts at once. A detailed example of cohort analysis is featured in the Results section below.
Results
The objective here is to demonstrate the power of our visual analytic system for discovering temporal patterns in cooccurrence event sequences. We first describe the EHR data and the medical challenges in understanding the development of Heart Failure (HF) in patients. Then we describe the text mining process on extracting Framingham symptoms from EHR data. Finally, we describe several use cases on mining HF symptom sequences using our proposed visual analytic system.
Study Population and Sources of Data
Data for this study were obtained from the Geisinger Clinic (GC) primary care practices’ EHRs. Geisinger Clinic is part of the Geisinger Health System that serves residents in central and northeastern Pennsylvania. The dataset used for this project consists of the full records for over 50,625 patients. A total of 4,644 incident HF cases were identified between 2003 and 2010. Up to ten control patients were selected for each case. Controls were clinic-matched, sex-matched, and age-matched to the corresponding case but did not meet operational criteria for HF on or before the corresponding case’s diagnosis date. Note that two different cases can share common controls, in this design. For this study, we extracted the clinical notes portion of the EHRs for 4,644 case patients and for 45,981 control patients. There are different types of HF cases: HF with preserved ejection fraction (HFpEF) and HF with reduced ejection fraction (HFrEF). HFpEF are patients with HF diagnosis and with ejection fraction ≥50%, while HFpEF are patients with HF diagnosis and with ejection fraction <50%. In this dataset, we have 1,200 confirmed HRrEF and 1,615 confirmed HFpEF cases, and the rest are HF cases without a confirmed subtype. In total, there are more than 3.3 million clinical notes, comprising over 4 gigabytes of text.
Framingham Symptoms
The clinical complexity and heterogeneity of HF has posed challenges to developing standardized criteria for its diagnosis. The Framingham HF criteria, originally published in 1971, were based on clinical data acquired in the 1950s and 60s15. In that study, 2 or more major criteria or 1 major and 2 or more minor criteria are used as the diagnostic criteria for HF. Understandably, there are limitations to applying these criteria today, as they predate technologies that are now routinely used in current practice (e.g., echocardiography and natriuretic peptide). In fact, of the 17 major and minor criteria (Table 1), several are impractical for routine care (e.g., serial forced vital capacity), are no longer performed (e.g., circulation time), or are less reliably determined by today’s physicians than those of the 1950s and 60s (e.g., S3 gallop). Nonetheless, the clinical construct inherent to the Framingham criteria makes them useful for diagnosis in primary care, especially since a majority of them are typically documented by primary care providers during routine encounters, well before more severe symptoms prompt the ordering of specific imaging or serum studies. However, there are a number of reasons why it is difficult for a physician to detect the clinical signal that is sensitive and specific to HF, even when there is documentation that Framingham criteria have been met. Based on the practical consideration of the availability, quality and severity, we selected nine out of 17 Framingham criteria as the focus in this study as indicated in Table 1.
Table 1:
Framingham signs and symptoms for HF
| Major Criteria | Short Name | Selected |
| Paroxysmal nocturnal dyspnea or orthopnea | PNDyspnea (PND) | |
| Neck vein distention | JVDistension (JVD) | |
| Rales | Rales (RALE) | Yes |
| Radiographic cardiomegaly | RCardiomegaly (RC) | Yes |
| Acute pulmonary edema | APEdema (APED) | Yes |
| S3 gallop | S3Gallop (S3G) | |
| Central venous pressure > 16 cm of H2O | ICVPressure (ICV) | |
| Circulation time of 25 seconds | (not extracted) | |
| Hepatojugular reflux | HJReflux (HJR) | Yes |
| Weight loss of 4.5 kg in 5 days, in response to Rx | WeightLoss (WTL) | |
| Minor Criteria | ||
| Bilateral ankle edema | AnkleEdema (ANKED) | Yes |
| Nocturnal cough | NightCough (NC) | Yes |
| Dyspnea on ordinary exertion | DOExertion (DOE) | Yes |
| Hepatomegaly | Hepatomegaly (HEP) | Yes |
| Pleural effusion | PleuralEffusion (PLE) | Yes |
| A decrease in vital capacity by 1/3 of max | (not extracted) | |
| Tachycardia (rate of ≥ 120/min) | Tachycardia (TACH) |
Need for Visual Analytic Tools
There are substantial challenges to identifying the earliest signs and symptoms of HF, as it rarely emerges as a solitary disease process. Other diseases co-occur in the natural history of HF and include COPD, renal insufficiency and venous stasis, among others. These co-morbidities can mimic and even mask the presence of HF and lead to false positive or negative decisions. In practice, the physician is left with the difficult task of basing their decision to act on the examination of a patient in the moment, not based on a sophisticated quantitative assessment of longitudinal patient data or even a manual review of such data.
The evolution of patterns of co-occurrence and symptoms specific to HF is both subtle and complex, which could be aided with the help of powerful visual analytic tools to make those subtle trends obvious. For example, during a given encounter it is usually difficult to separate the presentation of true and false positive HF signs and symptoms, especially in the face of concomitant diseases and variable states of wellness. However, if a visual analytic system can help physicians connect the dots using historical records from the patient and similar patients, emerging health risks can become discoverable.
Text Mining
Before the analysis, a clinical natural language processing (NLP) procedure was developed to help physicians to automatically identify the onset of Framingham criteria in longitudinal clinical notes9. This can accurately identify affirmations and negations of Framingham criteria, which can directly reduce workload for HF-related chart reviews. About 4.5 million criteria mentions were identified; 900 thousand were classified as positive criteria and the remaining 3.6 million were negative criteria. For this study, only affirmed criteria were analyzed. Of all of the HF cases, 97% (4,490 of 4,644) met Framingham diagnosis criteria for heart failure, whereas in the control cohort 8% (3725 of 45,981) met these diagnosis criteria.
Trend Discovery and Cross-cohort Patterns
Based on the scenario of the HF symptom analysis using the text mining results, we have gone through an iterative development process for building our visual analytic system through demonstration and discussion with four medical experts. In our following description of MatrixFlow, we describe trends the medical experts noted while using our system.
Figure 6a shows the evolution of co-occurrence matrices of positive Framingham symptoms in the HFrEF patients, where patients are aligned by their diagnosis date. Each matrix displays co-occurrence events in a 3-month window. The rightmost matrix corresponds to the window right before diagnosis and the leftmost one the window 15 to 18 months before the diagnosis. From left to right as time evolves, we can easily observe the percentage of patients having co-occurring Framingham symptoms is increasing, which confirms with the degrading clinical status of those patients. Notably, as patients gets closer to HF diagnosis, multiple Framingham symptoms starts to appear more frequently. A similar temporal pattern is observed in HFpEF patients (Figure 6b), which seems to suggest that despite the pathophysiological differences, both HF types seem to develop the same co-occurrence patterns on Framingham symptoms. On the other hand, control groups (Figure 6c) have much less obvious patterns, except a slight increase of prevalence on common symptoms like DOExertion and AnkleEdema, presumably due to the normal aging process.
Similar increasing trends on negative Framingham symptoms can be observed as shown in Figure 7. The reason is that as physicians suspect HF as a possible differential diagnosis, more frequent exams and documentations about the denials of those Framingham symptoms start to show up in the clinical notes. Those frequent negative mentions themselves are a subtle indicator that HF is in the horizon. We observe much larger percentage of patients with negative symptoms than positive symptoms.
Figure 7.
The temporal evolution of the negative Framingham symptoms in MatrixFlow for the HFrEF patient cohort.
By comparing Figures 6 and 7, one can notice that certain co-occurrences have a large number of both of positive and negative mentions. This is an important and surprising finding. The data suggests that doctors document more negative mentions when they suspect positive mentions on certain patients. For a single patient, a reversal pattern (i.e., positive mention of a symptom followed by a negative mention of the same symptom in a future encounter) is often observed as the patient approaches the HF diagnosis. When we plot the aggregated patterns over a patient cohort by a time window, the negative and positive mentions will co-occur due to the aggregation effect.
As described in our methods section, MatrixFlow uses hierarchical clustering to order the cells of the matrix visualizations, so that clusters of related co-occurring symptoms are evident. The ordering of clinical events remains the same for all time intervals of a cohort by finding the optimal ordering in the time interval closest to the diagnosis date and then is applied to earlier time periods. We use this strategy because if the hierarchical clustering method is applied independently to each time period, the ordering will often be slightly different across time intervals and makes comparisons difficult.
The value of hierarchical clustering is evident by comparing Figures 6 and 7. In the positive symptoms in Figure 6a–6b, DOExertion and AnkleEdema are next to each because many patients share them. However, the order of negative symptoms in Figure 7 is different. In fact, Rales becomes the most dominant among negative symptoms, which is clinically meaningful because Rales is the most apparent symptom of HF. Rales is also a symptom that requires physician to deliberately examine and document. Also, the fact that Rales is being checked more frequently over time is a strong indicator that HF is suspected by physicians.
Expert Evaluation
We have demonstrated our system to four medical experts (one cardiologist, two medical scientists who were formerly emergency room doctors, and one epidemiologist) in order to validate our hypotheses.
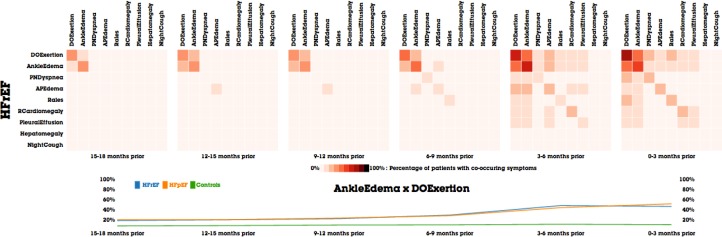
All four of the medical experts confirmed that temporal patterns of the progression of the disease were evident. One noted that it would take “years to distill such information out of following patients” to reach the same conclusions evidenced immediately by MatrixFlow. Another expert noted that leveraging the information displayed by MatrixFlow could “potentially help clinicians make earlier diagnoses, due to the large variability in diagnosis of diseases”. Similarly, another expert mentioned that visualizations of the historical information of similar patients can “help doctors prioritize a preventive strategy to avoid onset of heart failure” based on what strategy worked the best for those similar patients. Another expert remarked that the ability to compare cohorts was extremely useful, and noted that there is currently a research debate about whether HFpEF and HFrEF are the same or different diseases. They expert remarked that analyzing the cohorts in MatrixFlow may provide evidence for such debate (e.g., Figure 8 shows an example of the comparing the co-occurrence patterns of DOExertion and AnkleEdema among cohorts).
Figure 8.
Interactive line chart in MatrixFlow highlights the difference in co-occurrences of AnkleEdema and DOExertion between cohorts.
While MatrixFlow was initially designed to support population-level analysis, all of the medical experts quickly remarked on how MatrixFlow can also be used to perform individual patient analysis. One medical expert suggested that one can map an individual patient who is at risk of HF to a subcluster in MatrixFlow based on their existing Framingham symptoms and estimate the probability and the time period of developing HF. Another medical expert suggested that one could use MatrixFlow to look at post-diagnosis evolution of patients and to visually compare the effectiveness of different treatment plans.
Discussion and Conclusion
Based on our sessions with medical experts, we believe this is a rich space of future work to continue to enhance our visual analytics system. Currently, MatrixFlow is most effective in visualizing a small number of clinical events, as the matrix visualizations typically become more difficult to interpret as the size grows. There are several interactive visualization solutions to aid in this situation (e.g. the NodeTrix technique12) or an appropriate dimensionality reduction technique could also be used to handle large dimensional data. Currently, MatrixFlow only displays a single evolution pathway, but as patients split or merge into other meaningful cohorts, advanced visualization techniques could be utilized. We plan to investigate the right mechanism to support more complex evolution pathways.
In this paper, we present a visual analytic system, MatrixFlow, which converts clinical event sequences of patient cohorts into time-evolving networks and visualizes them as a temporal flow of matrices. MatrixFlow is an interactive visualization system that enables users to discover subtle patterns over time and across cohorts. We demonstrate the power of MatrixFlow in the context of analyzing Framingham symptom events extracted from a large cohort of HF cases and controls (n=50,625), We showcase the MatrixFlow system and conduct several interviews with medical experts to confirm its effectiveness in discovering temporal patterns. All the experts are extremely enthusiastic and positive towards the features provided by MatrixFlow, especially its capability to identify trends, clusters of interest, and perform cohort comparison.
References
- 1.Abello J, van Ham F. Matrix zoom: A visual interface to semi-external graphs. Proceedings of the 2004 IEEE Symposium on Information Visualization (INFOVIS’04); IEEE Computer Society; 2004. pp. 183–190. [Google Scholar]
- 2.Aigner W, Miksch S. CareVis: integrated visualization of computerized protocols and temporal patient data. Artificial intelligence in medicine. 2006;37(3):203–18. doi: 10.1016/j.artmed.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Ahn J, Plaisant C, Shneiderman B. A task taxonomy of network evolution analysis. University of Maryland; 2011. Technical Report HCIL-2011-09. [DOI] [PubMed] [Google Scholar]
- 4.Becker RA, Eick SG, Wills GJ. Visualizing network data. IEEE Transaction on Visualizations and Graphics. 1(1):16–28. [Google Scholar]
- 5.Byrd RJ, Steinhubl S, Sun J, Ebadollahi S, Stewart WF. Automatic identification of heart failure risk criteria, using text analysis of clinical notes from electronic health records. 2012. IBM technical report. [DOI] [PMC free article] [PubMed]
- 6.Cao N, Gotz D, Sun J, Lin Y, Qu H. SolarMap: Multifaceted Visual Analytics for Topic Exploration. IEEE International Conference on Data Mining (ICDM 2011). [Google Scholar]
- 7.Cao N, Gotz D, Sun J, Qu H. DICON: Interactive Visual Analysis of Multidimensional Clusters. IEEE Transactions on Visualization and Computer Graphics. 2011;17(12):2581–2590. doi: 10.1109/TVCG.2011.188. [DOI] [PubMed] [Google Scholar]
- 8.Ghoniem M, Fekete J-D, Castagliola P. A Comparison of the Readability of Graphs Using Node-Link and Matrix-Based Representations. IEEE Symposium on Information Visualization. (InfoVis 2004). [Google Scholar]
- 9.Gotz D, Sun J. IEEE VisWeek Workshop on Visual Analytics in Health Care 2010. SIGHIT Record. 2011;1(1):31–32. [Google Scholar]
- 10.Heer J, Perer A. Orion: A System for Modeling, Transformation and Visualization of Multi-dimensional Heterogeneous Networks. IEEE Conference on Visual Analytics Science and Technology (VAST 2011). [Google Scholar]
- 11.Henry N, Fekete JD. MatrixExplorer: a Dual-Representation System to Explore Social Networks. IEEE Transactions on Visualization and Computer Graphics. 2006;12(5):677–684. doi: 10.1109/TVCG.2006.160. [DOI] [PubMed] [Google Scholar]
- 12.Henry N, Fekete JD, McGuffin M. NodeTrix: a Hybrid Visualization of Social Networks. IEEE Transactions on Visualization and Computer. 2007 Nov-Dec;13(6):1302–1309. doi: 10.1109/TVCG.2007.70582. [DOI] [PubMed] [Google Scholar]
- 13.Herman I, Melancon G, Marshall MS. Graph visualization and navigation in information visualization: A survey. IEEE Transactions on Visualization and Computer Graphics (TVCG) 2000 Jan-Mar;6(1):24–43. [Google Scholar]
- 14.Klimov D, Shahar Y, Taieb-Maimon M. Intelligent visualization and exploration of time-oriented data of multiple patients. Artificial intelligence in medicine. 2010;49(1):11–31. doi: 10.1016/j.artmed.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 15.McKee PA, Castelli WP, McNamara PM, Kannel WB. The natural history of congestive heart failure: the Framingham study. N Engl J Med. 1971;285:1441–1446. doi: 10.1056/NEJM197112232852601. [DOI] [PubMed] [Google Scholar]
- 16.Moody J, McFarland D, Bender-deMoll S. Dynamic network visualization. American Journal of Sociology. 2005;110(4):1206–41. [Google Scholar]
- 17.Newman MEJ. Fast algorithm for detecting community structure in networks. Phys Rev E. 2004;69 doi: 10.1103/PhysRevE.69.066133. [DOI] [PubMed] [Google Scholar]
- 18.Wang TD, Plaisant C, Quinn A, Stanchak R, Shneiderman B, Murphy S. Aligning Temporal Data by Sentinel Events: Discovering Patterns in Electronic Health Records. Proceedings of the ACM SIGCHI Conference on Human Factors in Computing Systems (CHI 2008). [Google Scholar]
- 19.Wongsuphasawat K, Gómez JAG, Plaisant C, Wang TD, Taieb-Maimon M, Shneiderman B. LifeFlow: Visualizing an Overview of Event Sequences. Proceedings of the 2011 Annual Conference on Human Factors in Computing Systems (CHI 2011); pp. 1747–1756. [Google Scholar]
- 20.Yi J-S, Elmqvist N, Lee S. TimeMatrix: Visualizing Temporal Social Networks Using Interactive Matrix-Based Visualizations. International Journal of Human-Computer Interaction. 2010;26(11–12):1031–1051. [Google Scholar]