Abstract
The identification of high-risk patients is a critical component in improving patient outcomes and managing costs. This paper describes the Intelligent Care Delivery Analytics platform (ICDA), a system which enables risk assessment analytics that process large collections of dynamic electronic medical data to identify at-risk patients. ICDA works by ingesting large volumes of data into a common data model, then orchestrating a collection of analytics that identify at-risk patients. It also provides an interactive environment through which users can access and review the analytics results. In addition, ICDA provides APIs via which analytics results can be retrieved to surface in external applications. A detailed review of ICDA’s architecture is provided. Descriptions of four use cases are included to illustrate ICDA’s application within two different data environments. These use cases showcase the system’s flexibility and exemplify the types of analytics it enables.
1. Introduction
In many healthcare systems, costs and risk are not spread evenly across a population. Instead, a relatively small number of high-risk patients utilize more medical resources than their peers. Studies have shown that deficits in managing care for these patients can lead to even higher expenses and poorer outcomes [1]. Such findings have highlighted the need for systematic efforts that focus on identifying high risk patients and ensuring they receive the most efficient and effective care possible.
At the same time, adoption rates have been growing for electronic medical data (EMD) systems that store medical record, claim, lab, and pharmacy information [2]. This change is improving efficiency in many areas and is enabling a number of technologies that provide new insights into how a medical system is functioning. For example, dashboards can be developed to summarize overall institutional metrics directly from medical data, including patient re-admission rates or average length of stay.
In this paper, we describe Intelligent Care Delivery Analytics (ICDA), a predictive risk-assessment system. ICDA is designed to (1) ingest large volumes of heterogeneous electronic medical data, (2) analyze data for individual patients by comparing them to their peers using pluggable predictive analytics components which assess various forms of risk, and (3) provide an interactive environment through which clinical professionals can review and manage the population of patients identified as at-risk. ICDA is a scalable and general platform for predictive analytics and reporting that satisfies several key requirements:
Universal Standards-Based Analytics Environment. Given the wide range of data formats and siloed repositories in many medical institutions, ICDA provides a standards-based (i.e. ICD-9, NDC, CPT) analytics environment that represents data independently from the source data format. ICDA’s source-agnostic data model makes it deployable across a wide range of different data environments and legacy systems.
Open Data Access. Just as data sources are often fragmented across multiple systems, the analysis results produced by ICDA must be made accessible to an arbitrary set of existing user-facing systems. ICDA provides both (1) a web-based user interface for risky patient identification and review, and (2) web-based APIs that allow ICDA risk-assessment data to be accessed by external systems.
Pluggable Analytics Design. An analytics platform should be configurable to allow for deployments of selected sets of analytics to match an organization’s risk-assessment needs. This approach also allows for the deployment of new analytics to an existing installation. ICDA adopts a plugin-based analytics model to support this requirement where each type of risk assessment is implemented within an independent plugin. At runtime, the analytics plugins are orchestrated by ICDA and the results are disseminated through the ICDA platform.
Scalable for Large and Dynamic Data Sets. An analytics system must work reliably with high-volume data environments that change regularly as new patient information becomes available. The ICDA architecture is designed from the ground up to work in live deployment scenarios where large volumes of patient data must be regularly gathered and analyzed. This includes a focus on scalability and isolating long-running back-end data analysis processes from interactive user-facing components. We provide a detailed description of ICDA’s design and outline how our system architecture satisfies the above requirements.
We also discuss four specific risk-assessment use cases that we’ve enabled using ICDA through pilots with two different medical institutions. The pilot use cases include (1) risk assessment that a patient may be diagnosed with heart failure within a given time window, (2) treatment comparison for hyperlipidemia to identify patients at risk of receiving sub-optimal treatments, (3) utilization analytics that identify patients that over- or under-utilize medical resources, and (4) physician match analytics to identify patients at risk of a poor outcome due to their current physician assignment.
2. Related Work
Computational analysis of healthcare data has a long history and has been used to study a wide variety of diseases and conditions. This section provides an overview of the related work most relevant to ICDA.
Patient Population Analytics
Large population studies have been used for decades to investigate a range of medical topics. For example, randomized controlled trials (RCTs) conducted in the 1940s studied tuberculosis [3]. Meanwhile, longitudinal studies of people in Framingham, Massachusetts were used in the 1970s to develop the Framingham Criteria, a widely used risk assessment technique for heart failure [4]. Such studies are often considered the “gold standard” in medical research because they traditionally utilize data collected specifically for the study and produce statistically rigorous results. However, these same properties make this type of study relatively time consuming and expensive to conduct.
Researchers have more recently begun looking for ways to utilize existing clinical data and computers to speed the research process. For example, the i2b2 software platform [5] produced by the NIH-funded National Center for Biomedical Computing combines medical record data with genomic data and lets users create “mini-databases” for specific research projects. Another NIH-funded system is iDASH [6], which focuses on providing a secure, privacy-preserving environment for researchers to share and analyze data. iHealth Explorer [7], meanwhile, provides a web services model that lets users query databases to manually identify groups of patients. The system then provides a number of analytic algorithms that can be manually applied to perform tasks such as adverse drug reaction analysis. Addressing the challenge of unstructured data and conflicting coding schemes, the SHARPn platform [8] includes features that structure narrative text and normalize data to adhere to common standards.
Such systems can support the development of data-driven risk assessment techniques. For example, work on Hotspotting technology in Camden, New Jersey has helped identify so-called “super utilizers” [9] using EMD. Intervening to better manage these patients can help reduce costs and improve outcomes. Similarly, data-driven analyses have been used to estimate risk for developing certain carcinomas [10] and to generate survival estimates for transplant recipients [11]. These approaches are very useful because they use existing EMD collected during the delivery of care. The insights they produce can lead to important improvements in our medical understanding. However, unlike ICDA, these systems are not generally designed to support custom analyses for individual patients to identify specific personalized insights.
Personalized Insights for Individual Patients
The population-based analysis techniques described above are statistically powerful in part because they carefully control which patients are included in the study. However, this same property can limit their validity for many patients. Roughly 45% of primary care patients have multiple morbidities (with even higher percentages for older patients), making it a challenge for classical evidence-based medicine [12]. For this reason, there is a growing focus on using ad hoc, individualized analyses of large collections of medical data to inform complex decisions when no suitable trial can be found [13].
This approach often looks through existing data collected in EMD systems to find a custom set of patients that are clinically similar to the patient being treated. It then applies analytic algorithms to data from the similar patients to find insights that can inform the delivery of care [14, 15, 16]. Similar techniques have been use for real-time monitoring of physiological signals to predict adverse events [17]. While such techniques produce useful insights, they are computationally expensive to compute. ICDA provides a platform that can support a library of such personalized data-driven risk analytics and allows them to function at scale in an operational environment. In addition, ICDA makes this sort of analysis repeatable across multiple data environments and clinical use cases.
3. ICDA Architecture
ICDA is a scalable and flexible platform for predictive analytics and reporting. It provides a common data model and standard runtime that supports a toolbox of risk assessment components that can identify high-risk patients.
In supporting these capabilities, the ICDA architecture is motivated by four key requirements: (1) a standards-based analytics environment that allows individual components to be written once and used repeatedly within different clinical environments; (2) open data access so that risk assessments calculated within ICDA can be surfaced either though ICDA’s own user interface or though external third-party systems; (3) a pluggable analytics design so that deployments can be customized for specific use cases without changes to the platform; and (4) support for large scale and dynamic patient data to provide practical value to large care organizations.
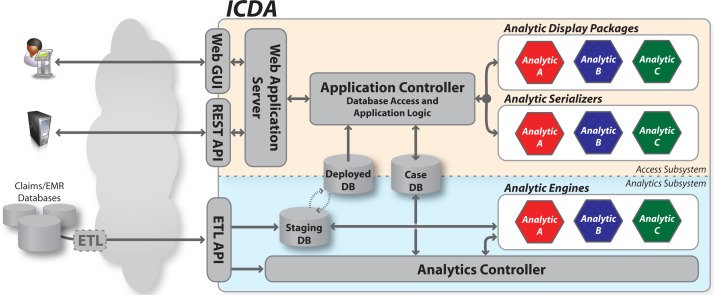
The ICDA architecture is illustrated in Figure 1. At the core of the system is a set of three databases. These databases serve as the bridge between the two main computational portions of ICDA: (1) the analytics subsystem, which calculates risk assessments for each patient in the database, and (2) the access subsystem, which provides access to the analytics results for both human users and external systems. As depicted by the color-coded components within the architecture diagram, an analytic plugin contains three distinct pieces which span both subsystems. The remainder of this section provides more detail for each of these architectural elements.
Figure 1:
The ICDA architecture consists of an analytics subsystem to coordinate analytic processing, and an access subsystem to expose the resulting risk assessments. Analytic plugins (each consisting of an analytic engine, a serializer, and a display package) can be deployed in arbitrary combinations to support specific use cases.
Databases
At the center of ICDA are three databases. Two patient databases—a staging database (DBS) and a deployed database (DBD)—contain detailed patient data including diagnoses, labs, medications, and procedures. Both use the same underlying data model which adopts widely used standards including ICD, CPT, and NDC. Data is ingested by ICDA into DBS where it is processed by analytics to identify at risk patients. When this process completes, a swap operation converts DBS to DBD, making the latest data available to users. This process is described in detail in Section 4. The dual database design allows new patient data to be incrementally loaded and analyzed (a process that can require a significant level of computation) in DBS without impacting the quality of service for users who access patient data from DBD.
A third case database (DBC) is used to store analytics results. A case file CPi is created in DBC for each high risk patient Pi identified by the analytics. The case file links together all risks associated with a given patient and contains metadata such as case status (new/open/closed) and a timestamp. Case files also capture the history of a given patient’s risk assessments which is used to ensure that DBC is kept synchronized with DBD as new data arrives and is processed by the system.
Analytics Subsystem
The analytics subsystem, shown in the lower portion of Figure 1, is responsible for managing the analysis of newly imported data by coordinating the execution of a set of one or more analytic engines. The set of analytic engines is specified in a configuration file which is processed by an analytics controller. The controller, launched each time new data arrives via the extract/transform/load (ETL) API, has four major responsibilities: to (1) launch each of the analytic engines, (2) monitor the engines’ status to detect and handle errors, (3) store status and result information within DBC, and (4) trigger the database swap operation to deploy DBS upon successful completion of all configured analytic engines.
Analytic engines are required to implement a specific API for launching, error reporting, and result production. This allows the controller to manage any analytic engine configured in the system without specific knowledge of how the engine works internally. The API is based on a command-line-executable-and-return-code model which provides the flexibility needed to minimally constrain analytic engine development. This allows for the incorporation of engines that are written using a wide variety of programming languages (e.g., Java or Python) and statistical analysis libraries (e.g., R or SPSS). Engines will typically query against DBS to access patient data during processing.
Access Subsystem
The access subsystem, shown in the upper portion of Figure 1, is responsible for providing individual users and external systems with interactive access to the analytics results. External access to ICDA is supported via a web application server that services both human users (via a browser-based user interface) and external systems (via a structured REST-based API). The user interface is shown in Figure 3 and described in more detail in Section 5.
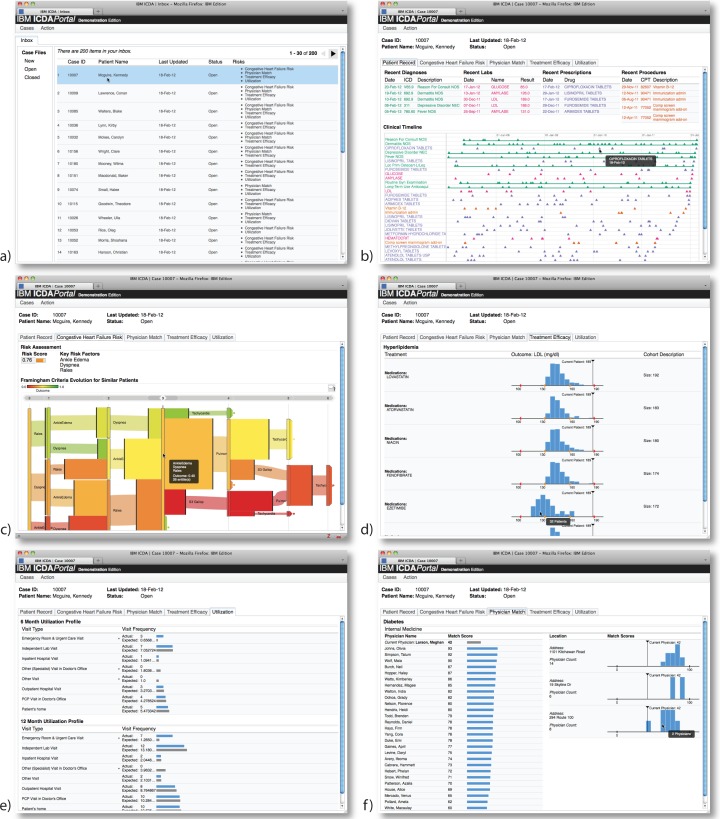
Figure 3:
The web interface for ICDA provides users with (a) an inbox showing patients flagged as high risk, (b) a patient summary view which provides a temporal summary of the medical record for an individual patient, and (c–f) targeted reports that convey detailed information supporting each of a patient’s identified risks. Zooming in on the images in electronic versions of this paper will reveal the full resolution of the screenshots.
The application controller contains the core platform logic for ICDA’s access subsystem. It processes requests that arrive via the web application server, manages database access, and provides the query and display logic for the base platform interfaces such as an inbox (containing a list of newly flagged at risk patients) and clinical summary screens that visualize the clinical record for an individual patient.
The controller also coordinates the operation of serialization and display packages. At startup, it processes a configuration file that specifies the set of analytic packages currently deployed in the system. At runtime, it coordinates the database queries and rendering routines required to display the current results for each risk factor contained in a given patient’s case CPi.
The query and rendering logic specific to an individual analytic type is not contained in the controller itself. Instead, this logic is captured in two key analytic-specific components: a serializer and a display package. These are shown as color-coded hexagons in Figure 1 and are two of the key pieces that make up an ICDA analytic plugin as defined later in this section. An analytic serializer contains the logic necessary to query DBD and/or DBC for the unique analysis results and supporting clinical information produced by the corresponding analytic engine. All executed queries are routed through the application controller which manages database connections and logging. Serializers then convert query results to an internal data structure. An analytic display package contains the logic required to convert a serializer’s data structure to a specified display format such as HTML (for the web-based GUI) or XML (for the REST API). These components work together to enable analytic-specific interface components which the controller ties together within a single overall web portal. Both serializers and display packages implement a specific ICDA API for instantiation and rendering that let the controller coordinate the user interface without specific knowledge about their internals.
Another responsibility for the access subsystem is safeguarding protected health information (PHI). The web server performs user authentication to ensure that only authorized users have access to ICDA data. In addition, all data access requests made through the application controller are logged along with the requesting user’s identification. This ensures HIPAA compliance.
Analytics Plugins
The components described above provide a base platform upon which specific risk assessment analytics can be built. However, the logic required for individual use cases is not part of the core platform. Instead, they are packaged as analytic plugins which bundle the three components described above: an analytic engine, a serializer, and a display package.
Engines, serializers and display packages all implement a set of predefined APIs which are then used at runtime by the core ICDA platform. Each plugin contains a manifest file that references the location of the software components for the three required components. For a given installation of the ICDA system, a global configuration file specifies which analytic plugins are actively deployed. The system then discovers and links to the required plugin components by processing the active plugins’ manifest files.
4. Data Flow
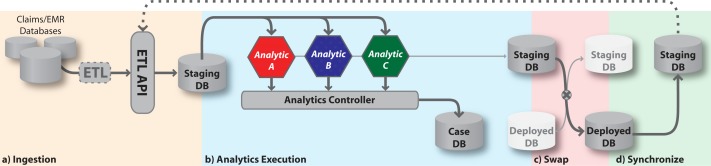
During normal operation, data is loaded into the staging database DBS by an ETL process before being analyzed by the analytics subsystem. When the analytics process completes, ICDA publishes the newly available analytics results and newly loaded patient data by performing a database swap that converts DBS to the deployed database DBD. The data is then served to requesting users and external systems via the access subsystem’s web GUI and REST API, respectively. This section describes this dataflow which is illustrated in Figure 2.
Figure 2:
Data flow within the ICDA system. (a) New patient data arrives via an ETL process and is stored in the staging database. (b) The analytic controller then runs the analytic engines and stores results in the case database. (c) When the results are ready, the deployed and staging databases are swapped. (d) Finally, the new staging database is synchronized to prepare for the next iteration of analytics.
Ingesting Data
Data is imported into ICDA via the analytic subsystem’s ETL API. For each external data source, an ETL module is used to extract information from the existing data repository (e.g., an electronic medical record system). The same module is responsible for a transformation step that maps between coding standards used in external data sources and those used within ICDA (e.g. ICD, CPT, NDC). Because data formats, system interfaces, and coding systems can vary widely, the ETL modules supporting data ingestion need to be custom designed for a given deployment. The ETL modules can be reused only to the extent that source data repository systems rely on standards (such as HL7).
Once the extraction and transformation steps are done, the ETL module loads the data into ICDA through the ETL API. This file-based API accepts ICDA-formatted data and incrementally updates DBS with new data which is stored in a schema optimized for performing computational analysis on large sets of patient data. When the ingestion process has completed, a signal is sent to the analytics controller to start an analytics run on the fresh data.
The ingestion of data is typically performed in batch mode on a regular schedule (e.g., nightly or hourly) to keep patient risk estimates up to date as patients’ conditions change. New ingestions cannot arrive too quickly, however. The ETL API will block the ingestion of new data if it is attempted prior to the completion of the prior analytics run.
Analytics Execution
After receiving the signal that an updated set of data is present in DBS, the analytics controller launches the set of active analytics engines, either serially or in parallel, based on dependency relationships defined in the global ICDA configuration file (dependencies will exist if one analytic engine requires as input the results produced by another). We refer to a single execution of the full set of analytics engines as a run. Each run is assigned a unique run ID by the controller which is used for logging and versioning of the results.
As they execute, each analytic engine pulls patient data from DBS and produces as output a file containing a list of patients that are considered at risk. The engine can also optionally write to a dedicated area in DBS to store intermediate results, to cache items such as statistical models between runs, or for any other purpose. Engines are also free to use resources outside of ICDA, such as external databases or even the global internet.
The controller processes the results files produced by the engines and stores the data in DBC. Note that this is the same DBC as the one being used by the access subsystem. However, new results are marked with the corresponding run ID as they are stored by the controller. The run ID value is then used to make sure that new results are not surfaced to the access subsystem until the corresponding analytics controller run has completed successfully. The changeover to a new run ID for the access subsystem is part of the database swap process described below.
Database Swap-and-Sync
If the analytics controller detects any unrecoverable failure during a run, the data flow halts and an administrator is notified. In contrast, if all engines complete successfully then the results are ready for deployment to the access subsystem. This happens via a carefully scripted database “swap-and-sync” operation.
This two-step process beings when the analytics controller notifies the application controller (via a HTTP-based API) that DBS with DBD have switched roles. This makes the latest patient data available to the access layer. At the same time, the analytics controller updates the active run ID on DBC, exposing the most recent case status and analytics results data. Any web sessions active at the time of this swap (for either the user-facing web GUI or the system-facing REST API) receive an exception indicating that the data has been refreshed and are redirected to the new DBD. The change is transparent to any new sessions which connect directly to the new DBD.
At this point, the access subsystem has full access to the latest patient data and analytics results. However, the new DBS, which was until this point the deployed database, does not contain the patient data received during the most recent ETL update. To complete the full swap-and-sync operation, the new DBS is synchronized with the new DBD. This is performed as a low-priority process to avoid degrading performance for DBD (which is now serving users’ interactive data requests). After the synchronization step has completed the ETL API is re-opened to accept the next batch of incremental patient data updates.
5. ICDA User Experience and Analytics Use Cases
ICDA includes a web-based user interface that provides access to detailed reports about the at-risk patients identified by analytic engines. Users connect to the system using a web browser and are asked to authenticate with a username and password. Once a user’s credentials have been verified, the user is shown an inbox (see Figure 3(a)) listing all at-risk patients that the user is permitted to see. The inbox shows the status for each patient’s case file (i.e., new, open or closed), the date that the case was last modified, and a list of identified risk factors.
Clicking on a patient’s name in the inbox takes the user to a view of the corresponding case file. This view shows a patient summary that includes a timeline-based display of the patient’s EMD (see Figure 3(b)). It also includes tabs that display detailed reports for each of the risks identified for the patient. The prototype deployment of our system includes four analytic engines which we describe below. All four of these engines run on the same ICDA platform despite being developed in the context of two very different data environments at two different institutions.
Heart Failure Risk Prediction
The goal of this plugin is to predict an individual patient’s risk of developing heart failure in a predefined future time window (e.g., 6 to 18 months). The novelty of this plugin compared to traditional risk assessment techniques is the focus on a finite (immediate) time window in terms of the prediction. Associating a prediction with a time window can help users better prioritize intervention strategies. In addition to the risk score, the engine provides a ranked list of contributing risk factors. This personalized set of risk factors can help providers customize a care plan for an individual patient. Risk factors include both co-morbid conditions (such as diabetes or hypertension) as well as symptoms (such as Framingham criteria [4]).
The analytic engine for this plugin uses a predictive modeling pipeline that leverages both structured and unstructured EMD (diagnoses, medications, lab results and physician notes). In our current prototype, the predictive model is trained using 7 years worth of data for 50K patients. The model is then used to estimate each patient’s risk of developing heart failure based on their recent encounters [15].
During each run of the analytics controller, the engine assesses heart failure risk scores for all patients and generates a list of patients with the highest estimated risk. The display package then renders a report, as shown in Figure 3(c), that includes a risk score, contributing risk factors, and a visualization of historical disease progression paths observed in similar patients [18].
Treatment Comparison
The objective of this plugin is to identify patients who are at risk of a sub-optimal outcome due to the treatments they have received for a specific condition. For example, one may want to compare alternative medications for a difficult-to-treat hyperlipidemia patient. Those who are receiving under-performing treatments are flagged as at risk along with data suggesting which alternatives (taken from guidelines) might perform better for the given patient.
The analytic engine for this plugin uses patient-similarity algorithms to identify a cohort of patients that are clinically most similar to a given target patient. The cohort is then subdivided based on treatments received and the past performance of alternative treatments are then compared. The similarity function is based on a Mahalanobis distance measure defined over patient characteristics such as co-morbidities, procedures, labs, and medications.
As part of each analytics controller run, the engine assesses all patients to determine the degree of risk they face due to sub-optimal treatment and produces a list of patients with the highest estimated risk. The plugin’s display package then renders a report for a patient, as shown in Figure 3(d), that allows for quick comparison of outcomes for alternatively treated similar patients.
Utilization Pattern Analysis
The goal of this plugin is to identify patients that are at risk due to unexpected utilization patterns. The plugin measures utilization in terms of the frequency of different visit types (e.g., specialist visits, primary care visits, emergency or urgent care encounters, etc.). Those patients that are either over- or under-utilizing resources may be at risk of poor outcomes, excessive costs, or both. Identifying these patients helps identify candidates for more careful care management.
The analytic engine for this plugin profiles each patient in a population based on the the frequency of various visit types. These profiles are then segmented into utilization clusters whose clinical features are used to train a classifier that can predict the expected utilization pattern for a given patient [16].
During each run, the engine assesses all patients to determine those with profiles that diverge most from expected levels. These patients are deemed at risk. The plugin’s display package then renders a report for each of these patients conveying both the patient’s actual and expected utilization rates. Especially abnormal visitation rates are marked visually to make it easy to identify why a given patient was flagged as at risk. For example, Figure 3(e) shows a patient with a higher-than-expected number of urgent care visits but no specialist visits. This patient may be an ideal candidate for a care management program.
Physician Matching
The objective of this plugin is to predict the best physician assignment for a given patient and to identify those patients that are at highest risk due to a poor match–given specific patient characteristics and physician experience—with their current physician. The algorithm both flags at-risk patients and provides a list of recommended physicians with high match scores. Care managers can use this data as input when recommending second opinions or assigning specialists.
The analytic engine builds a predictive model using characteristics from patients, physicians, and their interactions. The model maps these characteristics to medical outcomes using a dataset that segregates desired outcomes from undesired outcomes. The outcome measures are disease specific (e.g., A1C levels for diabetic patients). The model accounts for clinical differences between patients to allow prediction even when differences exist between the patient populations treated by different physicians [14].
During each run, the engine assigns a match score for each patient’s current physician as well as alternative physicians. A list of at-risk patients is produced that includes all patients whose current physician match score compares unfavorably to alternatives. The display package then renders a report, as shown in Figure 3(f), that includes the a patient’s current physician’s score, and a ranked list of the highest scored alternative physicians. The match scores are also grouped by practice location to highlight possible differences due to facility rather than practitioner.
6. Conclusion
We described Intelligent Care Delivery Analytics (ICDA), a predictive risk-assessment platform. The system is designed to (1) ingest large volumes of heterogeneous electronic medical data, (2) analyze data for individual patients by comparing them to their peers using pluggable predictive analytics components which access various forms of risk, and (3) provide an interactive environment through which clinical professionals can review and manage the population of patients identified as at-risk.
ICDA provides a universal standards-based analytics environment so that analytic engines for specific risk assessments can be written once and deployed in multiple settings with heterogeneous data environments. In addition, ICDA allows easy access to analysis results via both a web-based interface for users and a REST-based API for external system integration. The pluggable analytics API means that custom deployments of ICDA are possible for different installations. The design also allows new analytics to be deployed via relatively simple configuration changes. Finally, ICDA is designed specifically to work at scale for large and dynamic data environments making it practical to deploy ICDA-based analytics widely across an institution.
We also presented four specific risk assessment use cases that have been enabled by ICDA as part of pilot projects with two different medical institutions. These experiences highlight the ICDA platform’s ability to support portable and scalable risk assessment analytics that can help improve care delivery.
Given the promising results of these pilots, we plan to continue expanding ICDA’s capabilities. Directions for future work include the development of additional analytics engines, a comprehensive evaluation of ICDA’s performance in a large-scale clinical deployment, and an expansion of ICDA’s architecture to better support on-demand analytics for use cases where batch-based processing is insufficient.
References
- 1.Schoen Cathy, Osborn Robin, How Sabrina KH, Doty Michelle M, Peugh Jordon. In chronic condition: Experiences of patients with complex health care needs, in eight countries, 2008. Health Affairs. 2009 Jan;28(1):w1–w16. doi: 10.1377/hlthaff.28.1.w1. [DOI] [PubMed] [Google Scholar]
- 2.Jha Ashish K, DesRoches Catherine M, Kralovec Peter D, Joshi Maulik S. A progress report on electronic health records in U.S. hospitals. Health Affairs. 2010 Oct;29(10):1951–1957. doi: 10.1377/hlthaff.2010.0502. [DOI] [PubMed] [Google Scholar]
- 3.Streptomycin treatment of pulmonary tuberculosis. British Medical Journal. 1948 Oct;2(4582):769–782. [PMC free article] [PubMed] [Google Scholar]
- 4.McKee PA, Castelli WP, McNamara PM, Kannel WB. The natural history of congestive heart failure: the framingham study. The New England Journal of Medicine. 1971 Dec;285(26):1441–1446. doi: 10.1056/NEJM197112232852601. [DOI] [PubMed] [Google Scholar]
- 5.Murphy Shawn N, Weber Griffin, Mendis Michael, Gainer Vivian, Chueh Henry C, Churchill Susanne, Kohane Isaac. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) Journal of the American Medical Informatics Association. 2010 Mar;17(2):124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohno-Machado Lucila, Bafna Vineet, Boxwala Aziz A, Chapman Brian E, Chapman Wendy W, Chaudhuri Kamalika, Day Michele E, Farcas Claudiu, Heintzman Nathaniel D, Jiang Xiaoqian, Kim Hyeoneui, Kim Jihoon, Matheny Michael E, Resnic Frederic S, Vinterbo Staal A. iDASH: integrating data for analysis, anonymization, and sharing. Journal of the American Medical Informatics Association. 2011 Nov; doi: 10.1136/amiajnl-2011-000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAullay Damien, Williams Graham, Chen Jie, Jin Huidong, He Hongxing, Sparks Ross, Kelman Chris. A delivery framework for health data mining and analytics. Proceedings of the Twenty-eighth Australasian conference on Computer Science - Volume 38, ACSC ’05; Darlinghurst, Australia, Australia. 2005; Australian Computer Society, Inc; pp. 381–387. [Google Scholar]
- 8.Rea Susan, Pathak Jyotishman, Savova Guergana, Oniki Thomas A, Westberg Les, Beebe Calvin E, Tao Cui, Parker Craig G, Haug Peter J, Huff Stanley M, Chute Christopher G. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: The SHARPn project. Journal of Biomedical Informatics. 2012 Feb; doi: 10.1016/j.jbi.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner Jeffrey. Building an accountable care organization in camden, NJ. Prescriptions for Excellence in Health Care Newsletter Supplement. 2010 Jul;1(9) [Google Scholar]
- 10.Kurosaki Masayuki, Hiramatsu Naoki, Sakamoto Minoru, Suzuki Yoshiyuki, Iwasaki Manabu, Tamori Akihiro, Matsuura Kentaro, Kakinuma Sei, Sugauchi Fuminaka, Sakamoto Naoya, Nakagawa Mina, Izumi Namiki. Data mining model using simple and readily available factors could identify patients at high risk for hepatocellular carcinoma in chronic hepatitis c. Journal of Hepatology. 2012 Mar;56(3):602–608. doi: 10.1016/j.jhep.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Tang Hongying, Poynton Mollie R, Hurdle John F, Baird Bradley C, Koford James K, Goldfarb-Rumyantzev Alexander S. Predicting three-year kidney graft survival in recipients with systemic lupus erythematosus. ASAIO Journal (American Society for Artificial Internal Organs: 1992) 2011 Aug;57(4):300–309. doi: 10.1097/MAT.0b013e318222db30. [DOI] [PubMed] [Google Scholar]
- 12.Campbell-Scherer Denise. Multimorbidity: a challenge for evidence-based medicine. Evidence Based Medicine. 2010 Dec;15(6):165–166. doi: 10.1136/ebm1154. [DOI] [PubMed] [Google Scholar]
- 13.Frankovich Jennifer, Longhurst Christopher A, Sutherland Scott M. Evidence-Based medicine in the EMR era. New England Journal of Medicine. 2011 Nov;:1758–1759. doi: 10.1056/NEJMp1108726. [DOI] [PubMed] [Google Scholar]
- 14.Neuvirth Hani, Ozery-Flato Michal, Hu Jianying, Laserson Jonathan, Kohn Martin S, Ebadollahi Shahram, Rosen-Zvi Michal. Toward personalized care management of patients at risk: the diabetes case study. Proceedings of the 17th ACM SIGKDD international conference on Knowledge discovery and data mining, KDD ’11; New York, NY, USA. 2011; ACM; pp. 395–403. [Google Scholar]
- 15.Sun Jimeng, Luo Dijun, Byrd Roy, Hu Jianying, Edabollahi Shahram, Daar Zahra, Steinhubl Steven E, Stewart Walter F. Scalable predictive modeling pipeline and its application on early detection of heart failures using electronic health records. AMIA Annual Symposium Proceedings; 2012; submitted. [Google Scholar]
- 16.Hu Jianying, Wang Fei, Sun Jimeng, Sorrentino Robert, Edabollahi Shahram. A utilization analysis framework for hot spotting and anomaly detection. AMIA Annual Symposium Proceedings; 2012; submitted. [PMC free article] [PubMed] [Google Scholar]
- 17.Ebadollahi Shahram, Sun Jimeng, Gotz David, Hu Jianying, Sow Daby, Neti Chalapathy. Predicting patient’s trajectory of physiological data using temporal trends in similar patients: A system for Near-Term prognostics. American Medical Informatics Association Annual Symposium (AMIA); Washington, DC. 2010; [PMC free article] [PubMed] [Google Scholar]
- 18.Wongsuphasawat Krist, Gotz David. IEEE VisWeek Workshop on Visual Analytics in Healthcare. Providence, Rhode Island, USA: 2011. Outflow: Visualizing patient flow by symptoms and outcome. [Google Scholar]