Abstract
The highest concentrations of prostaglandins in nature are found in the Caribbean gorgonian Plexaura homomalla. Depending on its geographical location, this coral contains prostaglandins with typical mammalian stereochemistry (15S-hydroxy) or the unusual 15R-prostaglandins. Their metabolic origin has remained the subject of mechanistic speculations for three decades. Here, we report the structure of a type of cyclooxygenase (COX) that catalyzes transformation of arachidonic acid into 15R-prostaglandins. Using a homology-based reverse transcriptase–PCR strategy, we cloned a cDNA corresponding to a COX protein from the R variety of P. homomalla. The deduced peptide sequence shows 80% identity with the 15S-specific coral COX from the Arctic soft coral Gersemia fruticosa and ≈50% identity to mammalian COX-1 and COX-2. The predicted tertiary structure shows high homology with mammalian COX isozymes having all of the characteristic structural units and the amino acid residues important in catalysis. Some structural differences are apparent around the peroxidase active site, in the membrane-binding domain, and in the pattern of glycosylation. When expressed in Sf9 cells, the P. homomalla enzyme forms a 15R-prostaglandin endoperoxide together with 11R-hydroxyeicosatetraenoic acid and 15R-hydroxyeicosatetraenoic acid as by-products. The endoperoxide gives rise to 15R-prostaglandins and 12R-hydroxyheptadecatrienoic acid, identified by comparison to authentic standards. Evaluation of the structural differences of this 15R-COX isozyme should provide new insights into the substrate binding and stereospecificity of the dioxygenation reaction of arachidonic acid in the cyclooxygenase active site.
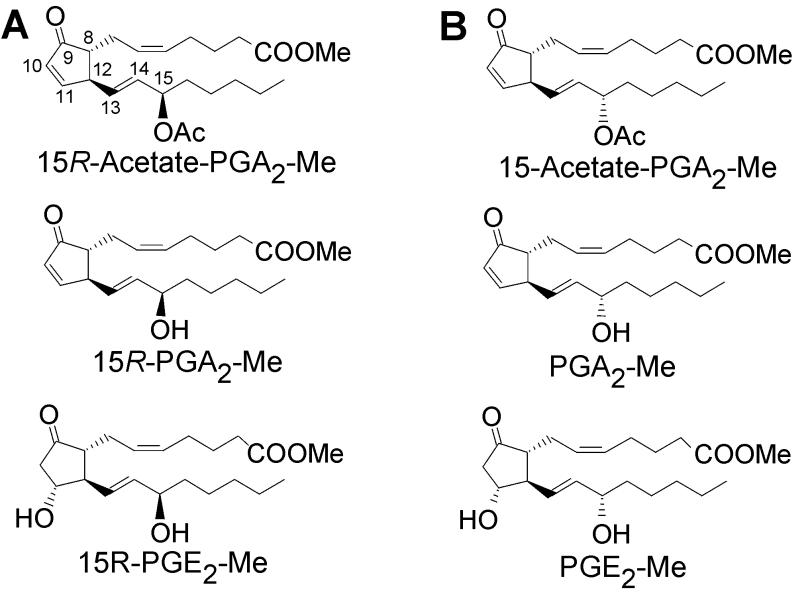
In 1969, shortly after the chemical structures of the prostaglandin (PG) hormones were elucidated, a rich natural source of prostaglandins was discovered in the Caribbean gorgonian Plexaura homomalla (1). This sea whip coral contains 2–3% of its dry weight as prostaglandin esters. In the original report, Weinheimer and Spraggins analyzed P. homomalla collected in the Florida Keys, and the main constituents were identified as 15R-PGA2 methyl ester acetate and 15R-PGA2 methyl ester, products that have the opposite configuration at C-15 to mammalian prostaglandins (Fig. 1A) (1). It was subsequently discovered that P. homomalla colonies collected in other Caribbean locations such as the Cayman Islands and Bahamas contain the same prostaglandin derivatives but with the normal 15S configuration (Fig. 1B) (2, 3). These later studies established that P. homomalla also contains PGE2 methyl ester (2, 3) as well as minor constituents, including the methyl ester 15-acetate derivatives of 5,6-trans-PGA2 and 13,14-cis-PGA2 (4). On account of the huge prostaglandin content, P. homomalla served as a source of prostaglandins for research until an efficient chemical synthesis was developed in the 1970s (5).
Figure 1.
Naturally occurring prostaglandins in the coral P. homomalla. (A) The major endogenous prostaglandins in the coral collected in the Florida Keys have the 15R configuration. (B) P. homomalla samples collected in the Cayman Islands and Bahamas contain prostaglandins with the typical 15S configuration.
The mechanism of biosynthesis of these prodigious quantities of prostaglandins has been investigated extensively over the years. Initial efforts suggested that the coral biosynthesis differed from the mammalian pathway and that the initial product was PGA2 (6, 7). Subsequent studies with improved analytical technology failed to detect any synthesis of prostaglandins during in vitro incubations of P. homomalla extracts (8). The biosynthesis in vitro was dominated by the strong 8R-lipoxygenase activity; this was coupled to allene oxide synthesis and further nonenzymatic reactions including hydrolysis and cyclization of the allene epoxide (8, 9). For a time, evidence from P. homomalla and other corals suggested that prostaglandin biosynthesis occurred analogously to jasmonic acid synthesis in plants via the lipoxygenase–allene oxide route (8, 10–12). Formation of an intermediate typical to mammalian prostaglandin synthesis in coral was established through our biochemical studies on the Arctic soft coral Gersemia fruticosa (13–15). This prostaglandin-containing coral converts radiolabeled arachidonic acid to the endoperoxide intermediate PGG2 in vitro. The presence of a cyclooxygenase (COX)-like enzyme in coral was established conclusively by molecular cloning from G. fruticosa cDNA encoding an enzyme with 50% identity to each of the mammalian COX isozymes, COX-1 and COX-2 (16). Here, we describe the molecular cloning and heterologous expression of a COX protein from P. homomalla. The active enzyme obtained has unique stereospecificity, catalyzing the transformation of arachidonic acid into 15R-prostaglandins.
Materials and Methods
PCR Cloning.
Total RNA was extracted from 2 g of frozen P. homomalla coral as previously described (17). The RNA (1 μg) was reverse transcribed into cDNA by using CLONTECH Marathon cDNA amplification kit. The PCR strategy used the same degenerate primers used to clone the COX cDNA from the coral G. fruticosa (16). The initial PCR fragment was obtained by using two sets of degenerate primers based on the conserved COX sequences AQHFTHQFFKT and YHWHP. The initial 559-bp PCR fragment was extended by 3′ and 5′ rapid amplification of cDNA ends (17). The full-length clone was obtained by using the proofreading Pfu DNA polymerase (Promega). Six full-length clones were fully sequenced.
Expression of Protein.
The coding regions of the COX enzymes (P. homomalla and G. fruticosa) were subcloned into the pFASTBAC1 donor vector, and recombinant baculovirus particles were isolated by using the Bac-to-Bac baculovirus expression system (Life Technologies, Grand Island, NY). Sf9 cells were cultured at 27°C in Sf-900II serum-free insect cell media. Cells with a density of 1.5–2 × 106 cells/ml were infected with recombinant baculovirus with a multiplicity of infection of 0.02. Cells were harvested 72 h after infection, washed with PBS, and stored as a pellet at −80°C.
Western Transfer Blotting.
The microsomal fraction of proteins was resolved by SDS/PAGE and transferred electrophoretically to a nitrocellulose membrane. The membrane was blocked with 1% dry milk in TBS for 1 h and incubated with 1:500 dilution of a mouse monoclonal antibody against rat COX-2 (PharMingen) in 1% dry milk in TBS (1 h). The membrane was washed thoroughly and incubated with 1:5,000 dilution of a goat-anti-mouse IgG alkaline phosphatase for 1 h. After washing, the membrane was visualized with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate in 100 mM Tris⋅HCl/10 mM NaCl/5 mM MgCl2, pH 9.5.
Enzyme Assay and Product Identification.
Cell pellets were sonicated briefly, suspended in 50 mM Tris⋅HCl (pH 8.0) containing 1 μM hematin plus 1 mM adrenaline or 500 μM phenol, and preincubated for 5 min at room temperature. Incubations with 50 μM or 100 μM [1-14C]arachidonic acid were performed for 15 min at room temperature by using the pellet from 1 to 5 million cells/ml of incubation mixture. For TLC analyses, reaction mixtures were acidified and the products extracted with ethyl acetate. TLC was performed using silica gel plates (Merck) and a solvent system of benzene/dioxane/acetic acid (5/2.5/0.25, vol/vol/vol). For product quantification, the TLC plates were cut into zones and extracted with methanol, and the radioactivity was measured with a liquid scintillation counter.
For HPLC analyses, the reaction was terminated with 2.5 vol of methanol and extracted with C18 Sep-Pak cartridges. In some experiments designed to reduce the prostaglandin endoperoxide in situ and to simplify the pattern of products, 0.5 mM SnCl2 was added to the incubation mixtures just before [1-14C]arachidonic acid. This gave an immediate reduction of the peroxy intermediates to the corresponding hydroxy compounds. The products formed were analyzed with reversed phase (RP)-HPLC as described in the legend to Fig. 4.
Figure 4.
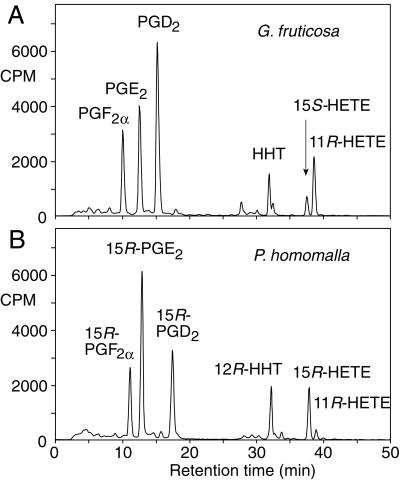
RP-HPLC analysis of [1-14C]arachidonic acid metabolites of G. fruticosa (A) and P. homomalla (B) COX expressed in Sf9 insect cells. The incubation products were run on a Waters Symmetry C18 column (25 × 0.46 cm) using a solvent of water/acetonitrile/acetic acid (62.5/37.5/0.01, vol/vol/vol, changed to the proportions 30/70/0.01 at 25 min) at a flow rate of 1 ml/min and using a Radiomatic Flo-One detector for on-line recording of radioactivity.
Chiral analyses (see Fig. 5) were carried out for P. homomalla COX products collected separately from RP-HPLC. The PGE2 was converted to PGB2 by treatment with 0.1 M methanolic KOH for 30 min at room temperature. The PGB2 and the peaks of hydroxyheptadecatrienoic acid (HHT), 11R-hydroxyeicosatetraenoic acid (11R-HETE), and 15R-HETE were methylated with diazomethane and purified individually before the chiral analysis. Authentic standards of prostaglandins, HETE, and HHT (Cayman Chemical, Ann Arbor, MI) were used for product identification. Racemic HHT was prepared by MnO2-catalyzed oxidation of HHT followed by NaBH4 reduction back to 12RS-HHT. An authentic standard of 15R-PGB2 was prepared by alkaline treatment of 15R-PGE2 (0.1 M methanolic KOH, 30 min at room temperature).
Figure 5.
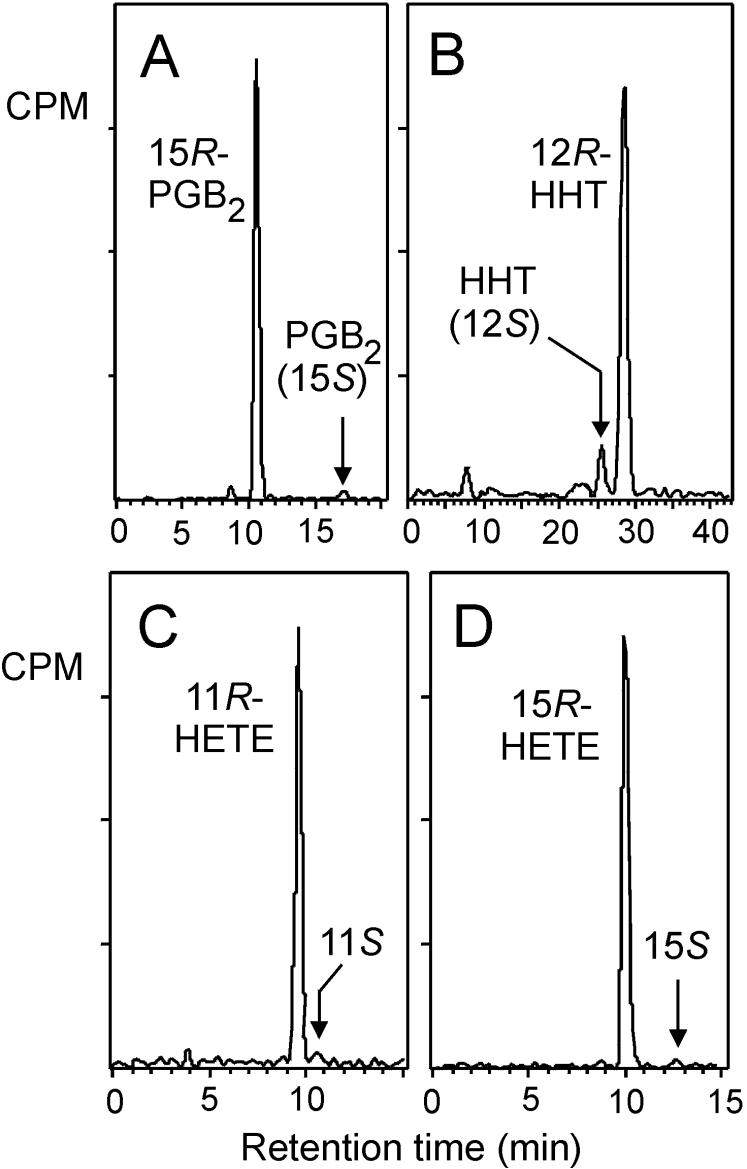
Chiral column HPLC of the compounds produced by P. homomalla COX expressed in Sf9 cells. PGE2 was analyzed after conversion to PGB2. Chiral analysis of individual radiolabeled products collected separately from RP-HPLC was performed using a Chiralpak AD column with a solvent of hexane/MeOH (100/2, vol/vol) for HETE methyl esters, the solvent proportions of 100/4, vol/vol, for HHT methyl esters and the solvent proportions of 100/5, vol/vol, for PGB2 methyl ester (21). Ratios of the stereoisomers were as follows: PGB2, 15R/15S, 97:3; HHT, 12R/12S, 90:10; 11-HETE, 11R/11S, 98:2; and 15-HETE, 15R/15S, 98:2. The proportion of S isomer in HHT was always greater than the percent of 15S prostaglandins formed in the same incubation. This difference may reflect a greater facility of the 15S endoperoxide to break down to HHT compared with its R diastereomer.
For NMR identification of 15R-PGF2α, a large-scale incubation (300 ml) with unlabeled arachidonic acid was carried out in the presence of SnCl2. The other conditions were the same as described in the enzyme assay.
Results
Homology-Based Reverse Transcriptase–PCR Cloning.
The cloning strategy was based on the design of degenerate primers representing amino acid sequences of conserved regions of mammalian COX isozymes. The PCR products were cloned into the pCR2.1 vector and sequenced. The full-length clone encoding the ORF plus extensions with BamHI restriction sites was amplified by PCR using Pfu DNA polymerase and cloned into the pGEM T-Easy vector system. Six positive clones were fully sequenced. Two different types of clones were found. The differences were in three amino acids, all located in the putative signal peptide region (Tyr versus His in position 6, Val versus Ile in position 12, and His versus Phe in position 17). Two clones (one of each type) were used for further studies.
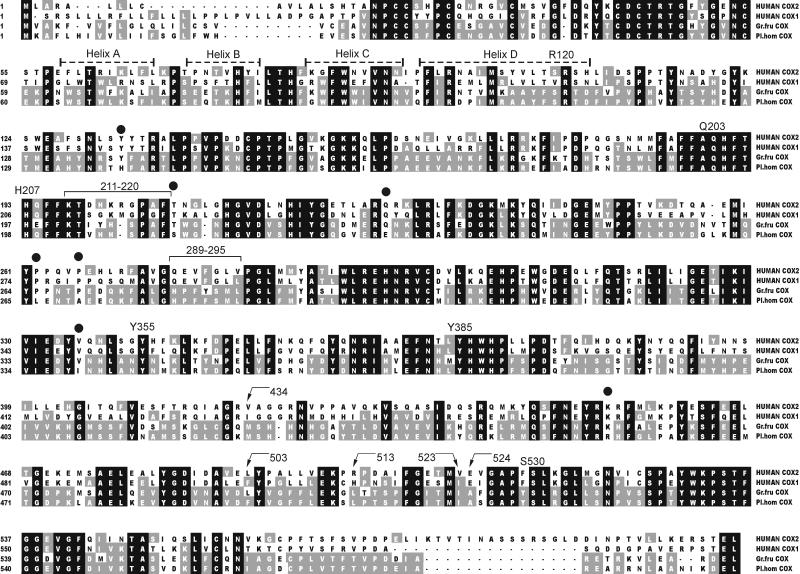
The ORF of 1,776 bp encodes a polypeptide (592 aa) with 50% amino acid identity and 68% similarity to each of mammalian COX-1 and COX-2. The sequence identity and similarity with the COX from the coral G. fruticosa is 80% and 92%, respectively (16). The deduced amino acid sequence alignment of P. homomalla COX with human COX-1 and COX-2 shows a good conservation of catalytically important amino acids (Fig. 2).
Figure 2.
Multiple sequence alignment of the deduced amino acid sequences of coral and human COX isozymes. The black background illustrates conserved regions. Residues of P. homomalla COX that are present in at least one of the other sequences are shaded gray. Catalytically important amino acids are given with numbers above the sequences (ovine COX-1 numbering is used for clarity). ●, sequences that are present only in P. homomalla COX, indicating possible amino acids responsible for the opposite stereospecificity at C-15 (comparison was carried out with all COX isozyme sequences available in the protein database).
Expression in Baculovirus Expression System.
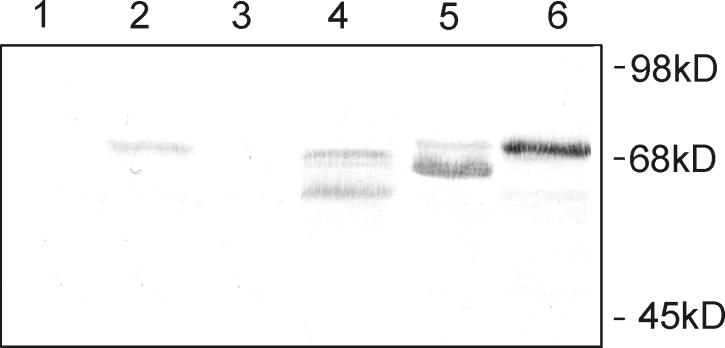
For expression, we used two fully sequenced clones of P. homomalla COX cDNA with differences in three amino acids. The different clones expressed the same enzymatic activity and gave an indistinguishable pattern of products. For comparison with the P. homomalla enzyme, the cDNA of the COX from the Arctic coral G. fruticosa was also expressed in the baculovirus system. The G. fruticosa enzyme required a lowering of the expression temperature (23°C) to obtain active enzyme. The result is in accordance with the Arctic nature of this coral (16). This was not necessary for the P. homomalla enzyme of Caribbean origin. The coral COX enzymes were found to crossreact with a commercial monoclonal antibody raised against murine COX-2. Immunostaining of the Western blot showed three closely migrating bands, most probably reflecting different states of glycosylation of the proteins (Fig. 3, lane 4) (18, 19). COX protein was not detected in an extract of P. homomalla (lane 1). The relatively low sensitivity of mammalian-specific antibodies against P. homomalla COX may account for this, together with the fact that in vitro prostaglandin synthesis has never been detected in P. homomalla, implying that there are low levels of the endogenous protein.
Figure 3.
Western blot of native coral COX enzymes and recombinant proteins expressed in Sf9 cells. Lanes: 1, acetone powder of native P. homomalla COX; 2, acetone powder of native G. fruticosa COX; 3, mock-infected Sf9 cells; 4, P. homomalla COX expressed in Sf9 cells; 5, G. fruticosa COX expressed in Sf9 cells; 6, ovine COX-1 as standard.
Incubations and Product Analysis.
The COX activities of the recombinant enzymes were compared by incubation with 100 μM [1-14C]arachidonic acid followed by TLC and/or HPLC analysis of the products. Suspended cell pellets of 3 million cells per ml were able to convert up to 30% of the arachidonic acid to prostaglandins and other products. The G. fruticosa COX converted [1-14C]arachidonic acid to prostaglandin endoperoxide, which degraded to a mixture of prostaglandins (PGF2α, PGE2, and PGD2) together with HHT and two other known COX by-products, 11R-HETE and 15S-HETE (Fig. 4A). This pattern of products is similar to that formed by mammalian COX-1 and COX-2. The P. homomalla COX produced a spectrum of products that were superficially similar (Fig. 4B), but on close examination it was found that the first three main peaks each resolved from authentic prostaglandins F2α, E2, and D2. They cochromatographed instead with authentic 15R-prostaglandins; this was especially evident on straight-phase HPLC systems in which the 15R- and 15S-prostaglandins are more widely resolved (data not shown).
P. homomalla is known to contain either 15R- or 15S-prostaglandins, depending on the site of collection (2, 3). Our sample came from a known location of 15R content, the Florida Keys. As evidence that the products are derived from a common 15R-prostaglandin endoperoxide, incubation with [1-14C]arachidonic acid was conducted in the presence of the mild reducing agent SnCl2. A single prostaglandin peak was detected on HPLC. This compound was formed by reduction of the prostaglandin endoperoxide, the primary product of the P. homomalla COX enzyme.
The identity of the reduced product with 15R-PGF2α was established by high-field (11.7 T) NMR spectroscopy. The carbon chemical shifts of the incubation product and 15R-PGF2α authentic standard were identical, as confirmed by the 20-line 13C NMR spectrum of their equimolar mixture in CDCl3 solution. Furthermore, a 3:4 mixture of the incubation product and 15S-PGF2α standard showed separate lines for nearly all of the carbons (Table 1). The 13C NMR study of the incubation product formed in the presence of SnCl2 shows conclusively that the P. homomalla COX forms a 15R-prostaglandin endoperoxide and that this gives rise to the 15R-prostaglandin products.
Table 1.
Comparison of 13C NMR chemical shifts of 15R- and 15S-PGF2α from their 3:4 mixture in CDCl3 solution
| PGF2α | C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | C-7 | C-8 | C-9 | C-10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 15R | 177.05 | 32.7 | 24.45 | 26.11 | 129.59 | 129.08 | 24.99 | 50.99 | 72.63 | 42.96 |
| 15S | 177.37 | 32.91 | 24.45 | 26.24 | 129.58 | 129.13 | 25.22 | 50.20 | 72.49 | 42.69 |
| 15R − 15S | −0.32 | −0.21 | 0 | −0.13 | 0.01 | −0.05 | −0.23 | 0.79 | 0.14 | 0.27 |
| C-11 | C-12 | C-13 | C-14 | C-15 | C-16 | C-17 | C-18 | C-19 | C-20 | |
| 15R | 77.87 | 55.65 | 132.4 | 134.59 | 72.96 | 37.05 | 25.13 | 31.7 | 22.61 | 14.03 |
| 15S | 77.61 | 55.34 | 132.61 | 134.95 | 73.13 | 36.87 | 25.22 | 31.7 | 22.61 | 14.03 |
| 15R − 15S | 0.26 | 0.31 | −0.21 | −0.36 | −0.17 | 0.18 | −0.09 | 0 | 0 | 0 |
A comparison of the chemical shifts from separate measurements cannot be taken as decisive because of very strong solvent and concentration effects in the 13C NMR spectra of prostaglandins. These dilution effects account for the slight differences between our observations and an earlier report of 13C NMR differences for 15R- and 15S-PGF2α (20).
Chiral analyses of several other products in the RP-HPLC chromatogram (Fig. 4B) were in accordance with this conclusion. The second major peak on RP-HPLC, on treatment with alkali, gave a product with identical HPLC and UV properties to PGB2. Alkaline treatment of PGE2 dehydrates the molecule and removes all but one chiral center at C-15. Chiral analysis of the B prostaglandin from P. homomalla revealed that it is almost exclusively 15R in configuration (Fig. 5A), thus identifying the original product as 15R-PGE2. Similarly, the peak in Fig. 4B corresponding to HHT was shown to be 12R-HHT, the mirror image of the G. fruticosa and mammalian HHT (Fig. 5B). Chiral analysis of the HETE peaks in the Fig. 4B chromatogram was also revealing. The absolute chemical configuration of the 11-HETE was almost exclusively 11R (Fig. 5C), which is identical to the result obtained for the mammalian COX-1, COX-2, and G. fruticosa 11-HETE by-products. This is significant because the chirality of the 11-HETE is known to reflect the chirality of the first oxygenation in the cyclooxygenase reaction, and this is expected to be identical in all COX enzymes, including in the P. homomalla COX. The 15-HETE peak in Fig. 4B is unusually prominent for a COX by-product, and the chiral analysis indicated it is almost pure 15R-HETE (Fig. 5D). The abundance and chiral purity of the 15-HETE are different from the results with other COX enzymes (22–24) and indicate that the P. homomalla COX enzyme has an active site that is set up to favor oxygenation in the 15R configuration.
Discussion
A COX Enzyme in P. homomalla.
In this article, we report molecular cloning and expression from P. homomalla of a type of COX enzyme with a unique 15R stereospecificity. Using the Swiss Model program (25) and RasMol 2.6, the predicted tertiary structure of the P. homomalla COX was compared with the crystal structures of mammalian COX-1 and COX-2 (26, 27). The characteristic structural units including the epidermal growth factor-like module, the membrane-binding motif, and the large globular catalytic domain with cyclooxygenase and peroxidase active sites (26–32) are present in the coral COX. Analysis of the primary and tertiary structures of P. homomalla COX shows strict conservation of the catalytically important amino acids in the cyclooxygenase active site. The catalytic Tyr-385, the target of aspirin attack Ser-530, and the carboxylate-binding Arg-120 and Tyr-355 are all present. (In line with convention, the amino acids of the coral protein are referred to here according to the residue numbers of ovine COX-1).
Despite the high overall homology with COX-1 and COX-2, several differences were found in the coral COX structure. Parts of the membrane-binding helices of the coral enzyme appear to lack the regular interchange of hydrophobic and hydrophilic residues, suggesting possible differences in the strength of membrane association between the mammalian and coral proteins. The peroxidase site of the coral COX, while preserving all of the catalytically vital residues, shows differences in the shield covering the active site and in the PGG2 binding site. Molecular modeling of residues 289–295 shows that there are more bulky amino acids over the heme in the coral enzyme, with potential ramifications for substrate access to the peroxidase active site. Differences were also found in the possible N-glycosylation sites. Only one of the conserved glycosylation sites in the COX isozymes, at Asn-144, is present in the P. homomalla enzyme. There are two additional consensus sequences for N-glycosylation, but their location differs from the consensus sequences in COX-1 and COX-2. Furthermore, a glycosylation site conserved in all known COX structures and located in the epidermal growth factor-like domain (at Asn-68) is missing altogether in the P. homomalla enzyme.
The main difference in the cyclooxygenase active sites of mammalian COX-1 and COX-2 is the presence in COX-2 of a bigger side pocket near the carboxylate-binding region (27, 28); an I523V substitution makes the COX-2 site accessible to larger substrates and COX-2-specific inhibitors (33). The other differences between COX-1 and COX-2 in this region are in amino acids 513 and 434 (Arg-513 and Val-434 in COX-2 versus His-513 and Ile-434 in COX-1). Altogether, these differences permit the selective inhibition of COX-1 and COX-2 (reviewed in ref. 34). The predicted size of the hydrophobic channel of the P. homomalla COX is more similar to COX-1, with more bulky amino acids, Ile-523 and Met-434, obstructing the side pocket. The effect of a substitution of polar amino acids His or Arg at the 513 position to neutral Leu is unclear and needs further investigation. Similarly, the conserved residue Glu-524, implicated in the binding of certain inhibitors, is represented by Ala in the coral enzyme.
Synthesis of 15R Prostaglandins.
Our results clearly establish that the main product of the P. homomalla COX is a 15R-configuration prostaglandin endoperoxide. The structural determinant conferring 15R-prostaglandin synthesis is intriguing and yet to be established. Presently, the only known COX-catalyzed reaction with purely 15R stereospecificity is the biosynthesis of 15R-HETE by aspirin-inhibited COX-2 (24, 35–38). Aspirin is known to inhibit COX enzymes by acetylating Ser-530 (39, 40). In the case of COX-1, this eliminates all oxygenase activity. In COX-2, although prostaglandin biosynthesis is abolished, the enzyme can still oxygenate arachidonic acid at C-15 (37, 38). It was shown recently that this reaction is initiated by abstraction of the pro-S-hydrogen from C-13 of arachidonic acid (41), the same reaction as in normal prostaglandin biosynthesis (22). A model was proposed in which arachidonic acid binds normally in the active site of aspirin-treated COX-2 with the exception that the omega carbon chain is folded over in an atypical conformation (42). In the new conformation, arachidonic acid is no longer oxygenated at C-11, thereby eliminating prostaglandin endoperoxide synthesis, and oxygenation occurs instead at C-15 in the R configuration.
It is likely that some aspects of this model occur during 15R-endoperoxide synthesis in the P. homomalla enzyme. Notably, the P. homomalla COX has a Ser-530 in common with all other cyclooxygenases, so presumably it is not directly implicated in this unusual 15R-endoperoxide synthesis. One would anticipate that some critical amino acid substitution(s) induce a folding over of the omega chain of arachidonic acid, but this change in conformation remains compatible with all of the earlier steps in endoperoxide synthesis. Comparison of the primary structure of the P. homomalla 15R-COX with the cyclooxygenases of 15S-stereospecificity shows seven strictly conserved amino acids that are different in the P. homomalla enzyme. One of them, Ile-349 in P. homomalla (Val-349 in all other COX emzymes), is located in the substrate-binding channel. Mutation of this residue to the Leu has been reported previously, although no changes in the stereospecificity of the prostaglandin synthesis were noted (41). The question of whether substitution to the more bulky Ile, the natural residue in the 15R-specific P. homomalla enzyme, could give rise to the changed specificity or whether it depends on several structural differences will hopefully be answered by further site-directed mutagenesis studies. Cloning of the COX enzyme from the 15S-specific P. homomalla would also be predicted to reveal the determinants. The answers should afford new insights into the substrate and inhibitor binding in the COX channel.
Origin of the P. homomalla Prostaglandins.
The Caribbean gorgonian P. homomalla is the richest source of prostaglandins in nature. The existence of the 15R-COX clearly explains the occurrence of the unusual 15R-prostaglandins in P. homomalla of Florida waters. The enzymatic basis for the synthesis of the P. homomalla prostaglandins was studied over the course of almost 30 years. The biochemical pathway remained unresolved largely because of the inability to develop a coral preparation capable of catalyzing prostaglandin biosynthesis in vitro (reviewed in refs. 43 and 44). The occurrence of unusual prostaglandin isomers in the coral only added to the mystique and contributed to speculations on the potential pathway of biosynthesis. Many of the “clues” to the biochemistry turned out to be red herrings. Our results now establish that a COX enzyme is expressed in P. homomalla, and this allows a rational explanation for many of the disparate pieces of evidence on the prostaglandin pathway.
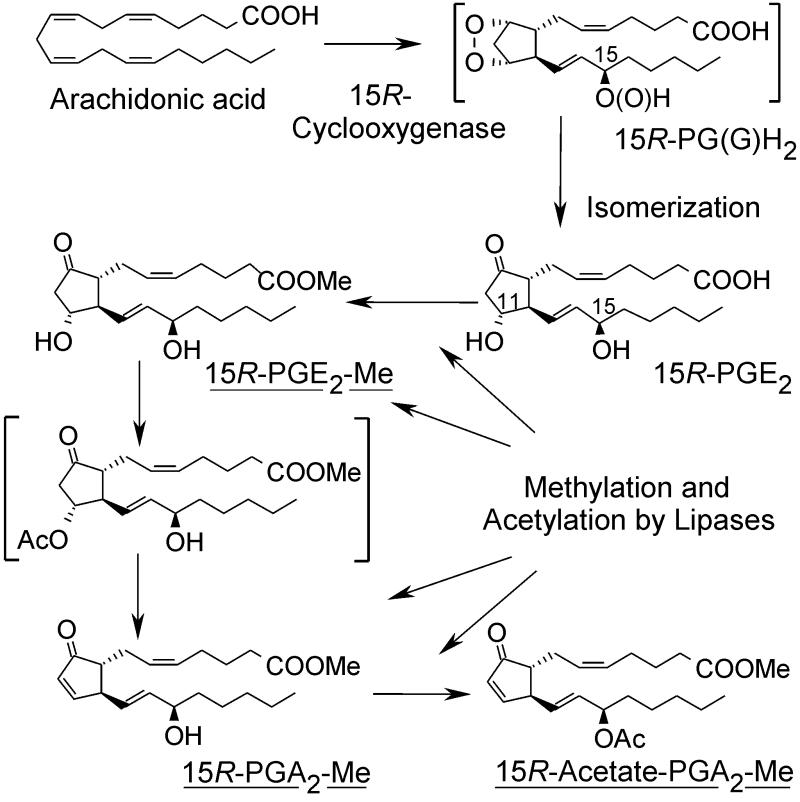
We speculate that biosynthesis via the COX pathway in the coral leads to the prostaglandin endoperoxide and then to PGE2 as the initial major product. We surmise that most of the PGE2 is converted to the methyl ester and acetylated enzymatically (Fig. 6). It is the acetylation that drives the conversion of PGE2 to PGA2 and thus leads to the accumulation of PGA2 methyl ester and its C-15 acetate. Acetylation at C-11 of PGE2 methyl ester is much faster than at C-15 (45). The 11-O-acetyl derivative of PGE2 is prone to elimination of acetic acid resulting in formation of the PGA2, and it may also be subject to lipase-catalyzed elimination (45). Further acetylation gives the PGA2 methyl ester 15-acetate, which then accumulates in the coral. This explains why the 15-acetate of PGA2 is found in P. homomalla, but PGE2 occurs only as the methyl ester (1–5).
Figure 6.
Proposed mechanism for prostaglandin formation in P. homomalla. Prostaglandin esters identified in fresh coral extracts are underlined. Unstable intermediates are in brackets.
Despite the enormously high content of endogenous prostaglandins in P. homomalla preparations, COX activity has never been detected in vitro. Western analysis also suggests that there is a relatively low level of COX protein (see Results and Fig. 3). It appears that P. homomalla does not have major stores of the COX enzyme, and the huge amount of prostaglandins can be explained by the long-term deposition of stable PGA2 esters in lipids. This fits with earlier evidence that there are far higher levels of prostaglandins in P. homomalla than there are stores of available arachidonic acid substrate (46).
Acknowledgments
We thank Omar Parve for helpful discussions, Anu Aaspõllu for assistance with sequencing, and Merike Meier and Monika Drews for help in insect cell culturing. This research was supported by Estonian Science Foundation Grant 3783 and by Fogarty International Center of the National Institutes of Health Grant TW00404 and Parent Grant GM53638. The Keys Marine Laboratory, Long Key, FL, contributed by collection of the coral.
Abbreviations
- COX
cyclooxygenase
- PG
prostaglandin
- HETE
hydroxyeicosatetraenoic acid
- HHT
hydroxyheptadecatrienoic acid
- RP
reversed phase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY004223).
References
- 1.Weinheimer A J, Spraggins R L. Tetrahedron Lett. 1969. 5185–5188. [DOI] [PubMed] [Google Scholar]
- 2.Schneider W P, Hamilton R D, Rhuland L E. J Am Chem Soc. 1972;94:2122–2123. doi: 10.1021/ja00761a060. [DOI] [PubMed] [Google Scholar]
- 3.Light R J, Samuelsson B. Eur J Biochem. 1972;28:232–240. doi: 10.1111/j.1432-1033.1972.tb01906.x. [DOI] [PubMed] [Google Scholar]
- 4.Schneider W P, Morge R A, Henson B E. J Am Chem Soc. 1977;99:6062–6066. doi: 10.1021/ja00460a037. [DOI] [PubMed] [Google Scholar]
- 5.Weinheimer A J. In: Prostaglandins from Plexaura homomalla: Ecology, Utilization and Conservation of a Major Medical Marine Resource. Bayer F M, Weinheimer A J, editors. Coral Gables, FL: Univ. of Miami Press; 1973. [Google Scholar]
- 6.Corey E J, Washburn W N, Chen J C. J Am Chem Soc. 1973;95:2054–2055. doi: 10.1021/ja00787a079. [DOI] [PubMed] [Google Scholar]
- 7.Corey E J, Ensley H E, Hamberg M, Samuelsson B. J. Chem. Soc. Chem. Commun. 1975. , 277–278. [Google Scholar]
- 8.Brash A R, Baertschi S W, Ingram C D, Harris T M. J Biol Chem. 1987;262:15829–15839. [PubMed] [Google Scholar]
- 9.Brash A R. J Am Chem Soc. 1989;111:1891–1892. [Google Scholar]
- 10.Corey E J, Lansbury P T, Yamada Y. Tetrahedron Lett. 1985. , 4171–4174. [Google Scholar]
- 11.Corey E J, d'Alarcao M, Matsuda S P T, Lansbury P T. J Am Chem Soc. 1987;109:289–290. [Google Scholar]
- 12.Koljak R, Boutaud O, Shieh B H, Samel N, Brash A R. Science. 1997;277:1994–1996. doi: 10.1126/science.277.5334.1994. [DOI] [PubMed] [Google Scholar]
- 13.Varvas K, Järving I, Koljak R, Vahemets A, Pehk T, Müürisepp A M, Lille Ü, Samel N. Tetrahedron Lett. 1993. 3643–3646. [Google Scholar]
- 14.Varvas K, Koljak R, Järving I, Pehk T, Samel N. Tetrahedron Lett. 1994. 8267–8270. [Google Scholar]
- 15.Varvas K, Järving I, Koljak R, Valmsen K, Brash A R, Samel N. J Biol Chem. 1999;274:9923–9929. doi: 10.1074/jbc.274.15.9923. [DOI] [PubMed] [Google Scholar]
- 16.Koljak R, Järving I, Kurg R, Boeglin W E, Varvas K, Valmsen K, Ustav M, Brash A R, Samel N. J Biol Chem. 2001;276:7033–7040. doi: 10.1074/jbc.M009803200. [DOI] [PubMed] [Google Scholar]
- 17.Brash A R, Boeglin W E, Chang M S, Shieh B H. J Biol Chem. 1996;271:20949–20957. doi: 10.1074/jbc.271.34.20949. [DOI] [PubMed] [Google Scholar]
- 18.Percival M D, Bastien L, Griffin P R, Kargman S, Ouellet M, O'Neill G P. Protein Expression Purif. 1997;9:388–398. doi: 10.1006/prep.1996.0685. [DOI] [PubMed] [Google Scholar]
- 19.Smith C J, Marnett L J. Arch Biochem Biophys. 1996;335:342–350. doi: 10.1006/abbi.1996.0515. [DOI] [PubMed] [Google Scholar]
- 20.Stork G, Isobe M. J Am Chem Soc. 1975;97:4745–4746. doi: 10.1021/ja00849a042. [DOI] [PubMed] [Google Scholar]
- 21.Schneider C, Boeglin W E, Brash A R. Anal Biochem. 2000;287:186–189. doi: 10.1006/abio.2000.4847. [DOI] [PubMed] [Google Scholar]
- 22.Hamberg M, Samuelsson B. J Biol Chem. 1967;242:5344–5354. [PubMed] [Google Scholar]
- 23.Hecker M, Ullrich V, Fisher C, Meese C O. Eur J Biochem. 1987;16:113–123. doi: 10.1111/j.1432-1033.1987.tb13587.x. [DOI] [PubMed] [Google Scholar]
- 24.Xiao G, Tsai A L, Palmer G, Boyar W C, Marshall P J, Kulmacz R J. Biochemistry. 1997;36:1836–1845. doi: 10.1021/bi962476u. [DOI] [PubMed] [Google Scholar]
- 25.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 26.Picot D, Loll P J, Garavito M. Nature (London) 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 27.Luong C, Miller A, Barnett J, Chow J, Ramesha C, Browner M F. Nat Struct Biol. 1996;3:927–933. doi: 10.1038/nsb1196-927. [DOI] [PubMed] [Google Scholar]
- 28.Kurumbail R G, Stevens A M, Gierse J K, McDonald J J, Stegeman R A, Pak J Y, Gildehaus D, Miyashiro J M, Penning T D, Seibert K, et al. Nature (London) 1996;384:644–648. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 29.Picot D, Garavito R M. FEBS Lett. 1994;346:21–25. doi: 10.1016/0014-5793(94)00314-9. [DOI] [PubMed] [Google Scholar]
- 30.Vane J R, Bakhle Y S, Botting R M. Annu Rev Pharmacol Toxicol. 1998;38:97–120. doi: 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- 31.Garavito R M, Picot D, Loll P J. Adv Prostaglandin Thromboxane Leukotriene Res. 1995;23:99–103. [PubMed] [Google Scholar]
- 32.Garavito R M, Picot D, Loll P J. Curr Opin Struct Biol. 1994;4:529–535. [Google Scholar]
- 33.Gierse J K, Hauser S D, Creely P, Koboldt C, Rangwala S H, Isakson P C, Seibert K. Biochem J. 1995;305:479–484. doi: 10.1042/bj3050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marnett A B, Goodwin D C, Rowlinson S, Kalgutkar A S, Landino L M. In: Comprehensive Natural Products Chemistry. Poulter C D, editor. Vol. 5. Oxford: Elsevier; 1999. pp. 225–261. [Google Scholar]
- 35.Holtzman M J, Turk J, Shornick L P. J Biol Chem. 1992;267:21438–21445. [PubMed] [Google Scholar]
- 36.Meade E A, Smith W L, DeWitt D L. J Biol Chem. 1993;268:6610–6614. [PubMed] [Google Scholar]
- 37.Lecomte M, Laneuville O, Ji C, DeWitt D L, Smith W L. J Biol Chem. 1994;269:13207–13215. [PubMed] [Google Scholar]
- 38.Rowlinson S W, Crews B C, Goodwin D C, Schneider C, Gierse J K, Marnett L J. J Biol Chem. 2000;275:6586–6591. doi: 10.1074/jbc.275.9.6586. [DOI] [PubMed] [Google Scholar]
- 39.Roth G J, Stanford N, Majerus P W. Proc Natl Acad Sci USA. 1975;72:3073–3076. doi: 10.1073/pnas.72.8.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalgutkar A S, Kozak K R, Crews B C, Hochgesang G P, Jr, Marnett L J. J Med Chem. 1998;41:4800–4818. doi: 10.1021/jm980303s. [DOI] [PubMed] [Google Scholar]
- 41.Schneider C, Brash A R. J Biol Chem. 2000;275:4743–4746. doi: 10.1074/jbc.275.7.4743. [DOI] [PubMed] [Google Scholar]
- 42.Thuresson E D, Lakkides K M, Smith W L. J Biol Chem. 2000;275:8501–8507. doi: 10.1074/jbc.275.12.8501. [DOI] [PubMed] [Google Scholar]
- 43.Gerwick W H, Nagle D G, Proteau P J. Top Curr Chem. 1993;167:117–180. [Google Scholar]
- 44.Gerwick W H. In: Comprehensive Natural Products Chemistry. Sankawa U, editor. Vol. 1. Oxford: Elsevier; 1999. pp. 207–254. [Google Scholar]
- 45.Parve O, Järving I, Martin I, Metsala A, Vallikivi I, Aidnik M, Pehk T, Samel N. Bioorg Med Chem Lett. 1999;9:1853–1858. doi: 10.1016/s0960-894x(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 46.Light R J. Biochim Biophys Acta. 1973;296:461–466. doi: 10.1016/0005-2760(73)90105-7. [DOI] [PubMed] [Google Scholar]