Abstract
Electronic Medical Records (EMRs) are valuable resources for clinical observational studies. Smoking status of a patient is one of the key factors for many diseases, but it is often embedded in narrative text. Natural language processing (NLP) systems have been developed for this specific task, such as the smoking status detection module in the clinical Text Analysis and Knowledge Extraction System (cTAKES). This study examined transportability of the smoking module in cTAKES on the Vanderbilt University Hospital’s EMR data. Our evaluation demonstrated that modest effort of change is necessary to achieve desirable performance. We modified the system by filtering notes, annotating new data for training the machine learning classifier, and adding rules to the rule-based classifiers. Our results showed that the customized module achieved significantly higher F-measures at all levels of classification (i.e., sentence, document, patient) compared to the direct application of the cTAKES module to the Vanderbilt data.
INTRODUCTION
Electronic medical record (EMR) systems provide a history of diagnoses, treatment, and response for patients. Longitudinal data stored in EMRs contain valuable medical information that can provide evidence for hypothesis generation and reveal associations between health problems, medications, and treatments. Unfortunately, detailed EMR patient information is often embedded in narrative reports, expressed in the form of fragmented, unstructured, and ungrammatical text. The medical informatics community has thus invested much effort to develop methods to abstract relevant information from clinical narratives. Clinical natural language processing (NLP) systems have been developed for both general concept identification purposes (e.g., MedLEE1, MetaMap2, KnowledgeMap3, and cTAKES4) and specific information extraction tasks such as medication and signature identification5–9.
Tobacco use is linked to diverse diseases such as cardiovascular disease (myocardial infarction, strokes), numerous types of cancers, infections, and gastrointestinal diseases. The World Health Organization (WHO) estimated that tobacco use has caused 100 million deaths over the 20th century10. However, patient smoking status information is often not recorded as a structured field in the EMR. Researchers often manually review clinical charts to determine the tobacco use of patients for clinical studies, which is costly and time-consuming. Hence, automatic identification of patient tobacco use becomes a challenge but also an opportunity for the clinical NLP community. The most common form of tobacco use is smoking, and consequently has been the major target of NLP efforts. The 2006 i2b2 (informatics for integrating biology and the bedside) Shared Task on NLP presented the Smoking Status Discovery challenge that raised the question: Can a patient’s smoking status be automatically and correctly identified from their clinical records?
The challenge asked participants to classify patient records into five predefined categories - past smoker (P), current smoker (C), smoker (S), non-smoker (N), and unknown (U)11. Past smokers are patients who have not smoked for at least one year. Current smokers include patients who have discharge summaries indicating that they are current smokers or have smoked within the past year. The smoker (S) category describes cases where not enough information is available in the medical records to classify a patient as either P or C. Non-smokers are the people who have never smoked and “unknown” includes patients with no smoking status mentions in records. A total of 11 teams participated in the smoking status detection challenge and each submitted up to 3 system runs, providing a total of 23 submissions. A comprehensive summary of the systems developed is provided by Uzuner et al.12. Among the submitted system runs, 12 runs achieved micro-averaged F-measures above 84% with the overall best performing system by Clark et al.13 achieving 90%. Most of the top performing systems have either filtered out the “unknown” documents before any classification or assigned “unknown” labels to documents when no smoking-related information is found13–18. Most systems employed machine learning techniques including Clark et al.13,14,17,19,20 and only one system used purely rule-based method21. Several teams combined rule-based and machine learning methods15,16,18, and others developed their own methods22,23.
As an entry system to the 2006 i2b2 challenge for patient smoking status identification, researchers from the Mayo Clinic developed a hybrid system by combining machine learning and rule-based methods16. The system was built within the IBM’s Unstructured Information Management Architecture (UIMA)24 and used components from the Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES)4. Later, Sohn et al.25 described extensions and improvements to the cTAKES smoking status classification component, which included improvements made to the negation detection for non-smokers, use of temporal resolution to distinguish a past smoker from current smoker, and advanced rules for recognizing ‘unknown’ categories. The extended system was reported to have a micro-averaged F-measure of 96.7%.25 In March 2009, cTAKES was released as an open-source system under the Open Health NLP Consortium (OHNLP)26 to allow researchers to build shareable NLP systems for information extraction from clinical text.
In this study, we applied the cTAKES smoking module to extract patient smoking status from clinical notes in the Vanderbilt University Medical Center (VUMC) EMR. The objective of this study is to assess the transportability of the smoking status detection module in cTAKES across institutions. More specifically, we want to answer two questions: 1) can the cTAKES smoking module developed at Mayo Clinic achieve satisfactory results on Vanderbilt data without any modification? and 2) what modifications are needed in order to reach the desired performance for the cTAKES smoking module? Furthermore, most existing studies aimed to determine smoking status at the document-level. However, a patient typically has many different types of clinical documents accumulated over a period of time. Patient-level smoking status is what needed in clinical research. It is not a trivial task to determine patient-level smoking statuses over a multitude of documents. The current smoking detection module in cTAKES25 attempted to assign patient-level smoking status via a rule-based classifier; however, they only reported system performance based on their annotated document-level labels. In this study, we evaluated the patient-level performance using the predicted document-level labels.
METHODS
The cTAKES smoking status detection module was evaluated on our data retrieved from the Vanderbilt Synthetic Derivative (SD) database, a de-identified version of the VUMC EMR27. In this study, we collected a cohort of 400 individuals from the SD and manually annotated them at sentence, document, and patient levels. These annotated data were then divided into training and test sets at each annotation level. The training sets were used in the development of the customized cTAKES smoking module for Vanderbilt data. We then evaluated both the original and the customized cTAKES modules using the independent test sets.
The 2006 i2b2 challenge asked participants to classify documents into five categories: past smoker (P), current smoker (C), smoker (S), non-smoker (N), and unknown (U). However, our data do not contain the S and U cases; thus, we only evaluated the cTAKES smoking status detection module on the classification of three categories: P, C, and N. Initial results have shown that direct application of the smoking detection module produced poor performance. In response, we modified the algorithm by analyzing the training data. Detail descriptions of the cTAKES module, our data set construction and module modifications are provided in the sections below.
Data sets
From an ongoing clopidogrel pharmacogenomics study, we collected a cohort of 400 individuals from the SD, where the patient level smoking status was annotated by domain experts. Our patient level annotation contained three categories only: past smoker (P), current smoker (C), non-smoker (N). Thus, we evaluated the cTAKES smoking status detection module on the classification of three categories: P, C, and N in this study.
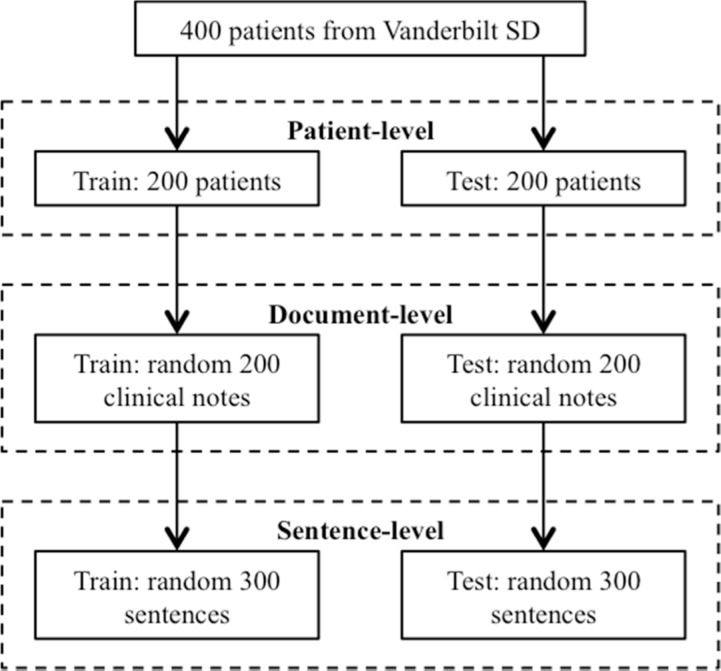
Figure 1 shows the process for constructing annotated data sets in this study. We divided the 400 patients into two groups: a training set with 200 patients and a test set with the remaining 200 patients. For each patient, we collected all types of clinical note about the patient from the SD. The patients in the training set had 27,702 clinical notes and patients in the test set had 32,021 notes. From those documents, we collected those contained at least one smoking related keywords (e.g., smoker) and randomly selected 200 documents from each set for document-level annotation. Lastly, for the sentence-level annotation, we randomly selected 300 sentences (containing smoking related keywords) from the 200 documents in the training or test set and manually annotated each sentence with a smoking status following the 2006 i2b2 challenge guidelines. All training sets were used for the development and improvement of the customized cTAKES smoking module and all test sets were kept independent and used for final evaluation only.
Figure 1.
The process of creating annotated data sets for training and testing at different levels.
The distributions of the sentence, document, and patient-level labels for both training and test dataset are shown in Table 1. To note, the rule-based document-level classification do not require annotation of the training set; thus, no status distribution is available for the document-level training set.
Table 1.
Gold standard smoking status distributions for the training and test datasets.
| Training Set | Test Set | |||||
|---|---|---|---|---|---|---|
| P | C | N | P | C | N | |
| Patient-level | 94 | 62 | 44 | 89 | 44 | 67 |
| Document-level | - | - | - | 80 | 41 | 79 |
| Sentence-level | 105 | 110 | 85 | 111 | 85 | 104 |
cTAKES Smoking Status Detection Module
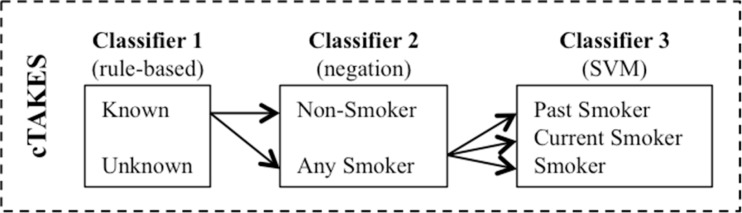
The cTAKES smoking status detection module makes predictions on three levels: sentence-level, document-level, and patient-level. Their sentence-level classification algorithm aims to label a sentence with past smoker (P), current smoker (C), smoker (S), non-smoker (N), or unknown (U). The sentence-level consists of three layers of classifiers (Figure 2). Classifier 1 is rule-based and aims to identify two categories of sentences - unknown and known (i.e. smoking-related). All smoking-related sentences are then passed to Classifier 2 that uses negation detection to distinguish non-smokers from any smokers. Finally, the sentences in the ‘any smokers’ category are passed to a support vector machine (SVM)28–30. The document-level classification uses a rule-based logic to assign document-level smoking statuses using all sentence labels from the sentence-level classifiers. Similarly, the patient-level smoking status classification is completed with a summarization logic applied over all patients’ documents. In this study, we used the version 1.1.0 of the cTAKES smoking module downloaded from http://sourceforge.net/projects/ohnlp/ in September, 2011. For initial evaluation, we applied the cTAKES module to the test set without any modification.
Figure 2.
The architecture of cTAKES smoking status detection module for sentence-level classification.
Customized cTAKES Smoking Status Detection Module
We applied the original cTAKES smoking module to the training sets and analyzed its errors. Our analysis showed that document-level and patient-level rules generally worked fine; but the sentence-level classification results were poor for Vanderbilt text. Therefore we proposed following modifications to improve its performance.
Sentence-Level Classification Two modifications were applied to this step. First, we modified the rule-based classifiers 1 and 2 by adding additional keywords collected from Vanderbilt clinical text. For example, we inserted new smoking-related keywords (i.e. anxiety, dependence, hx, positive, and social_history) and some ‘unknown’ sentences related words (i.e. raspy smoker’s laugh, smells of cigarette smoke). Second, we re-trained the SVM classifier with the updated feature list of smoking-related keywords for the annotated sentences from the training set. Since SVMs are very sensitive to the kernel parameter g and penalty parameter C, we optimized the parameters through a 5-fold cross-validation using the 300 training sentences. In this study, LIBSVM31 was employed and the highest average cross-validation accuracy of 93.49% was achieved by a RBF kernel with g = 0.8 and C = 60. After the optimal parameters were set, we retrained the SVM classifier with all 300 training sentences and evaluated on the 300 sentences in the test set.
Document-Level Classification We manually reviewed the existing rules implemented in the cTAKES smoking status detector for the document-level classification using our training data and found that they were generally working fine. However, there are some cases in our dataset that would not work well with the rules; thus, we inserted one new rule. The added rule gives precedence to non-smoker sentences over current smoker sentences if the document has both types. The reason for implementing this rule was due to sentence boundary issues where one sentence is broken down into two lines (separated by a carriage return); thus in case of negation, the smoking status detector won’t be able to pick up the negation for the second part of the sentence.
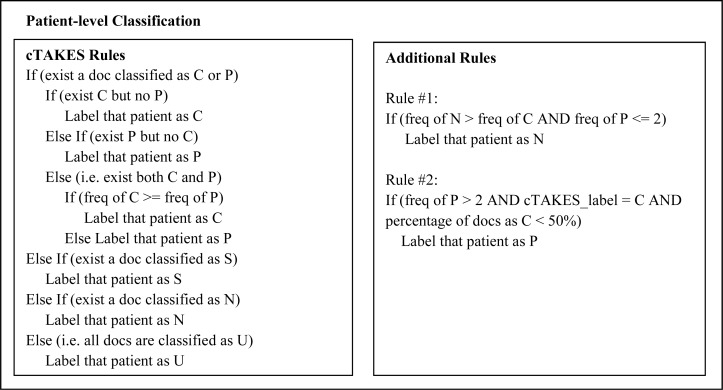
Patient-Level Classifications We adapted the patient-level rules from cTAKES first and applied them to the training data. By analyzing errors from the training set, we further developed two additional patient-level rules for Vanderbilt data (Figure 3). The first rule was to fix errors where N was incorrectly labeled as C, by comparing the counts of documents labeled as C and N. In the second rule, we calculated the percentage of documents labeled as C and used it to correct errors where P was incorrectly categorized as C by the cTEAKES rules. The contribution of these new rules is analyzed in Results.
Figure 3.
Patient-level classification rules
In addition, unlike the i2b2 challenge that used only discharge summaries, this study included many note types as sources to determine smoking status. The SD database contains many types of notes including discharge summary (DS), history & physicals (HP), problem list (PL), clinic visit (CV), clinical communication (CC), clinical forms (FORM), radiology notes (RAD), pathology notes (PATH), problem list (PL), and many others. Based on our observation on the results from the training sets, we noticed that the performance of cTAKES module was poor when all notes were used to determine patient smoking status. Therefore, through discussions with domain experts, we limited the notes to those most likely contain quality information about smoking status, namely PL, HP, and CV.
Evaluation
Systems’ outputs were compared with the gold standard in test sets. Precision, recall, and F-measure were calculated for each status category and used as metrics for this study (Equation 1). Precision is the proportion of true positives against all predicted positive results. Recall is the fraction of true positives among all positives. F-measure is the harmonic mean of precision and recall.
| (1) |
To evaluate overall system performance, we also computed micro-averages and macro-averages of each of the above metrics. Macro-averages calculate the evaluation metrics for each category first and then taking the average; thus it discounts the performance of better-populated categories. In contrast, micro-averages are calculated first using a global count of each class and then averaging these sums; so they are dominated by those categories with the greatest number of samples. We reported both macro-averages and micro-averages of each metric. Followings are the formulas for macro-average (Equation 2) and micro-average (Equation 3) of each metric.
| (2) |
| (3) |
where M is the number of categories and Pi, Ri, Fi, TPi, and FNi are precision, recall, F-measure, true positive, and false negative for each status category, respectively.
Using the test set, we measured the performance of smoking status detectors at three different levels: sentence, document, and patient. Prediction results were reported accumulatively, meaning a latter classifier takes the prediction from the previous classifier as its input. For example, the predicted labels of each sentence were passed through rule-based logics for document-level classification, and then document-level predictions were used to make smoking status predictions at patient level. The training data were only used for training the SVM classifier and developing new rules, and the test data were used for evaluation.
RESULTS
The original cTAKES smoking status detection module and the customized module were evaluated on our test dataset for sentence, document, and patient-level classifications. For the sentence-level classification, precision, recall, and F-measure produced by the cTAKES module and the customized module over the test set are summarized in Table 2. As shown, when applied unchanged, the smoking status detector in cTAKES could only identify current and past smokers with F-measures around 60% while the F-measure for predicting non-smokers was 97%. After altering the model as described above and retraining the SVM classifier with Vanderbilt data, F-measures for current smokers and past smokers reached 92% and 94%, respectively. Both macro- and micro-average F-measures also increased significantly from 75% to 94%.
Table 2.
Sentence-level classification evaluation on the test set
| cTAKES Module | Customized Module | |||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-measure | Precision | Recall | F-measure | |
| Current Smoker (C) | 0.58 | 0.81 | 0.68 | 0.90 | 0.94 | 0.92 |
| Past Smoker (P) | 0.76 | 0.49 | 0.59 | 0.96 | 0.92 | 0.94 |
| Non-smoker (N) | 0.96 | 0.97 | 0.97 | 0.96 | 0.97 | 0.97 |
| Macro Average | 0.77 | 0.76 | 0.75 | 0.94 | 0.94 | 0.94 |
| Micro Average | 0.78 | 0.75 | 0.75 | 0.94 | 0.94 | 0.94 |
Results comparing the original cTAKES smoking status detection module and the customized module at the document-level classification are displayed in Table 3. The customized module had higher F-measures for all three labels. The micro-average F-measure increased 14% and macro-average F-measure increased 15%.
Table 3.
Document-level classification evaluation on the test set
| cTAKES Module | Customized Module | |||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-measure | Precision | Recall | F-measure | |
| Current Smoker (C) | 0.48 | 0.90 | 0.63 | 0.76 | 0.93 | 0.84 |
| Past Smoker (P) | 0.90 | 0.54 | 0.67 | 1.0 | 0.79 | 0.87 |
| Non-smoker (N) | 0.93 | 0.85 | 0.88 | 0.89 | 0.97 | 0.93 |
| Macro Average | 0.77 | 0.76 | 0.73 | 0.88 | 0.89 | 0.88 |
| Micro Average | 0.82 | 0.74 | 0.75 | 0.90 | 0.89 | 0.89 |
The patient-level classification results exhibit similar trend (Table 4) where F-measures of all smoking status categories including the micro- and macro-average F-measures improved significantly in the customized module. We also reported performance for identifying the presence and absence of smoking because some observational studies only consider whether a patient has ever smoked or not. In this case, the original cTAKES module produced 84% and 46% in F-measures for presence and absence, respectively. On the other hand, the customized module had F-measures of 93% and 87% for presence and absence, respectively.
Table 4.
Patient-level classification evaluation on the test set
| cTAKES Module | Customized Module | |||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-measure | Precision | Recall | F-measure | |
| i2b2 Classification | ||||||
| Current Smoker (C) | 0.30 | 0.84 | 0.45 | 0.67 | 0.92 | 0.78 |
| Past Smoker (P) | 0.82 | 0.51 | 0.63 | 0.93 | 0.73 | 0.82 |
| Non-smoker (N) | 0.91 | 0.31 | 0.47 | 0.85 | 0.89 | 0.87 |
| Macro Average | 0.68 | 0.55 | 0.52 | 0.82 | 0.85 | 0.83 |
| Micro Average | 0.74 | 0.52 | 0.54 | 0.85 | 0.82 | 0.83 |
| Ever/Never Classification | ||||||
| Presence (current or past smokers) | 0.74 | 0.98 | 0.84 | 0.95 | 0.92 | 0.93 |
| Absence (non-smokers) | 0.91 | 0.31 | 0.46 | 0.85 | 0.89 | 0.87 |
Moreover, we analyzed the contribution of the two added rules on patient-level classification by comparing to the performance of original cTAKES rules, when the same document-level inputs were used. The improvements in F-measures were significant where the micro-average F-measure increased from 65% to 83% (Table 5).
Table 5.
Effect of additional rule on patient-level classification
| cTAKES Rules | Customized Rules | |||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-measure | Precision | Recall | F-measure | |
| i2b2 Classification | ||||||
| Current Smoker (C) | 0.42 | 0.92 | 0.58 | 0.67 | 0.92 | 0.78 |
| Past Smoker (P) | 0.93 | 0.67 | 0.78 | 0.93 | 0.73 | 0.82 |
| Non-smoker (N) | 0.91 | 0.31 | 0.47 | 0.85 | 0.89 | 0.87 |
| Macro Average | 0.75 | 0.62 | 0.63 | 0.82 | 0.85 | 0.83 |
| Micro Average | 0.82 | 0.60 | 0.65 | 0.85 | 0.82 | 0.83 |
| Ever/Never Classification | ||||||
| Presence (current or past smokers) | 0.78 | 0.98 | 0.87 | 0.95 | 0.92 | 0.93 |
| Absence (non-smokers) | 0.91 | 0.31 | 0.47 | 0.85 | 0.89 | 0.87 |
DISCUSSION
Despite the success in developing NLP system for clinical data extraction, transferability of these systems to different institutions is still in question. Hripsak et al.32 conducted a study on the transferability of MedLEE, a general purpose NLP system, to other institution’s reports and observed a small drop in performance. Transporting a general purpose system to a different institution may be different from transferring a specific NLP module to a different institution. This study demonstrates that direct application of the cTAKES smoking status module at Vanderbilt did not produce satisfactory performance. The drop in performance was much larger compared to the observation by Hripcsak et al.32 in transferring a general purpose system to a different institution. However, higher performance of the existing module was achieved to Vanderbilt corpora with reasonable effort, by modifying the cTAKES algorithm to include a selection of relevant note types, annotation of local data to retrain the SVM classifier in recognizing past and current smokers, and addition of precedence rule logics.
The retrained SVM classifier could identify past and current smokers at the sentence level with F-measures of 94% and 92% respectively compared to 59% and 68% with the original module. Similarly, the document and patient level classifications for the past and current smokers also improved significantly (i.e., 20% and 21% increases in F-measure for past and current labels respectively at the document level and 19% and 33% increases in F-measure for past and current smokers respectively at the patient level). The micro-average F-measures increased 19% for sentence-level, 14% for document-level, and 29% for patient-level classifications. An interesting property we noticed was that the improvement for non-smoker identification was not as large as other two smoking status labels. With the additional rule at the document level, there was only 5% increase in F-measure for non-smokers. This may imply that the cTAKES logic rules for document-level classification were well defined and more resistant to change in data than machine learners. For the patient-level classification, direct application of the original cTAKES rules generated poor micro-average F-measure of 65%. After applying two additional rules (Figure 3), the micro-average F-measure improved significantly to 83%; however, it is in low eighties. This may suggest that the smoking status detection task for the patient-level over a multitude of documents is not trivial, which requires further exploration. As a future work, we plan to develop more sophisticated algorithms to automatically learn rules.
In Sohn et al.25, the cTAKES smoking status detection module was reported to produce F-measures of 70.6% for current smokers, 85.7% for past smokers, and 96.1% for non-smokers at the document-level. After adapting the system to our data, the customized module generated F-measures of 84% for current smokers, 87% for past smokers, and 93% for non-smokers, which were similar to Sohn et al.’s results. It is important to note that the patient-level classification in Sohn et al.25 used manually annotated document-level labels as input, and they reported results of identifying all patients’ (N=36) smoking statuses correctly. In this study, we used predicted document labels to determine patient level smoking status (N=200), and our results showed much lower performance, especially for current and past smokers (see Table 4). Much of the lower performance in these two groups resulted from misclassification of a smoker as either a past or current smoker because a patient may go through rounds of quitting and restarting smoking, which would complicate the classification task. Thus, when using a binary classification scheme of ever/never smoker, the algorithms performed much better (F-measure 0.93).
It was also observed that restricting the types of notes significantly improved the results. When all notes were applied to determine patient smoking status, our customized module produced 42% and 61% in F-measures for current and past smokers, respectively. On the other hand, when limited sets of notes were applied, F-measures significantly improved to 78% and 82% for current and past smokers, respectively. Additional clinical notes may contain unseen patterns of smoking text, for which the NLP system would interpret it incorrectly, thus impacting the final status. For example, some letters to patients (e.g., discharge letters) contain general comments about smoking, e.g., “Smoking is injurious to health”. The system would classify it as a present smoker in this case, which was wrong. Therefore, our experimental results showed that limiting the note types to certain types of clinical notes yielded a much better performance. As EMR systems become more robust and include more diverse types of interactions between the patient and healthcare system, we expect that this phenomenon will increase. Future NLP efforts in a variety of fields may also benefit from restricted document sets. The “optimal” document set for NLP is an open question in informatics.
Moreover, we examined the cTAKES smoking status detection module errors. One source of errors was the presence of abbreviated smoking-related keywords in our data. For example, the sentence “Tob neg” was incorrectly labeled as current smoker by the cTAKES module because it does not recognize “neg” as negation. In another example, the cTAKES module classified “He has hx of smoking” as current smoker rather than past smoker because its keyword list does not contain “hx” for history. We added these words to the customized module. Another source of errors was observed to be from the SVM. For example, in the sentence “smoking six years ago”, the word “ago” is in the keyword list, but the SVM classifier still classified it as current smoker. For the sentence “70 year old male smoker with lung mass”, the cTAKES module appears to need present-tense verbs like “is” to identify it as current smoker. In those cases, we found that including such sentences in the training data would help the SVM classifier to correctly identify them as past smokers.
When we analyzed the errors produced by the customized smoking status detection module, sentence boundary detection was observed to be an important factor. In our data, many full sentences were split incorrectly into multiple ones by forced carriage returns, which may lead to missed negation words. For instance, the sentence “Denies EtOH, smoking, illicit drug use” was split into two sentences: “Denies” and “EtOH, smoking, illicit drug use”. Thus the smoking status detection module classified the latter sentence incorrectly as current smoker. The rule we added for the document-level classification only worked in situations where there is a non-smoker sentence in the same document. If no ‘non-smoker’ sentences exist in the same document as the wrongly classified current smoker sentence, the wrong classification would not be corrected. Due to those reasons, the performance for identifying current and past smokers at patient level was still not optimal, which requires further investigation.
Finally, this study adds to the growing knowledge about transportability of patient feature extraction algorithms. Recent work has demonstrated that deterministic, rule-based algorithms using billing codes, laboratory values, and medication data, and NLP can port well to diverse EMR implementations33,34. Other works has similarly demonstrated that logistic regression algorithms can work effectively in different healthcare settings, different EMRs, and different NLP engines35. These results highlight the importance of local site validation and optimization, for some algorithms. Machine learning approaches may be especially sensitive to local environments.
CONCLUSION
This study examined the portability of the smoking status detection module in cTAKES on individuals derived from the VUMC EMR. Results demonstrated that modifications were necessary to achieve adequate performance. The modifications included filtering of the notes, annotation of new data for training the SVM classifier, and addition of rules for the rule-based classifiers. Outside of the cost of annotation, however, these changes required a relatively modest input of effort. Our results showed that the customized smoking status detection module achieved significantly higher F-measures at all levels of classification (i.e., sentence, document, patient) compared to the direct application of the cTAKES module to Vanderbilt data.
Acknowledgments
This study was supported in part by grants from the National Cancer Institute (NCI) R01CA141307. The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s Synthetic Derivative which is supported by institutional funding and by the Vanderbilt CTSA grant 1UL1RR024975-01 from NCRR/NIH.
REFERENCES
- 1.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994 Mar-Apr;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010 May-Jun;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denny JC, Smithers JD, Miller RA, Spickard A., 3rd “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc. 2003 Jul-Aug;10(4):351–362. doi: 10.1197/jamia.M1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savova GK, Kipper-Schuler K, Buntrock JD, Chute CG. UIMA-based clinical information extraction system. LREC 2008: Towards enhanced interoperability for large HLT systems: UIMA for NLP. 2008 [Google Scholar]
- 5.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010 Jan-Feb;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):514–518. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spasic I, Sarafraz F, Keane JA, Nenadic G. Medication information extraction with linguistic pattern matching and semantic rules. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):532–535. doi: 10.1136/jamia.2010.003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):524–527. doi: 10.1136/jamia.2010.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meystre SM, Thibault J, Shen S, Hurdle JF, South BR. Textractor: a hybrid system for medications and reason for their prescription extraction from clinical text documents. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):559–562. doi: 10.1136/jamia.2010.004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO report on the global tobacco epidemic, 2008. World Health Organization; [Google Scholar]
- 11.Uzuner O, Szolovits PS, Kohane I. i2b2 workshop on natural language processing challenges for clinical records. Paper presented at: Fall Symposium of the American Medical Informatics Association; 2006. [Google Scholar]
- 12.Uzuner O, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008 Jan-Feb;15(1):14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark C, Good K, Jezierny L, Macpherson M, Wilson B, Chajewska U. Identifying smokers with a medical extraction system. J Am Med Inform Assoc. 2008 Jan-Feb;15(1):36–39. doi: 10.1197/jamia.M2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aramaki E, Imai T, Miyo K, Ohe K. Patient status classification by using rule based sentence extraction and BM25 kNN-based classifier. Paper presented at: i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- 15.Cohen AM. Five-way smoking status classification using text hot-spot identification and error-correcting output codes. J Am Med Inform Assoc. 2008 Jan-Feb;15(1):32–35. doi: 10.1197/jamia.M2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savova GK, Ogren PV, Duffy PH, Buntrock JD, Chute CG. Mayo clinic NLP system for patient smoking status identification. J Am Med Inform Assoc. 2008 Jan-Feb;15(1):25–28. doi: 10.1197/jamia.M2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szarvas G, Farkas R, Ivan S, Kocsor A, Fekete RB. Automatic extraction of semantic content from medical discharge records. Paper presented at: i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- 18.Wicentowski R, Sydes MR. Using implicit information to identify smoking status in smoke-blind medical discharge summaries. J Am Med Inform Assoc. 2008 Jan-Feb;15(1):29–31. doi: 10.1197/jamia.M2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrero F, Hidalgo JG, Puertas E, Mana M, Mata J. Quick prototyping of high performance text classifiers. Paper presented at: i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- 20.Pederson T. Determining smoker status using supervised and unsupervised learning with lexical features. Paper presented at: i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- 21.Guillen R. Automated de-identification and categorization of medical records. Paper presented at: i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- 22.Heinze DT, Morsch ML, Potter BC, Sheffer RE., Jr Medical i2b2 NLP smoking challenge: the A-Life system architecture and methodology. J Am Med Inform Assoc. 2008 Jan-Feb;15(1):40–43. doi: 10.1197/jamia.M2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rekdal M. Identifying smoking status using Argus MLP. Paper presented at: i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- 24.Unstructured Information Management Architecture (UIMA) http://uima-framework.sourceforge.net.
- 25.Sohn S, Savova GK. Mayo clinic smoking status classification system: extensions and improvements. AMIA Annu Symp Proc. 2009;2009:619–623. [PMC free article] [PubMed] [Google Scholar]
- 26.Open Health Natural Language Processing (OHNLP) Consortium http://www.ohnlp.org.
- 27.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008 Sep;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keerthi SS, Shevade SK, SBhattacharyya C, Murthy KRK. Improvements to Platt’s SMO algorithm for SVM classifier design. Neural Computation. 2001;13(3):637–649. [Google Scholar]
- 29.Platt J. Advances in Kernel Methods - Support Vector Learning. MIT Press; 1998. Machines using sequential minimal optimization. [Google Scholar]
- 30.Witten IH, Frank E. Data Mining: Practical machine learning tools and techniques. 2nd ed. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- 31.Fan RE, Chen PH, Lin CJ. Working set selection using second order information for training support vector machines. Journal of Machine Learning Research. 2005 Dec;6:1889–1918. [Google Scholar]
- 32.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods of information in medicine. 1998 Jan;37(1):1–7. [PubMed] [Google Scholar]
- 33.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011 Oct 7;89(4):529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012 Mar 1;19(2):212–218. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012 Feb 28; doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]