Abstract
The development of Electronic Health Record (EHR)-based phenotype selection algorithms is a non-trivial and highly iterative process involving domain experts and informaticians. To make it easier to port algorithms across institutions, it is desirable to represent them using an unambiguous formal specification language. For this purpose we evaluated the recently developed National Quality Forum (NQF) information model designed for EHR-based quality measures: the Quality Data Model (QDM). We selected 9 phenotyping algorithms that had been previously developed as part of the eMERGE consortium and translated them into QDM format. Our study concluded that the QDM contains several core elements that make it a promising format for EHR-driven phenotyping algorithms for clinical research. However, we also found areas in which the QDM could be usefully extended, such as representing information extracted from clinical text, and the ability to handle algorithms that do not consist of Boolean combinations of criteria.
Introduction and Motivation
The growing adoption of electronic health records (EHRs) is a crucial enabling step for the large-scale re-use of clinical data for research projects and for healthcare quality measurement. For clinical research, there is great interest in using EHRs to automatically identify patients that match clinical study eligibility criteria, making it possible to leverage existing patient data to inexpensively and automatically generate lists of patients that possess desired phenotypic traits by using EHR-driven phenotyping algorithms.1,2 For quality measurement, there is a strong push to use EHR data for generating much more comprehensive patient population metrics than is feasible with paper records and manual review.3 Yet the development of EHR-based phenotyping algorithms and quality measures is a non-trivial and highly iterative process involving domain experts and data analysts.4 It is therefore desirable to make it as easy as possible to re-use them across institutions in order to minimize the degree of effort involved, as well as the potential for errors due to ambiguity or under-specification. Part of the solution to this issue is the adoption of an unambiguous and precise formal specification language, imposing a standard representational syntax and semantics. In the domain of healthcare quality measurement, the non-profit National Quality Forum (NQF)5 has recently developed and released the Quality Data Model (QDM),6 which is an information model for representing EHR-based quality “eMeasures”.3 The primary objective of our current study is to investigate the suitability of the QDM for also representing EHR-driven phenotyping algorithms designed for clinical research.
Our experience in the development of phenotyping algorithms stems from work performed as part of the electronic Medical Records and Genomics (eMERGE)7 consortium, a network of seven sites using data collected in the EHR as part of routine clinical care to detect phenotypes for use in genome-wide association studies. The successful first phase of eMERGE involved the development and validation of 14 phenotyping algorithms, which were shared among five separate sites with widely diverse EHR systems.8 Execution of these phenotypes has resulted in new genetic discoveries using data from one and across multiple sites.9,10 The second phase of eMERGE, beginning in 2011, added two additional sites and has the goal of developing and disseminating a total of 35 EHR-based phenotyping algorithms. It has become abundantly clear to those involved in this effort that the adoption of an appropriate formal representation language for the algorithms would provide practical benefits for both portability and presentation of results. The work in this paper is framed by a pragmatic attempt to realize these benefits in the context of lessons learned from the eMERGE project.
A variety of alternative possibilities exist for representing phenotype definition criteria,11 including the HL7 Arden Syntax,12 the SAGE guideline model,13 and the GELLO clinical decision support language.14 However, as described by Weng et al.11 and in our own prior work4 there is currently no definitive formal language to select for the purpose of representing EHR-based phenotyping algorithms. Consequently, we investigate the suitability of the QDM for this purpose, given current momentum behind its adoption due to the implementation of Meaningful Use reporting requirements put into place by the Centers for Medicare & Medicaid Services (CMS).15–17 From our perspective, it is evident that there is substantial overlap between the representational needs of EHR-based quality measures and the needs of EHR-based phenotyping algorithms developed for clinical research. Quality measures and phenotyping algorithms both share the requirement of delineating sets of structured codes for defining inclusion and exclusion criteria, the requirement of combining such elements using complex Boolean logic, and the requirement for a temporal expression language that can express timing constraints on clinical events and states. The QDM provides these necessary core elements (among others), making it a promising representational scheme and information model for clinical research phenotyping algorithms.
In order to evaluate the suitability of the QDM for expressing and sharing phenotype selection algorithms developed for clinical research, we selected nine algorithms that have previously been developed as part of the eMERGE project and represented them in terms of the QDM. We have done so by making use of the web-based Measure Authoring Tool (MAT) that has been provided by the NQF to support the creation of QDM-based eMeasures.18 In the following sections, we describe our experiences performing this exercise, including an analysis of both the QDM and the MAT. Our analysis of the QDM highlights the elements of the model that are particularly useful to our endeavor. We follow up this analysis with a discussion of areas in which the QDM could be usefully augmented, paying particular attention to the problem of representing the extraction of information stored in clinical text, and to the issue of extending the QDM to types of classification models that are not based solely on Boolean combinations of inclusion and exclusion criteria. We conclude with a discussion on issues related to the mapping from formal QDM specifications to executable code, including the possibility of automating these mappings.
Background
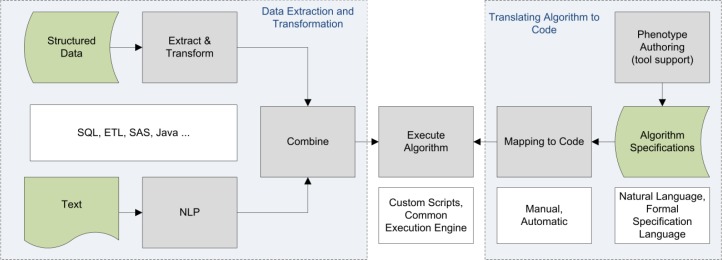
The growing adoption of EHR systems19,20 opens the door to secondary uses of large quantities of clinical data for quality improvement efforts and for clinical research on patient populations. These two use cases share some common requirements, including at the most basic level the need to extract relevant cohorts of patients using selection criteria based on information stored in the EHR. For quality improvement efforts, these cohorts serve as inputs to various quality metrics and reporting tools. For clinical research, these cohorts can be linked to biorepository data,7 and can be used as input to statistical analysis tools for data mining purposes.21 The basic flow of extracting cohorts from the EHR is essentially the same across these use cases, and can be thought of abstractly in terms of the schematic workflow shown in Figure 1.
Figure 1.
Schematic workflow for executing EHR-based phenotyping algorithms.
Executing a phenotyping algorithm broadly involves two steps: (1) translating an algorithm specification into executable code, and (2) extracting data from multiple information sources (EHRs, data warehouses and other clinical information systems) and converting it into the format required by the algorithm. In the first phase of the eMERGE project, algorithms were specified using free text descriptions, tables, flowcharts, and lists of structured codes.4 Mappings to executable code involved the work of data analysts and informaticians at each individual site where the algorithm was implemented, a time consuming process that had to be repeated for every site and every algorithm. Members of the eMERGE consortium had the advantage of being able to collaborate and consult across institutional boundaries with other eMERGE members when questions arose regarding implementational details. This advantage would not accrue to external institutions attempting to implement these algorithms, highlighting the need for a more structured approach to this task.
The second algorithm execution step involves the extraction of data from data source systems, transforming these data into the required format, and feeding them into the implemented algorithm. At a high level, there are two main sources of EHR data available: structured codes (diagnoses, medications, lab values, etc.) and unstructured clinical text (radiology reports, encounter notes, discharge summaries, etc.). The process of extracting and transforming data typically includes queries against a relational or SAS database, and natural language processing (NLP) software for extracting structured information from text. A formally defined target specification language would benefit this data extraction and transformation process by allowing for more consistent patterns of data extraction and transformation that could be at least partially automated.
Given these evident advantages, the adoption of a standardized formal representation for phenotyping algorithms is a logical next step to pursue. This step is particularly salient in the context of eMERGE phase 2, which involves the collaboration of seven institutions and the development of many new phenotyping algorithms. The need to adopt a standard formal representation in the context of the eMERGE project was discussed in our prior work (Conway et al. 2011),4 which this paper builds upon. In order to determine which features a formal representational format would need to support, the Conway et al. paper analyzed 14 eMERGE algorithms in terms of their structure, the types of data elements they used, and the types of logic employed. Conway et al. found that these algorithms commonly used Boolean logic (86%), nested Boolean operators (64%), temporal constraints on criteria (71%), structured codes from external vocabularies (100%), and NLP or indicative keywords (57%). In the current study, we investigate whether the QDM and the NQF’s measure authoring tool (MAT) can adequately represent these types of features. The QDM has already been heavily tested by the NQF, which supported the re-tooling and release of 113 NQF-endorsed quality measures into the QDM format. Here we determine if the QDM can be easily extended to represent the eMERGE phenotyping algorithms that have been specifically developed for clinical research.
Materials and Methods
We selected 9 of the 14 phenotyping algorithms analyzed in Conway et al.4 to be translated into the QDM format (listed in Tables 1 and 2). To perform this task, we used the NQF web-based Measure Authoring Tool (MAT)18 as the user interface. Several authors (WKT, LVR, JAP, PLP, JP) registered for and were granted access to the MAT, and subsequently reviewed the MAT documentation. These algorithms were originally developed and evaluated as part of the eMERGE network, and are publicly available on the web.22 We chose these nine algorithms for the current study due to their diverse combinations of logical operators, temporal criteria, and data element types. While the focus of this study is on the ability of the QDM to represent the various constructs utilized in phenotyping algorithms, our evaluation of the MAT also informs others of the applicability of this tool to develop such algorithms.
Table 1.
Use of QDM Value Sets within eMERGE Case/Control algorithms.
| Algorithm | Clinical Information | Terminology | No. of Value Sets Used |
|---|---|---|---|
|
| |||
| Diabetic retinopathy | |||
| Cases | Diagnosis | ICD-9 | 5 |
| ICD-10 | 1 | ||
| SNOMED-CT | 1 | ||
| Grouped | 1 | ||
| Keywords | 2 | ||
| Controls | Diagnosis | ICD-9 | 5 |
| ICD-10 | 1 | ||
| SNOMED-CT | 1 | ||
| Grouped | 1 | ||
| Keywords | 1 | ||
| Procedure | CPT | 3 | |
| Grouped | 1 | ||
|
| |||
| Peripheral arterial disease | Diagnosis | ICD-9 | 4 |
| Laboratory | LOINC | 1 | |
| Medication | RXNORM | 1 | |
| Procedure | CPT | 4 | |
| ICD-9 | 4 | ||
| Grouped | 4 | ||
| Physical Exam | Grouped | 1 | |
| Keywords | 2 | ||
|
| |||
| Resistant hypertension | |||
| Cases | Diagnosis | ICD-9 | 1 |
| Diagnostic Study | UMLS CUI | 1 | |
| Laboratory | LOINC | 1 | |
| Medication | RXNORM | 10 | |
| Physical Exam | LOINC | 2 | |
| Controls | Diagnosis | ICD-9 | 2 |
| ICD-10 | 1 | ||
| SNOMED-CT | 1 | ||
| Grouped | 1 | ||
| Medication | RXNORM | 10 | |
| Grouped | 1 | ||
| Physical Exam | LOINC | 2 | |
| Grouped | 2 | ||
|
| |||
| Type 2 diabetes | Diagnosis | ICD-9 | 3 |
| Laboratory | LOINC | 3 | |
| Medication | RXNORM | 1 | |
| Encounter | CPT | 7 | |
| Grouped | 1 | ||
| Patient Char. | HL7 | 1 | |
|
| |||
| Cataract | |||
| Cases | Diagnosis | ICD-9 | 1 |
| UMLS CUI | 1 | ||
| Patient Char. | LOINC | 1 | |
| Controls | Diagnosis | ICD-9 | 1 |
| Procedure | CPT | 1 | |
| Patient Char. | LOINC | 1 | |
Table 2.
Use of QDM Value Sets within eMERGE continuous measure algorithms.
| Algorithm | Clinical Information | Terminology | No. of Value Sets Used |
|---|---|---|---|
|
| |||
| Height | Diagnosis | ICD-9 | 12 |
| Grouped | 2 | ||
| Laboratory | LOINC | 1 | |
| Medication | RXNORM | 5 | |
| Grouped | 1 | ||
| Patient Char. | SNOMED-CT | 1 | |
| Physical Exam | LOINC | 1 | |
| SNOMED-CT | 1 | ||
| Grouped | 1 | ||
|
| |||
| Serum lipid level | Diagnosis | ICD-9 | 4 |
| ICD-10 | 2 | ||
| SNOMED-CT | 2 | ||
| Grouped | 3 | ||
| Laboratory | LOINC | 4 | |
| Grouped | 1 | ||
| Medication | RXNORM | 8 | |
| Grouped | 2 | ||
|
| |||
| Low HDL cholesterol level | Diagnosis | ICD-9 | 4 |
| ICD-10 | 2 | ||
| SNOMED-CT | 2 | ||
| Grouped | 3 | ||
| Keywords | 1 | ||
| Laboratory | LOINC | 1 | |
| Grouped | 1 | ||
| Medication | RXNORM | 7 | |
| Grouped | 1 | ||
|
| |||
| QRS duration | Diagnosis | ICD-9 | 2 |
| Diagnostic Study | UMLS CUI | 1 | |
| Laboratory | LOINC | 3 | |
| Medication | RXNORM | 2 | |
| Physical Exam | SNOMED-CT | 1 | |
The selected algorithms were translated to the QDM by at least one author using the MAT, and shared with the other authors for review via the MAT. If the MAT did not seem capable of representing a component of a phenotyping algorithm, it was brought up to the group for review where a final determination was made if the QDM as a model would support the concept, or if it was a limitation of the MAT itself. Thus, for each algorithm an evaluation was made for the ability of the QDM to represent each component, and the ability of the MAT to graphically create the algorithm.
Once developed, the algorithms were exported from the MAT as XML documents. These documents were then analyzed for various metrics used as proxies for the complexity and richness of the algorithm representation. We automated this document analysis with a Python script that read in each of the XML documents, iterated through the nodes of the QDM tree, and calculated the various metrics. The MAT uses value sets (collections of code values from terminologies) as a core building block, and allows the value sets to be shared across algorithms. For each algorithm, the number of value sets was counted. The MAT also supports grouped value sets – collections of value sets which may represent multiple vocabularies (e.g., all ICD-9 and all ICD-10 diagnostic codes for the same disease), or a higher-level grouping of more granular value sets (e.g., value sets for specific severity codes of diabetic retinopathy may be grouped into a diabetic retinopathy value set). We used both kinds of grouped value sets in our QDM representations. For each algorithm, the number of grouped value sets was collected. Counts for value set metrics (Tables 1 and 2) are tabulated separately by the type of clinical information (diagnosis, procedure, medication, laboratory test) and by specific terminologies. The NLP component of algorithms most often defined lists of keywords, or UMLS CUIs (common output from many NLP systems) that should be detected. The MAT did not, however, provide UMLS CUIs or keywords as a “terminology” to associate with value sets. Instead, attempts were made to create a reasonable representation using available terminologies as placeholders. These are listed in Tables 1 and 2 as “Keywords” and “UMLS CUI” terminologies, but were manually curated separately from the algorithm definition and added here for discussion purposes.
An important part of algorithm development also includes the use of temporal logic to define a window of time in which certain conditions must be met, and to relate the order in which events occurred (e.g., Diagnosis X before Medication Y). For all the algorithms, we determined whether or not the QDM could express the types of temporal relationships specified in the eMERGE algorithms, and the number of temporal relationships in the resulting QDM documents was counted.
Once the basic components of an algorithm are defined, they are then combined using Boolean logical operators (AND, OR, NOT). The number of AND and OR operators was counted for each algorithm, in addition to the maximum depth of nested Boolean conditions. For example, “x AND y” has a nesting depth of one, while “x AND (y OR z OR (a AND b))” has a depth of three. We excluded the unary operator NOT from this metric.
Uses of NLP were also assessed from the original algorithms. Following the convention adopted in Conway, et al.4, a “Yes” or “No” indication was made for each algorithm if NLP was explicitly mentioned within the algorithm definition. Some institutions utilized NLP as a component of their data warehouse (i.e. a repository of extracted medications), however this was not counted unless explicitly mentioned in the algorithm. In addition, a “Yes” or “No” indication was made if any use of NLP looked for the presence of negation23 (e.g., the document must say “the patient did not have condition X”), or used negation as part of the found results (e.g., the patient has no documents which indicate condition X). While subtle, the difference in the use of negation has a potentially significant impact on the results returned by an algorithm. Finally, a “Yes” or “No” indication was made if any use of NLP was restricted to a particular section of a clinical document.24
Results
The analysis of value sets used by eMERGE algorithms can be seen in Tables 1 and 2. Table 1 shows the results for the algorithms that employed a case selection logic, assigning patients a “Case=True” or “Case=False” status. Three of the algorithms (Diabetic Retinopathy, Resistant Hypertension, and Cataracts) also have logic for assigning patients a “control” category. Table 2 shows the results of our analysis of values sets for algorithms that generate a continuous output measure for each patient, such as height or serum lipid level. All algorithms in both of these tables utilized more than one type of clinical information, which in turn meant each algorithm used more than one terminology. The number of value sets varied from 6 to 35 (avg=18.3) across all algorithms. Grouped value sets were used in 7 of 9 algorithms (78%).
The analysis of Boolean operators and temporal expressions is shown in Table 3. All of the selected algorithms used Boolean operators, while 6 out of 9 (67%) used nesting of greater than depth 1. Eight out of 9 algorithms also used temporal operators (89%). We found the QDM temporal expression capability to be adequate to cover the temporal relationships used in our eMERGE algorithm set. For example, the QRS duration algorithm utilized the most temporal relationships (8), which center on the identification of a “normal” electrocardiogram (ECG) and relate inclusion and exclusion events around that event.
Table 3.
Use of Boolean and temporal relationships within eMERGE algorithms implemented in the QDM
| Algorithm | Boolean Operators | Max Depth | Temporal Relationships |
|---|---|---|---|
|
| |||
| Diabetic retinopathy | |||
| Cases | 8 | 2 | 0 |
| Controls | 6 | 2 | 0 |
|
| |||
| Height | 8 | 1 | 4 |
|
| |||
| Serum lipid level | 6 | 1 | 5 |
|
| |||
| Low HDL cholesterol level | 6 | 1 | 5 |
|
| |||
| Peripheral arterial disease | 17 | 4 | 1 |
|
| |||
| QRS duration | 28 | 4 | 8 |
|
| |||
| Resistant hypertension | |||
| Cases | 172 | 5 | 2 |
| Controls | 26 | 4 | 5 |
|
| |||
| Type 2 diabetes | 15 | 3 | 5 |
|
| |||
| Cataract | |||
| Cases | 9 | 3 | 2 |
| Controls | 3 | 1 | 2 |
The analysis of algorithm use of NLP is shown in Table 4. Seven of the 9 algorithms specifically described the execution of an NLP algorithm, the use of keyword detection, or regular expressions (78%). A requirement of term negation was described in 2 of the 9 algorithms (22%). Two of 9 (22%) algorithms also required absence of given concepts (e.g., “no evidence of heart disease”), and restrictions to specific clinical document sections (22%).
Table 4.
Use of Natural Language Processing (NLP) as specified within eMERGE algorithms. Results indicate if the algorithm mentions the use of NLP at all, required that negation be factored into the inclusion/exclusion criteria (e.g., presence/absence of “Patient does not have condition X”), requires that no evidence of a concept be found (e.g., no presence of “Patient has X”), or if the NLP required searching in specific document sections.
| Algorithm | Uses NLP | Term Negation | No Evidence Of | Document Section Restriction |
|---|---|---|---|---|
| Diabetic retinopathy | Yes | Yes | No | Yes |
| Height | Yes | No | No | No |
| Serum lipid level | No | - | - | - |
| Low HDL cholesterol level | Yes | No | Yes | No |
| Peripheral arterial disease | Yes | No | No | No |
| QRS duration | Yes | Yes | Yes | Yes |
| Resistant hypertension | Yes | No | No | No |
| Type 2 diabetes | No | - | - | - |
| Cataract | Yes | No | No | No |
Discussion
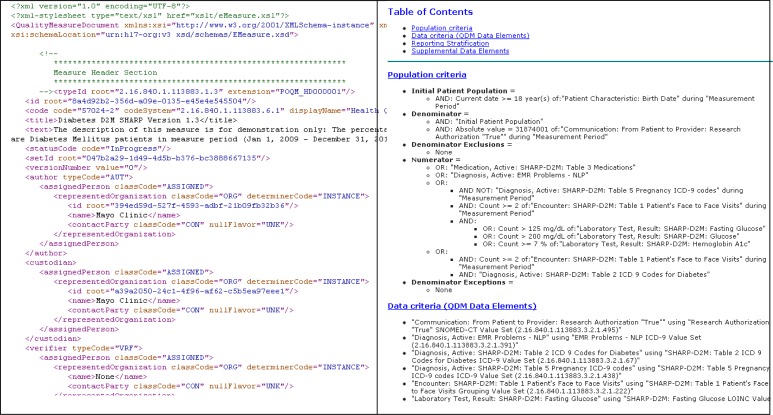
Implementing the selected eMERGE phenotyping algorithms using the MAT yielded insights on the suitability of this tool and the QDM overall for representing phenotyping algorithms for clinical research. In our estimation one of the core strengths of the QDM is its ability to represent phenotyping algorithms for both machine and human consumption. In addition to supporting the export of structured XML file descriptors of algorithms, the MAT also allows users to export a style sheet for executing an XSL transform of the XML source into human-readable HTML (Figure 2). The source XML permits the algorithm developer to add structured human-readable visual aids such as tables and lists that become part of the generated HTML, as well as hyperlinks to external images, flowcharts, and other supporting documents. In the eMERGE project we found these types of documentation to be an invaluable means for communicating algorithm logic to external institutions,4 and the ability of the MAT to support this is very welcome.
Figure 2.
QDM document for the Type 2 Diabetes algorithm in XML and transformed into HTML using XSLT.
The representation of value sets within the QDM as lists external to the measure definition, which are assigned a globally unique identifier, allows for the re-use of value sets. Furthermore, the MAT facilitates the sharing of value sets across algorithms by providing a centralized, searchable repository of value sets for authors. This capability was employed in several of the eMERGE algorithms, indicating the usefulness of this feature for algorithm development. Shared value sets were used in several instances for the case vs. control population definitions within a single phenotyping algorithm (e.g., the Diabetic Retinopathy case and control populations shared diagnosis value sets). Sharing also occurred at a more significant level, such as diagnosis value sets between the Type 2 Diabetes and Diabetic Retinopathy algorithms, and laboratory value sets between the Low HDL and Serum Lipid Levels algorithms. We were also able to re-use pre-existing value sets that the MAT allowed us to discover with its search capability. For example, the Resistant Hypertension algorithm re-used a pre-existing grouped value sets for “ACE Inhibitor/ or ARB medications” from the American Medical Association - Physician Consortium for Performance Improvement, and the Serum Lipids algorithm re-used the HDL, LDL, Total Cholesterol, and Triglycerides laboratory tests value sets from the National Committee for Quality Assurance. This kind of re-use is another major benefit of using the QDM and MAT. One author alone used pre-existing value sets for more than a third of the total used across four phenotyping algorithms. In addition, the QDM requires strict definitions of value sets – code sets, code set version, a code and description for each entry – which are not imposed in the current free-text document-based representation of the eMERGE algorithms. These strict definitions impose high standards on the algorithm developer to perform more work up front to provide precise definitions, which in turn decreases potential downstream errors due to ambiguity and under-specification. It also provides the potential for future automation of the algorithms.
A limitation of the QDM, however, is the lack of support for sharing logic (whether partial components or entire algorithm) between algorithms. An example is within the eMERGE Diabetic Retinopathy algorithm, which utilized the output of the Type 2 Diabetes algorithm. This requires algorithm developers to completely re-implement logic, which makes maintainability and portability difficult.
An immediate observation during the implementation of the algorithms surrounded a key difference in the way that quality measures and research-oriented phenotyping algorithms are defined: the former often use proportion or ratio relations among sets of patients, while the latter are often defined in terms of non-overlapping case and control populations. The MAT, developed for quality measure authoring, enforced stricter rules around eligible populations that did not give us the freedom to define a “case” population and a “control” population in the phenotyping algorithm specification. However, the underlying XML format and schemas do not prohibit us from defining the desired case and control populations. In this situation, the MAT is imposing restrictions that go beyond what is required by the underlying format, and a modification of the tool would help resolve the issue.
The use of relationships (Table 2) varied in complexity across algorithms. However, both the QDM and MAT easily handled the nesting of Boolean conditions. A nice feature of MAT is the ability to view the nested Boolean logic in graphical form, almost like a flowchart. We have found graphical representations like this to be very useful for eMERGE sites to communicate the overall logic of an algorithm. However, we experienced a limitation to the QDM when algorithms go beyond basic Boolean combinations of criteria. For example, in phenotyping algorithms such as PAD and Resistant Hypertension, groups of criteria are defined and combined using a counting rule (e.g., “at least 2 of the 4 criteria must be true”). This type of algorithm cannot currently be directly defined within the QDM, necessitating a much more complex and redundant formulation using Boolean operators. At a more fundamental level, Boolean combinations of inclusion and exclusion criteria are just one way to express classification algorithms that rely on EHR data. There are a multitude of other approaches, such as decision trees, logistic regression, neural networks, and support vector machines. As a concrete example of this in the clinical domain, a successful rheumatoid arthritis algorithm based on logistic regression has recently been developed and ported to multiple institutions.25,26 In the eMERGE network, logistic regression was also used for an alternate form of the rule-based PAD algorithm analyzed in this paper. While representing these more advanced types of classification algorithms is undeniably a thorny issue, it should be kept in mind that Boolean rules are just one of many ways to approach the implementation of EHR-based phenotyping algorithms, and may not be the most accurate.
The use of temporal relationships was also supported by the QDM and MAT, but did exhibit some limitations. The Resistant Hypertension algorithm, for example, required the definition of a “qualification event” – defined as the earliest date a set of criteria were met. Given that the criteria were in turn temporally related to each other, there is a need within phenotyping algorithms to easily chain together events temporally. While the QDM does support this, it was not as easily or directly accomplished using the MAT. In addition, the Resistant Hypertension algorithm required the detection of multiple medications being taken concurrently, and that this concurrent definition be met multiple times. Although the QDM does provide the constructs to define concurrency, counting and temporal relationships, it was not clear to the authors if the definition would be interpreted as such. Given that the QDM does support a way to represent the algorithm, additional work may be needed in describing the implementation to ensure a consistent interpretation.
Given the prevalence and thus importance of NLP within the algorithms (Table 3), support for NLP constructs is a critical component of a formal specification. As it was lacking UMLS as a valid code set, the MAT did not directly support value sets of UMLS CUIs. In addition, requiring an explicit code set made it difficult to directly translate lists of keywords into a value set. Behind the MAT, however, the QDM specification would allow the representation of these value sets. In addition, the requirement of QDM value sets to be assigned a specific category (e.g., Procedure, Condition/Diagnosis/Problem) could help algorithm developers clarify aspects of their NLP requirements. The Cataract algorithm, for example, contained an NLP term list, which mixed procedures and diagnosis terms as indicators that a cataract is or was present. While the algorithm is valid as defined, splitting the term list into two procedure and diagnosis categories and then combining them as a grouped value set can not only impose a stricter definition, but may potentially improve the chance of reusing these code sets (e.g., a new algorithm seeking just terms related to cataract procedures). Another example of this was PAD, which uses lists of keywords indicating both diagnosis (atherosclerotic disease) and anatomical location (arteries of the lower extremities). We also see a need for richer constructs within an authoring tool and the QDM for NLP-specific purposes. The distinction between types of negation (Table 3) is a prime example. While the MAT and the QDM currently support a simple negation (“NOT” function), it is ambiguous as to how that level of negation is applied to a list of keywords.
We note some limitations to our study: Our metrics demonstrate the implementation of the algorithm as released in the eMERGE Phenotype Library, and do not purport to be “optimized” for the QDM. For example, it may be possible to reduce the number of nested Boolean conditions by modifying the algorithm; however this was outside the scope of this study. In addition, the authors purposely entered the study from a naïve point of view as to the capabilities of the MAT to construct measures. It is possible that consultation with an expert on the tool may yield additional suggestions on how to implement the algorithms. Conversely, the naïve point of view may raise potential areas of ambiguity that an expert may not otherwise note. Moreover, it is also important to note that such meaningful modifications must be undertaken with care, as alterations in the algorithm could affect accuracy.
Conclusions and Future Work
In this study we used the NQF Measure Authoring Tool to represent nine phenotyping algorithms within the NQF Quality Data Model. Despite differences in the intent between electronic phenotyping for research and quality measurement from EHRs, we believe there are significant overlapping technical requirements between the algorithms to warrant convergence on a common data model. Our findings suggest that the QDM may provide such a common platform for sharing electronic phenotyping algorithms across diverse sites, with some modifications. We identified potential areas for improvement in both the QDM and MAT to support robust representation of phenotyping algorithms, which we anticipate may also support representation of related quality measures. In particular we identify the need for improved handling of unstructured data and flexible methods to incorporate non-Boolean logic. In future work, we will aim to adapt and create common libraries of downloadable phenotype algorithms. By authoring these as formal structured representations, such algorithms could easily be shared across multiple institutions, “implementation ready”. Furthermore, as mentioned in the Introduction, our eventual goal is to create an environment that can facilitate the translation of the QDM represented phenotyping algorithms into executable codes and scripts that can be implemented on existing EHR systems. To this end, our group is currently investigating several open-source technologies, including the JBoss® Drools27 business logic integration platform.28
Acknowledgments
The eMERGE Network was initiated and funded by the National Human Genome Research Institute, with additional funding from the National Institute of General Medical Sciences: Northwestern University (1U01 HG006388-01), Marshfield Clinic (1U01 HG006389-01), Vanderbilt University (1U01 HG004603), the Mayo Clinic (1U01 HG006379). Support for this research at the Mayo Clinic was also provided by the Office of the National Coordinator for Health Information Technology (90TR0002) through the Strategic Health IT Advanced Research Projects Area 4 (SHARPn) Program.
References
- 1.Powell J, Buchan I. Electronic health records should support clinical research. J Med Internet Res. 2005;7:e4. doi: 10.2196/jmir.7.1.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy EC, Ferris FL, 3rd, O’Donnell WR. An electronic medical records system for clinical research and the EMR EDC interface. Invest Ophthalmol Vis Sci. 2007;48:4383–9. doi: 10.1167/iovs.07-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kallem C. Transforming clinical quality measures for EHR use. NQF refines emeasures for use in EHRs and meaningful use program. J AHIMA. 2011;82:52–3. [PubMed] [Google Scholar]
- 4.Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2011. pp. 274–83. [PMC free article] [PubMed] [Google Scholar]
- 5.National Quality Forum (Accessed at http://www.qualityforum.org.)
- 6.Quality Data Model (Accessed at http://www.qualityforum.org/QualityDataModel.aspx.)
- 7.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3:79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. JAMIA. 2012;19:212–8. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–42. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: a literature review. Journal of biomedical informatics. 2010;43:451–67. doi: 10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hripcsak G. Arden Syntax for Medical Logic Modules. MD Comput. 1991;8:76, 8. [PubMed] [Google Scholar]
- 13.Tu SW, Campbell JR, Glasgow J, et al. The SAGE Guideline Model: achievements and overview. Journal of the American Medical Informatics Association : JAMIA. 2007;14:589–98. doi: 10.1197/jamia.M2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sordo M, Ogunyemi O, Boxwala AA, Greenes RA. GELLO: an object-oriented query and expression language for clinical decision support. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2003. p. 1012. [PMC free article] [PubMed] [Google Scholar]
- 15.Buntin MB, Jain SH, Blumenthal D. Health information technology: laying the infrastructure for national health reform. Health Aff (Millwood) 2010;29:1214–9. doi: 10.1377/hlthaff.2010.0503. [DOI] [PubMed] [Google Scholar]
- 16.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med. 2010;363:501–4. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 17.Blumenthal D. Promoting use of health IT: why be a meaningful user? Conn Med. 2010;74:299–300. [PubMed] [Google Scholar]
- 18.National Quality Forum Measure Authoring Tool (Accessed at http://www.qualityforum.org/MAT/.)
- 19.DesRoches CM, Campbell EG, Rao SR, et al. Electronic health records in ambulatory care--a national survey of physicians. N Engl J Med. 2008;359:50–60. doi: 10.1056/NEJMsa0802005. [DOI] [PubMed] [Google Scholar]
- 20.Jha AK, DesRoches CM, Campbell EG, et al. Use of electronic health records in U.S. hospitals. N Engl J Med. 2009;360:1628–38. doi: 10.1056/NEJMsa0900592. [DOI] [PubMed] [Google Scholar]
- 21.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.PheKB: A Knowledge Base for Discovering Phenotypes from Electronic Medical Records (Accessed at http://PheKB.org.)
- 23.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. Journal of biomedical informatics. 2009;42:839–51. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denny JC, Spickard A, 3rd, Johnson KB, Peterson NB, Peterson JF, Miller RA. Evaluation of a method to identify and categorize section headers in clinical documents. JAMIA. 2009;16:806–15. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–7. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. Journal of the American Medical Informatics Association : JAMIA. 2012 doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.JBoss Drools Business Logic Integration Platform (Accessed at http://www.jboss.org/drools.)
- 28.Li D, Shrestha G, Murthy S, et al. Modeling and Executing Electronic Health Records Driven Phenotyping Algorithms using the NQF Quality Data Model and JBoss® Drools Engine. AMIA Symposium; 2012. [PMC free article] [PubMed] [Google Scholar]