Abstract
Human studies are one of the most valuable sources of knowledge in biomedical research, but data about their design and results are currently widely dispersed in siloed systems. Federation of these data is needed to facilitate large-scale data analysis to realize the goals of evidence-based medicine. The Human Studies Database project has developed an informatics infrastructure for federated query of human studies databases, using a generalizable approach to ontology-based data access. Our approach has three main components. First, the Ontology of Clinical Research (OCRe) provides the reference semantics. Second, a data model, automatically derived from OCRe into XSD, maintains semantic synchrony of the underlying representations while facilitating data acquisition using common XML technologies. Finally, the Query Integrator issues queries distributed over the data, OCRe, and other ontologies such as SNOMED in BioPortal. We report on a demonstration of this infrastructure on data acquired from institutional systems and from ClinicalTrials.gov.
Introduction
Human studies, encompassing interventional and observational studies, are one of the most important sources of evidence for advancing our understanding of health, disease, and therapy. However, human studies information is currently widely dispersed in institutional review board, clinical trial management, trial registry and other systems. There is a growing interest in sharing raw data1,2 and study design metadata to facilitate large-scale data mining and synthesis, and to promote transparency and accountability.3 It is unlikely that such sharing will be accomplished by aggregating all data into a single database. Instead, the most feasible data sharing approach is to “federate” queries over locally controlled databases whose metadata are standardized to a common model of clinical research.
The Human Studies Database (HSDB) Project is a consortium of CTSA research institutions that has been developing semantic and data sharing technologies to federate descriptions of human studies design.4 In 2008, we canvassed eight participating CTSA institutions to identify priority use cases to motivate the HSDB project. These use cases addressed investigator needs (e.g., finding studies based on analytic method, decision support for study design), administrative needs (e.g., providing a profile of an institution’s research that includes methodology and level of evidence), analytic needs (e.g., decision support for evidence analysts), and meta-study needs (e.g., advanced and faster assembly of systematic reviews). These use cases induce the following requirements:
An ontology of clinical research to serve as the reference semantics for data sharing
Ensuring respect of investigators’ intellectual investment and property (both legal and perceived)
Ability to ingest and instantiate study metadata in an ontology-compliant form, from multiple manual and electronic sources (e.g., institutional review board (e-IRB) systems, clinical trial management systems (CTMS), and other relational and XML-accessible databases)
Ensuring internal coherence of study instances, by leveraging axioms in the ontology
Ability to query ontology-conformant study instances within or between institutions, using concepts from the ontology as well as from controlled vocabularies
Ability to scale data acquisition and query performance
Ease of acquisition and maintenance of local databases
Previously, we have described the Ontology of Clinical Research (OCRe) and its use in classifying studies.5 In the HSDB project, we use OCRe as the reference ontology for describing study instances for federated querying. For requirements #3 and #4 above, however, OCRe’s axiomatic formulation in OWL is optimized for logical reasoning and not the acquisition and maintenance of large volumes of instance data. Instead of instantiating instances directly in OWL, we developed a representation of OCRe in XSD (XML Schema Definition), a commonly understood and relatively easy to use format that is nevertheless conformant with the logical structure of OCRe.
In this paper, we demonstrate how, using the XSD representation of OCRe, we created an end-to-end informatics infrastructure that accommodates the processes from study ingestion through multi-center querying. We were able to automatically ingest study data from an institutional e-IRB, to ingest the entire ClinicalTrials.gov repository, to perform inter-institutional ontology-based queries at high granularity, and to identify inconsistencies in instance data using OCRe’s axioms. These accomplishments are a proof of demonstration of the value of OCRe and the feasibility of our general-purpose federated query infrastructure.
Background
Ontology-based data access
With the emergence of Semantic Web, increasing amount of machine-interpretable data are becoming queryable across the Internet. The use of SPARQL6 to query RDF data is the most commonly used technique. A recent development is ontology-based data access (OBDA), which refers to the mediation of access to data repositories through ontologies that provide a high-level conceptual view of the data. This mediation – which bridges the logical and physical views of data – is accomplished by using mappings to specify the semantic correspondence between an ontology and the data stored at the (usually distributed) sources. The objective is to enable users to access data without the need to know how data are actually organized and where they are stored. One example is Mastro, a tool that allows the specification of queries in the form of conjunction of atoms whose predicates are the classes and properties of the ontology, and uses a mapping mechanism to translate those expressions into SQL queries of the underlying relational data sources.7 While OBDA is promising, it is not yet mature for use across multiple query techniques and data representations (e.g., mixing queries to XML and RDF data). Another approach is to integrate results of queries made to sources that may use different query techniques and data formats. For example, the DXBrain distributed query system permits distributed queries over ontologies and XML datasets annotated with terms from those ontologies.8 DXBrain is a precursor to the Query Integrator we use in HSDB, and has been applied to distributed querying over functional imaging datasets.9
BRIDG and other clinical research models and data models
The Biomedical Research Integrated Domain Group (BRIDG) model captures dynamic and static semantics for the domain of protocol-driven research and its associated regulatory artifacts.10 It is an industry standard model developed collaboratively by the Clinical Data Interchange Standards Consortium (CDISC), the HL7 Regulated Clinical Research Information Management Technical Committee (RCRIM TC), the National Cancer Institute (NCI) and its Cancer Biomedical Informatics Grid (caBIG®), and the US Food and Drug Administration (FDA). The CDISC Study/Trial Design Model (STDM) model is harmonized with BRIDG, while other clinical research models also borrow from BRIDG modeling. Because of its more operational focus, the BRIDG model does not deeply model concepts that are central to scientific analysis of clinical research, such as study design type and the relationship between outcomes and analyses. OCRe’s focus on supporting scientific analysis complements BRIDG. Where OCRe’s administrative data elements overlap with those of BRIDG, we officially harmonized them in early 201211 to BRIDG version 3.1.12 OCRe also borrows BRIDG’s modeling of epochs, arms, and activities.
ClinicalTrials.gov
ClinicalTrials.gov is the world’s largest trial registry, with over 120,000 entries. Registration is mandated by law.13 ClinicalTrials.gov entries are available for bulk download via an Application Programming Interface (API). Despite increasingly stringent quality assurance,14 issues related to data structure, nomenclature, changes in data collection over time, and incomplete data continue to limit the usability of ClinicalTrials.gov for scientific data analysis.15,16 The underlying data representation problems are not resolved by transforming the data to RDF without substantial curation (e.g., the LinkedCT project17). The HSDB project aims to leverage the size of ClinicalTrials.gov’s database with the representational clarity and accuracy of OCRe to support large-scale scientific query of human studies.
Methods
This section addresses the several methods used to scope the problem, to redefine OCRe in XSD, to perform the inter-institutional queries, and to ingest large amounts of study metadata.
Scoping
Clinical research is defined by the NIH to be “research with human subjects,” ranging from patient-oriented research to epidemiologic studies, outcomes research, and health services research.18 In this era of translational research and precision medicine,19 researchers will increasingly want to query across multiple clinical domains and across the full range of clinical research, from mechanistic studies to outcomes research using electronic health records.
To accommodate this broad range of study type and domain interests, we scoped this phase of the HSDB project to include any study collecting or analyzing data about individual humans, in whole or in part, living or dead. This scope includes studies such as autopsy, chart review, expression profiles of surgical pathology, shopping habits, utility assessment, scaled instrument development using data from individual humans, etc. In more abstract terms, the scope of HSDB covers all interventional and observational human studies of any design (e.g., randomized, cross-sectional), any intent (e.g., therapeutic, preventive), in any clinical domain, and with any type of data (e.g., quantitative, qualitative, imaging, genomics, proteomics). By contrast, studies on whole populations, organisms other than humans, decision strategies, and the like are considered out of scope. The current goal of the HSDB project is to federate study design information of in-scope human studies (e.g., study design type, interventions, outcomes) as a prelude to the more complicated task of sharing results data.
Ontology of Clinical Research
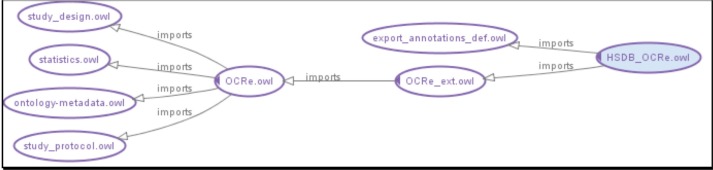
OCRe is an OWL 2.0 ontology focused on the design and analysis of human studies. OCRe is designed to be a foundational ontology that models the entities and relationships of human studies without catering to the requirements of specific applications. Its scope goes beyond HSDB’s in that it places human studies in relationship to studies on populations, organisms other than humans, decision strategies, and the like. OCRe is organized as a set of modular components with the main modules being study protocol, study design, statistics, and core OCRe related by their import relationship as shown in Figure 1. OCRe_ext, further described below, defines new classes and properties that are fully defined in terms of the underlying OCRe constructs, to serve specific uses at an appropriate granularity of modeling. In our case, the specific use is for human studies data federation in the HSDB project. To add additional annotations (described below) for the HSDB project, we defined HSDB_OCRe which imports OCRe_ext.
Figure 1.
OCRe Import Model
OCRe modules are independent of any clinical domain because the clinical content is expressed through external ontologies and terminologies such as SNOMED. This expression is achieved by relating OCRe entities (e.g., outcome phenomenon) to external concepts (e.g., acute myocardial infarction) and their associated terminology codes (e.g., SNOMED code for acute myocardial infarction). In this next section, we describe the major parts of OCRe.
Study Design Typology
Although the most apparent use-case need is for sharing patient-level data from human studies, providing researchers with patient-level data alone is not useful because the study data must be interpreted within the context of how those data were collected (e.g., interventional versus observational study) and the purpose for which they were intended (e.g., public health versus regulatory filing). Federated querying of clinical research data would be scientifically useful only if researchers can precisely query study design features as well as the results data.
It is therefore vital that OCRe contain a rich model of human study designs. We postulated that there exist a small number of high-level study design types that represent distinct approaches to human investigations, and that we could reliably classify all human studies into these design types. Since each study type is subject to a distinct set of biases and interpretive pitfalls, a study’s design type would strongly inform the interpretation and reuse of its data and biosamples. Through iterative consultation with statisticians and epidemiologists, we defined a typology of study designs based on discriminating factors that define mutually exclusive and exhaustive study types.
OCRe’s study design module of OCRe is a design typology represented as an OWL hierarchy. The typology classifies studies hierarchically into four interventional or four observational high-level design types. Additional descriptors elaborate on secondary design features that introduce or mitigate additional interpretive features (e.g., randomization, blinding). A preliminary evaluation of this typology with 35 protocols collected from four institutions showed a Fleiss’ kappa value 0.442, indicating initial promise for reliably classifying study design types in human research.5 A larger validation study of an improved typology is in progress.
Eligibility Criteria
OCRe uses Eligibility Rule Grammar and Ontology (ERGO) Annotation20 to capture the clinical content of eligibility criteria in machine-readable form. ERGO Annotation is a syntax for coding the central clinical meaning of eligibility criteria, informed by both the complexity of natural language and the requirements for computability.
Study Arms and Interventions
OCRe models both interventional and observational studies. NIH defines a study intervention as “manipulations of the subject or the subject’s environment that are performed for research purposes.”18 In OCRe interventions or combinations of interventions (i.e., regimens) are attached to arms and study participants are assigned to these arms. In addition, for each participant the intervention(s) actually received over time is modeled. Depending on the objective of the analysis, subjects can then be grouped (independent variable) by the intervention they were originally assigned to (intention-to-treat analysis) or the intervention they received (as-treated analysis). In observational studies, the independent variables for study analyses called exposures, to indicate that they are not manipulated (i.e., assigned) by the study investigator.
Study Outcomes and Analyses
In OCRe’s conceptualization, the study protocol specifies the study activities (e.g., intervention assignment, data collection) to be carried out to achieve the study’s scientific objectives. These activities are related to the generation of observations, which are then analyzed to support or refute study hypotheses. To model these relationships, OCRe first defines a study phenomenon as “a fact or event of interest susceptible to description and explanation.” Study phenomena are represented by one or more specific study variables that may be derived from other variables. For example, the study phenomenon of cardiovascular morbidity may be represented as a composite variable derived from cardiovascular death, myocardial infarction (MI), and stroke variables. Each variable can be further described by its type (e.g., dichotomous), coding (e.g., death or not), time points of assessment (e.g., 6 months after index MI), and assessment method (e.g., death certificate). All variables are associated with participant-level and study-level observations (observations aggregated across subjects). A study protocol may specify several analyses, each having dependent and independent variables that represent various study phenomena. Variables may play the role of dependent or independent variables in different analyses. If the study protocol designates a primary analysis, the dependent variable of that analysis represents what is conventionally known as the primary outcome of the study. To our knowledge, OCRe is the first model to disambiguate study phenomena of interest from the variables that code observations of those phenomena, and from the use of those variables in study analyses. This clarity of modeling should provide a strong ontological foundation for scientific query and analysis in HSDB.
OCRe_ext
OCRe is designed to be a foundational ontology that models the entities and relationships of human studies without catering to the requirements of specific applications. To enable the efficient acquisition and federated querying of a large volume of study information, the HSDB project requires shortcuts in navigating OCRe entities and relationships. For example, OCRe reifies Funding as a class of administrative relationship associated with studies, where the reified relationship is identified by some funding number and has some person or organization as actors. OCRe_ext contains a Funder class and a ‘has funder’ object property that has Study and Funder classes as domain and range, respectively. Funder is defined as equivalent to Person or Organization that ‘is an actor of’ some Funding that ‘is funding relation of’ some Study. Similarly, we relate the OCRe_ext ‘has funder’ object property to the underlying ‘has funding relation’ and ‘has actor’ properties in OCRe through the use of a property chain. As described in the next section, to accommodate non-ontological extensions (e.g., annotations) necessary for the HSDB project, we defined HSDB_OCRe.owl, which imports annotation properties necessary for deriving an XML-schema based data model.
HSDB_OCRe and Derivation of HSDB XSD
HSDB_OCRe provides an ontology of human studies suitable for the HSDB project. To support instantiating and organizing data of specific studies, a shared data model (schema) is also needed. Ideally, the ontology and the data schema are closely related. The most straightforward approach is to use the ontology directly as a data model, but complications arise immediately. For example, an OWL-based ontology makes the open-world assumption (OWA): an assertion is possibly true unless it can be explicitly shown to be false. One consequence of OWA is that the completeness of data entry cannot be checked unless we add closure axioms to say that only explicitly asserted data elements exist. A data model, on the other hand, usually makes the closed-world assumption (i.e. a Person cannot have a middle name unless property ‘has middle name’ is explicitly declared to be part of the model for describing a Person). Another approach is to derive a data model in a common format (e.g., UML or XSD) from an ontology. Here again, hidden subtleties and complexities arise. For example, a property may have Study as its domain (e.g. ‘has recruitment in the future’), but the HSDB data model does not need to include that relationship. Therefore the choice of OCRe elements to include in the data model must be explicitly specified.
Why XSD
The choice of data model for HSDB was not initially obvious. Together with the HDSB pilot institutions (UCSF, The Rockefeller University, Johns Hopkins, Duke, and Mayo), we iteratively explored approaches to manual and automated instantiation of OCRe-compliant study instances from institutional sources in UML, XSD, and RDF. Each approach was critiqued for feasibility, sustainability, and alignment with each institution’s priorities and data management culture. In the end, we adopted the XML schema to represent templates for instantiating descriptions of human studies for the following reasons: XML is easy to use, widely understood, and tools are commonly available for curation and maintenance. In addition, data from relational databases (the most common source database for human study data) are easily exported in XML because both relational and XML schemas share the closed world assumption (i.e., these schemas are templates). RDF, by contrast, is open world and does not provide a template for targeted export from relational databases. UML was deemed too cumbersome for mapping from OWL to UML, and also required more specialized expertise than is easily found in our pilot institutions.
To specify the common XML schema, we defined an XSD file augmented with Semantic Annotations for SWDL (SASWDL) technology to index the data elements and types to classes, properties, and value sets in HSDB-OCRe through URI references to HSDB_OCRe residing in NCBO’s BioPortal (Figure 2).
Figure 2.
Example of XSD element ( ScientificTitle) indexed to OCRe entity OCRE900213 via a URI to BioPortal
After deciding on XSD as the template formalism, we identified a set of 28 administrative and study design data elements for HSDB federation based on the pilot institutions’ use case priorities.4 The administrative data elements include (notation is of types in HSDB XSD) ScientificTitle, PrincipalInvestigator, RecruitmentSite, ContactForPublicQueries, PlannedSampleSize, among others. The study design data elements include StudyDesign, PlannedProcedureSpecification (which describes interventions in an interventional study, and OutcomeVariable with its Priority (primary or secondary) and EffectiveTime (time points). These data elements are specified in the XSD and indexed back to their respective OCRe entities in BioPortal. The data elements do not include eligibility criteria.
Automated generation of HSDB_XSD
To maintain maximum synchrony between the OWL and XSD representations as HSDB_OCRe evolves over time, we developed a data model extractor to automatically derive HSDB_XSD directly from the OWL format. Then HSDB_OCRe changes can be automatically propagated to the schemas of local databases, where programmatic approaches may be possible to update the XML instance data to conform to the revised XML.
As discussed above, the choice of OCRe elements to include in the data model must be explicitly stated. To guide the data model extractor, we added annotations to the ontology that specify the single root class for the XSD, the properties that should become element tags in an XSD complex type, the parent class to export in case a class has multiple parents, and how value sets for property values should be generated from ontology elements (e.g., treating individuals or subclasses of class as enumerations of values). These annotations are comments on ontological entities in HSDB_OCRe, and are entirely separable from the domain model itself as captured in core OCRe.
The data model extractor is written in Java and uses the OWL-API21 to access the OWL entities in HSDB_OCRe. A simplified sketch of the extraction procedure is as follows:
A class in the ontology that is to have a corresponding template in the data model will include ‘data element’ annotations for every ontological property to be included as a tag in the template.
We create an XSD complex type for the class with a list of nested XSD elements, one for each property referenced via a ‘data element’ annotation (except in the special case of value sets). We determine by examining the ‘has subclass’ and ‘has single parent’ annotation properties if this complex type should extend another type or are the parent of another extending type. Those types are created if they do not already exist. If the class represents a value set (as indicated by the annotation ‘has value set type’), a value set is extracted from the ontology based on the type of value set declared (i.e. values based on all instances of this class, values based on all subclasses of this class, all leaf subclasses of this class, etc.).
For each element created in the previous step, we must create at least one corresponding XSD type (if it does not exist already). The type to be created is determined by examining the property range in the ontology or, if present, the local restrictions. Sometimes there is more than one class in the range and we may need to create more than one type. For example, we may create an XSD choice type to represent a disjunction and then an XSD complex type for each of the disjuncts.
The types mentioned in #3 are created from the classes in the property range or restriction by repeating steps #1-#3 iteratively until we cannot reach any more of the ontology. This combination of properties from the domain model and annotation properties together define a connected subgraph that can be walked by the data model extractor to generate the derived XSD.
The end product of this process is an XSD schema definition for a hierarchical XML data model. The above process must be seeded with an initial starting location within the connected subgraph that will represent the root in the XSD. This is indicated within HSDB_OCRe by the ‘xsd_root’ annotation property, which indicates an ontological property from which to construct the corresponding XSD root element.
Manual and Automated Data Acquisition of HSDB XML Instances
Data acquisition of HSDB XML instances can occur in different ways, from manual entry to bulk extract-transform-load (ETL). The infrastructure is purposefully designed to be agnostic to mode of data acquisition. The only requirement is that the data comply with the XML schema as specified by HSDB XSD. We explored three ways of acquiring instances, two institutional-based, and one bulk ETL of ClinicalTrials.gov entries.
Institutional acquisition of data instances
Two institutions (UCSF and Hopkins) encountered difficulties in obtaining data from their institutional databases. The difficulties were socio-technical (IRB data stored in a proprietary database accessible only via customized reports) and administrative (permission had to be sought from investigators because of resistance to sharing their study protocols). These institutions therefore chose to test the manual instantiation of selected institutional interventional and observational studies based on ClinicalTrials.gov entries and publicly available protocols. We used the Oxygen XML editor to manually create a data file compliant with the XSD. Then we annotated interventions and outcomes with their corresponding SNOMED codes. This test verified the usability of the XML for instantiating real studies.
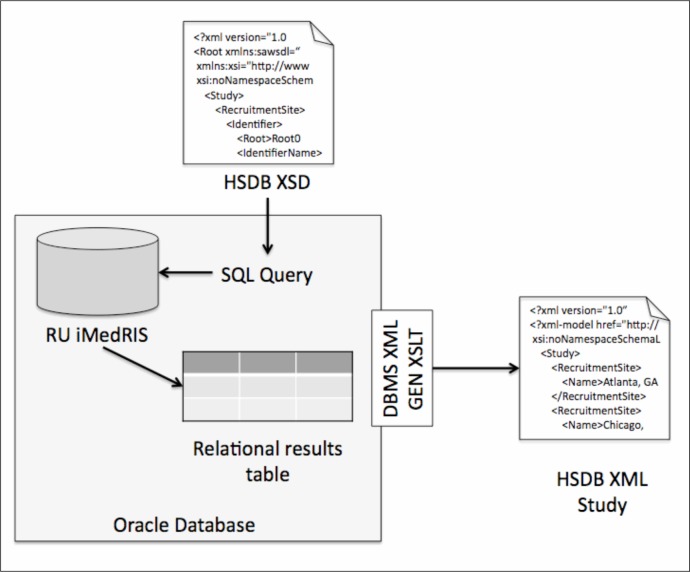
With initial usability of the XML verified, Rockefeller University (RU) piloted the automated acquisition of study design information from their iMedRIS® e-IRB system, which is stored in an Oracle relational database. First, we used HSDB XSD to define the SQL query that mapped constructs of the schema to the relational tables. The XSD has nested elements with complex structures represented in the form of child and parent relationships. These features were captured by the SQL query in the form of primary and foreign keys of the tables. Once the base (SQL) query was formulated, we used Oracle’s DBMS_XMLGEN PL/SQL supplied package22 to convert the results of the SQL query “on the fly” into XML instances. An XSLT stylesheet (see below) was used to transform canonical XML into the XSD compliant format. We then used the Oxygen editor in post processing to perform data cleanup and XML validation (Figure 3).
Figure 3.
Rockefeller University data acquisition workflow
Transforming ClinicalTrials.gov into HSDB XML Instances
A large source of human studies data is ClinicalTrials.gov. Complementing institutional acquisition of human studies data, we performed bulk ETL (Extract, Transform and Load) of this public domain data to test HSDB_XSD and to better demonstrate the power of ontology-based query of human studies.
We first mapped elements from ClinicalTrials.gov’s newest XSD from December 201123 to corresponding elements in HSDB_XSD. One element, ClinicalTrials.gov’s official_title, mapped directly to HSDB_XSD’s ScientificTitle, thus allowing direct transfer of instance values between the XSDs. One category of data elements required relatively simple programmatic transformations. For example, ClinicalTrials.gov classifies a sponsor as lead_sponsor or collaborator. We mapped lead_sponsor to HSDB XSD the Actor of a SponsoringRelation with Priority = primary and collaborator to the Actor of a SponsoringRelation with Priority = secondary. Another category of elements required more complex transformation rules, to account for different semantic granularity or a different structure between the two XML schemas. For example, in ClinicalTrials.gov interventions are defined outside of arms and then linked to the relevant arm(s) using a label, while in HSDB interventions are defined within the arm to which they are assigned.
We used an XSL Stylesheet Transform, or XSLT to implement the mapping. The benefit of using XSLT rather than Java or Perl, for example, is that XSLT is declarative, is commonly used for this purpose, and enables us to stay within the XML paradigm. ClinicalTrials.gov entries were downloaded from their API as separate files, and each file was transformed into HSDB_XML files using the Saxon XML processor and XSLT.
Query Integrator for Querying XML Instances
Once human study instances are posted to the web in a common data schema like HSDB_XML, those data are amenable to federated querying. The query system for the HSDB infrastructure is the Query Integrator (QI), a web-based application that permits distributed queries to be written over multiple web-based sources (such as OCRe and the HSDB_XML instances), saved in a query database, and then treated as sources that themselves may be queried.24,25 Queries may be written, edited, saved, discovered, and executed in several languages including XQuery and SPARQL, where each such language is handled by a separate query engine. A REST Query Execution Service (QES) inside of the QI allows the addressing of a query via a URL, such that the results of one query can be consumed as the source for another, thereby allowing chained queries potentially across different query languages. Prototypes of the QI have been used for quality control on neuroimaging datasets26 and value set generation.27
In the HSDB infrastructure, local databases of XML instances compliant with HSDB_XSD are exposed on the web and their URL is entered into an HSDB registry. Each registry entry can be the address to one study, or a collection of studies. The collection of sources can then be treated as one large virtual XML source, which can be queried using XQuery. In addition, QI can issue SPARQL queries to any of the ontologies in BioPortal, including HSDB_OCRe and SNOMED. With the query-chaining feature, arbitrarily complex queries can be built up exploiting OCRe’s logical structure and SNOMED’s taxonomic hierarchies over the federated instance data.
We chose the QI architecture after extensive study of alternatives, including caBIG, I2B2 and BIRN. We found that these alternatives were rather “heavyweight”, requiring considerable resources to setup and maintain. In contrast, the QI is “lightweight”, requiring little effort to create an integration query over one or more sources, or to reuse a query saved in other instances of the QI. Our approach is to provide end users with custom GUIs that will compose and execute QI queries appropriate to the sources and the relevant query languages. Detailed information about the QI, including other tradeoffs and its relation to other data integration methods, is provided in a recent journal article.28
Results
Demonstration of Federated Querying of HSDB XML Instances
At the CTSA All-Hands meeting in Oct. 2011, we demonstrated an end-to-end demo of OCRe-based querying across human studies descriptions from and housed at three separate institutions (UCSF, Rockefeller, Hopkins).
As described above, four studies were manually instantiated in HSDB_XML at UCSF; two studies were manually instantiated at Hopkins; Rockefeller instantiated 186 studies using a partially automated process
UCSF data was stored in AWS (Amazon S3). Hopkins and Rockefeller data were in institutional servers behind firewalls. The URLs were added to an HSDB-specific QI registry to expose the data to the query engine.
Using the QI user interface,25 we wrote and issued XQuery queries that executed in real-time over the three data sources as well as over OCRe and SNOMED in BioPortal.
An example query was “Find all placebo-controlled trials in which a macrolide (a type of antibiotic) was used as an intervention.” This query executes over the appropriate HSDB_XML element for subclasses of interventions as defined in OCRe. It combines that query with a call to SNOMED in BioPortal for all antibiotics that are children of macrolides, and returns all studies where at least one intervention is an antibiotic classified as a macrolide. Executing this query over the data sources returns the study HIVNET 024 (NCT00021671), since it includes subjects randomized to erythromycin, a macrolide.29
Note that querying ClinicalTrials.gov for “intervention = macrolide” does not return NCT00021671, because ‘macrolide’ does not appear in the intervention, title, keyword, or MeSH tags in that study’s record. The ClinicalTrials.gov search engine also lacks precision, because searching for “intervention = macrolide” returns NCT01489878, an observational study where macrolide use is both an inclusion criteria and a component of the primary and secondary outcomes. In HSDB modeling, however, each intervention is associated with a SNOMED CUI, which allows the Query Integrator to make use of SNOMED’s semantic hierarchy and return all studies where one intervention is an antibiotic classified as a macrolide in SNOMED.
Bulk transform of ClinicalTrials.gov
The XSLT 2.0 code for bulk transform of ClinicalTrials.gov took about 15 minutes to process 120,828 files (all the studies in the registry) downloaded from the ClinicalTrials.gov API on 2/10/2012. Of these files, 81.5% (98,457) were interventional studies, 18.0% (21,808) were observational studies, and the rest were Expanded Access and other study types not covered by HSDB_XSD. The bulk transform revealed some problems with ClinicalTrials.gov data. First, the data model is confusing: observational studies can have “assigned interventions” even though participants in observational studies are never assigned to any intervention. Second, data values frequently violate the ClinicalTrials.gov XSD specification: e.g., despite separate tags for first, middle, and last name, the whole name is stored in the last name tag. Third, value set specifications are not adhered to: the study_type element can have a value of “N/A,” which is not specified in the data element definitions document.30
Logical Curation of HSDB_XML Instance Data
One major benefit of using a logically consistent underlying semantic model is that automated, scalable processes can be defined to systematically identify inconsistencies in instance data. To perform this logical curation directly using OCRe’s modeling in OWL, the instance data must be in RDF, which is not the case for HSDB. Furthermore, OWL’s OWA makes checking completeness of data entries cumbersome. Therefore, we formulate integrity constraints that we want to check as queries over the HSDB_XML instances. We demonstrate OCRe-based logical curation to flag problems in data consistency regarding study design, arms, and allocation of interventions.
For illustrative purposes, we enumerated three integrity constraints that should apply to descriptions of a human study. If a study is described as: (1) a parallel group or crossover study, it must have at least 2 arms; (2) having randomized allocation, it must have at least 2 arms; and (3) having randomized allocation, it must be a parallel group or crossover study. These integrity constraints are represented as axioms in OCRe. We formulated these axioms as XQuery queries stored in the QI metadata repository, and ran them against 1356 interventional studies first submitted to ClinicalTrials.gov in 2012, and against the studies collected from Rockefeller, UCSF, and Hopkins (total of 211). We limited the ClinicalTrials.gov scope to recently submitted studies, because our focus was on testing our approach to logical curation and not on assessing ClinicalTrials.gov’s data quality.
In the ClinicalTrials.gov data set, the queries identified 11/968 studies (1.1%) whose design was declared to be parallel group or crossover but had fewer than two arms. 8/1356 studies (0.6%) of studies designated as randomized had fewer than two arms. 49/1356 studies (3.6%) were designated as randomized but had a study design other than parallel group or crossover. Manual review of a sample of the flagged studies confirmed these problematic entries. For example, the last query revealed a number of ostensibly single group studies that had ‘random allocation,’ which is non-sensical. No studies from the institutional data sources were found to violate these integrity constraints.
Discussion
The HSDB project is a multi-institutional collaboration that has developed and demonstrated an end-to-end informatics infrastructure for ontology-based data access to descriptions of human studies, using OCRe as the reference semantics, SNOMED as the controlled clinical vocabulary, and incorporating BioPortal, Query Integrator, and multiple other ontology and semantic web technologies. We showed that this system can accommodate instances acquired manually or automatically from institutional data sources, and via bulk ETL from ClinicalTrials.gov. We showed the ability to issue distributed queries that are more semantically precise (e.g., study design types are clearly defined and modeled), conceptually precise (e.g., only interventions with macrolides, not outcomes), and able to exploit the semantic hierarchies in SNOMED (e.g., retrieving studies on all macrolide antibiotics). Finally, we showed the value of OCRe in enabling logical curation of instance data. These capabilities give HSDB a greater ability to meet the scientific query and analysis needs of translational researchers that are not currently met by ClinicalTrials.gov or other human studies data sharing initiatives.
With our successful demonstration of the overall design of the HSDB infrastructure, the remaining work and challenges concern the ontology, and data acquisition and annotation. Building on the modeling of interventions/exposures, outcomes, and analyses described above, we started to work on extending OCRe to capture participant-level and study-level results. These extensions then need to be mapped to HSDB_XSD and a new XLST stylesheet, followed by mapping to institutional data sources and ClinicalTrials.gov.
Data acquisition poses more technical and socio-technical challenges. Among some of our pilot institutions, data acquisition from electronic source systems (eIRB, CTMS) was difficult due to contract provisions (data is held by the company and the institution does not have direct access to the data schema or the raw data), organizational hurdles (agreement needed to be secured from many groups in the institution, e.g., IRB, research management, IT governance), and technical hurdles (source data stored at a very coarse granularity in text blobs, and extensive customized installations precluding a turnkey approach even to widely used commercial systems, e.g., Velos). With this experience, we conclude that the barriers are quite high to extracting data out of eIRB systems and CTMSs, even if all systems used the BRIDG model to facilitate interoperation, which is not currently the case. Nevertheless, we have begun harmonizing OCRe with BRIDG for this purpose.
A potentially better approach is being pursued by the Mayo Clinic. Mayo is re-engineering its systems and processes to increase the standardization of clinical research data for programmatic access, and hence to facilitate the efficient management of this data with reduced administrative burden. Mayo is evaluating the mapping of existing study metadata to OCRe representation, and the adoption of standardized OCRe data elements into the re-engineered infrastructure. Integrated OCRe elements would in turn be directly enabled and available for multi-institutional collaboration through open HSDB tools (e.g. Query Integrator) and representation formats (e.g. RDF, XML).
Once data is acquired in an HSDB-compliant format, it is available for more precise and granular queries on study design and methodology afforded by OCRe’s modeling. We are designing QI query templates and a user interface for specifying parameters for those templates. To get the most value out of the data, however, the data instances need to be coded to a controlled vocabulary such as SNOMED so that clinically precise and granular queries can also be made. The data elements for which SNOMED coding is particularly needed pertain to eligibility criteria, interventions/exposures, and outcomes. We are currently integrating MetaMap in our process to annotate interventions and outcomes. The ERGO project20 and others31 are using annotation and natural language processing (NLP) methods to annotate eligibility criteria with controlled vocabulary terms. The descriptions of interventions and outcomes are often in large text blobs in eIRBs, CTMSs, and ClinicalTrials.gov, that requires NLP to disambiguate which named entity in the text should be annotated as the intervention or outcome. ClinicalTrials.gov assigns MeSH terms for interventions (in the intervention_browse tag in the downloaded XML), but they caution that “relevancy is determine(d) programmatically, and can be wrong.”23 The challenge of annotating source data to controlled vocabularies is not unique to the HSDB project.
Conclusion
In conclusion, we used OCRe as the reference semantics along with common XML technologies to develop an end-to-end informatics infrastructure that enables data acquisition, logical curation, and federated querying of human studies with precise and granular queries. This scalable infrastructure provides a strong foundation for supporting large-scale analysis and synthesis of human studies data to advance research and evidence-based practice.
Acknowledgments
This publication was made possible by Grant Numbers RR026040, and UL1RR025005 (Johns Hopkins), UL1RR024143 (Rockefeller), UL1RR024131 (UCSF), UL1RR025767 (UTHSC-San Antonio), UL1 RR024150 (Mayo), UL1RR025014 (UW), and UL1RR024128 (Duke) from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
References
- 1.Kaiser J. Making clinical data widely available. Science. 2008 Oct 10;322(5899):217–8. doi: 10.1126/science.322.5899.217. [DOI] [PubMed] [Google Scholar]
- 2.Hrynaszkiewicz I, Norton ML, Vickers AJ, Altman DG. Preparing raw clinical data for publication: guidance for journal editors, authors, and peer reviewers. Trials. 2010 Jan 29;11:9. doi: 10.1186/1745-6215-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sim I, Chan AW, Gülmezoglu AM, Evans T, Pang T. Clinical Trial Registration: Transparency is the watchword. Lancet. 2006;367(9523):1631–1633. doi: 10.1016/S0140-6736(06)68708-4. [DOI] [PubMed] [Google Scholar]
- 4.Sim I, Carini S, Tu S, Wynden R, Pollock BH, Mollah SA, Gabriel D, Hagler HK, Scheuermann RH, Lehmann HP, Wittkowski KM, Nahm M, Bakken S. The human studies database project: federating human studies design data using the ontology of clinical research. AMIA Summits Transl Sci Proc. 2010 2010 Mar 1;:51–5. [PMC free article] [PubMed] [Google Scholar]
- 5.Carini S, Pollock BH, Lehmann HP, Bakken S, Barbour EM, Gabriel D, Hagler HK, Harper CR, Mollah SA, Nahm M, Nguyen HH, Scheuermann RH, Sim I. Development and evaluation of a study design typology for human subjects research. AMIA Annu Symp Proc; 2009 Nov 14; pp. 81–5. [PMC free article] [PubMed] [Google Scholar]
- 6.Prud’hommeaux E, Seaborne A. SPARQL Query Language for RDF. 2008. Available from: http://www.w3.org/TR/rdf-sparql-query/
- 7.Calvanese D, De Giacomo G, Lembo D, Lenzerini M, Poggi A, Rodriguez-Muro M, Rosati R, Ruzzi M, Fabio Savo D. The Mastro ystem for ontology-based data access. Semantic Web Journal (SWJ) 2012 To appear. Available from: http://www.semantic-web-journal.net/sites/default/files/swj103_1.pdf. [Google Scholar]
- 8.Detwiler LT, Suciu D, Franklin JD, Moore EB, Poliakov AV, Lee ES, Corina DP, Ojemann GA, Brinkley JF. Distributed XQuery-based integration and visualization of multimodality brain mapping data. Front Neuroinform. 2009;3:2. doi: 10.3389/neuro.11.002.2009. Epub 2009 Jan 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner JA, Mejino JL, Brinkley JF, Detwiler LT, Lee HJ, Martone ME, Rubin DL. Application of neuroanatomical ontologies for neuroimaging data annotation. Front Neuroinform. 2010 Jun 10;:4. doi: 10.3389/fninf.2010.00010. pii: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridsma DB, Evans J, Hastak S, Mead CN. The BRIDG project: a technical report. J Am Med Inform Assoc. 2008 Mar-Apr;15(2):130–7. doi: 10.1197/jamia.M2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakraborty S, Sandberg S, Carini S, Tu S, Sim I, Nahm M. Harmonization of the HSDB administrative data elements with the BRIDG model. Translational Science (ACRT/SCTS/AFMR Joint Annual) Meeting; April, 2012; Washington DC. (Abstract) [Google Scholar]
- 12.The BRIDG Model [Internet] Available from: http://www.bridgmodel.org/
- 13.U. S. Public Law 110-85 (Food and Drug Administration Amendments Act of 2007)
- 14.Zarin DA, Tse T, Williams RJ, Califf RM, Ide NC. The ClinicalTrials.gov results database--update and key issues. N Engl J Med. 2011 Mar 3;364(9):852–60. doi: 10.1056/NEJMsa1012065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tasneem A, Aberle L, Ananth H, Chakraborty S, Chiswell K, McCourt BJ, Pietrobon R. The database for aggregate analysis of ClinicalTrialsgov (AACT) and subsequent regrouping by clinical specialty. PLoS One. doi: 10.1371/journal.pone.0033677. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhavnani SK, Carini S, Ross J, Sim I. Network analysis of clinical trials on depression: Implications for comparative effectiveness research. AMIA Annu Symp Proc; 2010 Nov 13; 2010. pp. 51–5. [PMC free article] [PubMed] [Google Scholar]
- 17.Hassanzadeh O, Kementsietsidis A, Lipyeow L, Miller R, Wang M. LinkedCT: A linked data space for clinical trials. CoRR abs/0908.0567 2009.
- 18.National Institutes of Health Glossary and Acronym List [Internet] Available from: http://grants.nih.gov/grants/glossary.htm.
- 19.National Research Council . Toward precision medicine: Building a knowledge network for biomedical research and a new taxonomy of disease. Washington, DC: The National Academies Press; 2011. [PubMed] [Google Scholar]
- 20.Tu SW, Peleg M, Carini S, Bobak M, Ross J, Rubin D, Sim I. A practical method for transforming free-text eligibility criteria into computable criteria. J Biomed Inform. 2011 Apr;44(2):239–50. doi: 10.1016/j.jbi.2010.09.007. Epub 2010 Sep 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The OWL API [Internet]. Available from: http://owlapi.sourceforge.net/
- 22.DBMS XMLGEN [Internet]. Available from: http://www.orafaq.com/wiki/DBMS_XMLGEN
- 23.National Library of Medicine, National Institutes of Health XML Schema for ClinicalTrials.gov public XML [Internet] Version: 2011.12.14. Available from: http://clinicaltrials.gov/ct2/html/images/info/public.xsd.
- 24.Detwiler LT, Shaw M, Brinkley JF. Ontology view query management. Proc AMIA Symp; 2010. p. 1023. [Google Scholar]
- 25.The Query Integrator [Internet]. Available from: http://www.si.washington.edu/projects/QI
- 26.Nichols N, Detwiler LT, Franklin JD, Brinkley JF. Distributed queries for quality control checks in clinical trials. AMIA Summits Transl Sci Proc. 2011:115. [Google Scholar]
- 27.Brinkley JF, Detwiler LT. Query chains for dynamic generation of value sets. Proc AMIA Symp; 2011. p. 1699. [Google Scholar]
- 28.Brinkley JF, Detwiler LT. A Query Integrator and Manager for the Query Web. J Biomed Inform. 2012 doi: 10.1016/j.jbi.2012.03.008. http://dx.doi.org/10.1016/j.jbi.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Human Studies Database wiki: Demo QI Queries [Internet] Available from: http://hsdbwiki.org/index.php/Queries
- 30.ClinicalTrials.gov Protocol Data Element Definitions (DRAFT) August 2011 [Internet] Available from: http://prsinfo.clinicaltrials.gov/definitions.html
- 31.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc. 2011 Dec;18(Suppl 1):i116–24. doi: 10.1136/amiajnl-2011-000321. Epub 2011 Jul 31. [DOI] [PMC free article] [PubMed] [Google Scholar]