Abstract
Sudden Unexpected Death in Epilepsy (SUDEP) is a poorly understood phenomenon. Patient cohorts to power statistical studies in SUDEP need to be drawn from multiple centers due to the low rate of reported SUDEP incidences. But the current practice of manual chart review of Epilepsy Monitoring Units (EMU) patient discharge summaries is time-consuming, tedious, and not scalable for large studies. To address this challenge in the multi-center NIH-funded Prevention and Risk Identification of SUDEP Mortality (PRISM) Project, we have developed the Epilepsy Data Extraction and Annotation (EpiDEA) system for effective processing of discharge summaries. EpiDEA uses a novel Epilepsy and Seizure Ontology (EpSO), which has been developed based on the International League Against Epilepsy (ILAE) classification system, as the core knowledge resource. By extending the cTAKES natural language processing tool developed at the Mayo Clinic, EpiDEA implements specialized functions to address the unique challenges of processing epilepsy and seizure-related clinical free text in discharge summaries. The EpiDEA system was evaluated on a corpus of 104 discharge summaries from the University Hospitals Case Medical Center EMU and achieved an overall precision of 93.59% and recall of 84.01% with an F-measure of 88.53%. The results were compared against a gold standard created by two epileptologists. We demonstrate the use of EpiDEA for cohort identification through use of an intuitive visual query interface that can be directly used by clinical researchers.
Introduction
Sudden Unexpected Death in Epilepsy (SUDEP) is a poorly understood phenomenon, where the mechanisms of death are unknown and effective prevention strategies are yet to be defined[1]. SUDEP research studies are constrained by low rate of reported incidents, about 1% annually in the United States. This requires multi-center studies to identify an appropriately sized patient cohort, such as the Prevention and Risk Identication of SUDEP Mortality (PRISM) project, funded by the National Institute of Neurological Disorders and Stroke (NINDS), which is carried out as part of a SUDEP Centers Without Walls initiative. The PRISM project aims to recruit 1200 patients from the Epilepsy Monitoring Units (EMUs), which has the highest number of potential SUDEP patients, at four centers: University Hospitals Case Medical Center (UH CMC Cleveland), Ronald Reagan University of California Los Angeles Medical Center (RRUMC-Los Angeles), The National Hospital for Neurology and Neurosurgery (NHNN, London, UK), and the Northwestern Memorial Hospital (NMH Chicago).
Following the recommendations of a 2008 NINDS-sponsored meeting[2], a new approach is being adopted for SUDEP studies that uses multi-modal patient parameters, including electroencephalography (EEG), electrocardiography (ECG), cardiovascular, biochemical, and genetic factors. These patient parameters are primarily recorded in the discharge summaries generated by EMUs, which are used by the PRISM project for cohort identification. Currently, discharge summaries are manually reviewed by investigators in the participating EMUs to recruit potential patients. This is not only a tedious approach, but also will not scale to the requirements of the PRISM project (with about 4000 patients per EMU). Automatic processing of discharge summaries for cohort identification is significant challenge due to multiple reasons and requires a dedicated epilepsy natural language processing (NLP) system. A key challenge is the extensive occurrence of clinical free text in the discharge summaries, which is a well known issue in processing of patient information in many clinical domains [3,4,5,6]. In addition, the EMUs patient discharge summaries include highly specialized epilepsy and seizure-specific terminology for describing multi-modal data features in EEG, EKG, and magnetic resonance imaging (MRI) reports. Hence, in contrast to other sources of clinical free text, such as death certificates and billing information, epilepsy discharge summaries present unique set of challenges to existing NLP tools.
To address these challenges, we introduce the Epilepsy Data Extraction and Annotation (EpiDEA) system as a focused, flexible, and effective text analysis tool for epilepsy and seizure-related clinical free text. EpiDEA uses a novel Epilepsy and Seizure Ontology (EpSO) as the knowledge resource for processing specialized epilepsy terms. EpSO is being developed as part of the PRISM project and is the first formal representation of the epilepsy and seizure classification system recommended by the International League Against Epilepsy (ILAE) in 2010[7]. EpiDEA extends the clinical Text Analysis and Knowledge Extraction System (cTAKES)[8] developed at the Mayo Clinic. The EpiDEA system also incorporates a visual interface for cohort identification that can be directly used by clinical researchers. EpiDEA is used to identify patients using constraints deciding seizure semiology, EEG and MRI patterns, and anti-epileptic drug medication, which are of particular interest in the study of SUDEP.
1. Background
Parsing and analyzing clinical narratives presents a unique set of challenges that distinguish it from the broader biomedical natural language processing (NLP) approaches[5]. There has been extensive work in creating clinical NLP systems that focus on information extraction from free text in specific disease domains. For example, the Cancer Text Information System (caTIES) developed at the University of Pittsburgh extracts coded information from surgical pathology reports using terms from the National Cancer Institute (NCI) Thesaurus[9]. Similarly, the SymText NLP system has been used to detect acute bacterial pneumonia from chest x-rays[10].
The Medical Language Extraction and Encoding System (MedLEE) was developed at the Columbia University to extract information from radiological reports for detecting tuberculosis in patients[11]. The MedLEE has been extended to process mammography reports[12] and to additional domains including pathology[13]. The SPECIALIST system developed at the U.S. National Library of Medicine (NLM) has been used to identify anatomical information in coronary catheterization reports, where the Unified Medical Language System (UMLS) is used as an intermediate knowledge resource[14].
The EpiDEA system described in this paper draws on background knowledge of two resources. The first resource is the cTAKES tool developed at the Mayo clinic, which is an open source NLP system for extracting information from clinical narratives in electronic medical records[8]. The second is a set of patient discharge summaries generated by the UH CMC EMU. We provide a description of these resources to prepare for the work described in the subsequent sections.
cTAKES
This is a multi-component pipeline, which combines rule-based and machine learning approaches for knowledge extraction from clinical free text[8]. The cTAKES builds on the Apache Unstructured Information Management Architecture (UIMA) framework[15] and OpenNLP toolkit[16]. cTAKES consists of six components, namely (1) Sentence boundary detector, (2) Tokenizer, (3) Normalizer, (4) Part-of-speech (POS) Tagger, (5) Shallow parser, and (6) Named Entity Recognition (NER) annotator, which are invoked sequentially to create an annotated dataset. The downloadable version of cTAKES was trained on GENIA, Penn TreeBank (PTB), and a corpus derived from the Mayo Clinic Electronic Medical Record (EMR) consisting of 273 clinical notes[8]. cTAKES is built using an extensible architecture, which allows implementation of specialized NLP systems focusing on specific clinical domains.
EMU patient discharge summaries
The EMU patient discharge summaries used in this paper were generated for patients evaluated at the UH CMC. The patients are continuously monitored over a period of 120 hours and a range of multi-modal parameters are collected, including sleep, EEG, EKG, evoked potential, autonomic, biochemical, endocrine and respiratory measurements. The discharge summaries include image files corresponding to segments of EEG signals, which are found to be clinically relevant. The discharge summaries are created using a document-based template and are divided into four sections, namely:
Epilepsy Classification: Describes the etiology, seizure semiology, epileptogenic zone, and co-morbidities of the patient.
History and Exam: This section describes the seizure types, evolution and frequency of seizure, risk factors and family history, medications, results of physical and neurological examination, and psychosocial history of the patient.
Evaluation: Results of EEG, Magnetic Resonance Imaging (MRI), and sleep study of the patient are described in this section.
Conclusions and Recommendations: The recommendation of the attending physician is recorded in this section.
The four sections have interleaving unstructured free text and semi-structured “attribute-value” text. The final version of the report is stored as either a PDF document or an image file.
2. Methods
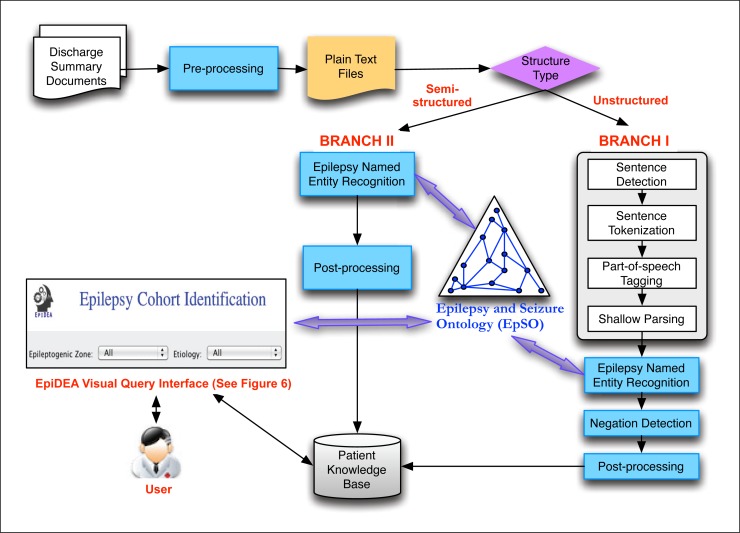
EpiDEA is a flexible and adaptable system, which allows users to selectively apply its components according to variations in the structure and syntax of discharge summaries. In the PRISM project, EpiDEA is deployed with two distinct branches (Figure 1), where Branch I is used to process unstructured free text and Branch II is used to process the less complex semi-structured sections of the discharge summary reports. The EpiDEA system has been implemented using a pipeline-based architectural approach and individual components are invoked sequentially. The final result set from both the branches is used to populate the PRISM patient knowledge base, which is queried using a visual query interface.
Figure 1:
Overview of the EpiDEA system. (Branch I is used to process unstructured free text and Branch II is used to process the less complex semi-structured sections of the discharge summary reports.)
The Epilepsy and Seizure ontology
EpSO is the core resource of the EpiDEA system and is critical for accurately identifying the most relevant epilepsy and seizure-related entities in the discharge summaries. Epilepsy, affecting 50–60 million individuals worldwide, is an extremely heterogeneous condition both etiologically as well as in its clinical manifestations. Traditionally, classification of epilepsy and seizure terminology has been a difficult task due to the inherent complexity of epilepsy, its symptoms, the etiology, and the diverse community of users[7]. Hence, EpSO is novel work in formalizing the new epilepsy and seizure classification system[17] that was proposed by the International League Against Epilepsy (ILAE) classification and terminology commission (CTC)[7] in 2010. EpSO is modeled using the description logic-based Web Ontology Language (OWL) and is a collaborative effort between epileptologists, clinicians, members of the ILAE CTC, and computer scientists[17]. Hence, unlike previous paper-based version of the ILAE classification system, including the classification of seizures and epilepsies, EpSO can be used seamlessly with informatics tools such as EpiDEA.
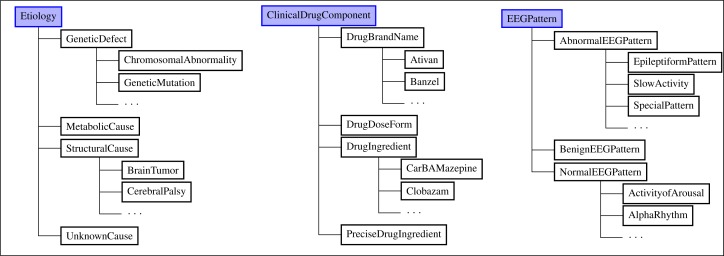
The current version of EpSO has more than 590 classes and their associated properties to represent the ILAE classification system and additional terms needed to represent information generated during a patient’s stay in the EMU. These include classes (Figure 2) modeling the etiology of epilepsy, which are categorized into four broad categories according to the ILAE classification system[7]: “genetic” (e.g. chromosomal abnormality), “metabolic” (e.g. substance abuse), “structural” (e.g. cerebral palsy), and “unknown” (e.g. childhood febrile seizures). Anti-epileptic drugs are modeled using the RxNorm approach to capture drug ingredient, the associated brand names, the strength, and dosage types. EEG patterns recorded during a patients evaluation in EMU are important to successful diagnosis and treatment of epilepsy patients, hence EpSO models the three variations of EEG patterns namely normal, abnormal, and benign (benign patterns are not normal but they are also not clinically significant). EpSO also models brain anatomy information by re-using FMA classes, and parameters associated with measurement of seizures (e.g. scalp electrodes versus intracranial electrodes for EEG recordings) by re-using classes defined in the Neural ElectroMagnetic Ontologies (NEMO). Figure 2 shows the EpSO class hierarchies for “Etiology,” “ClinicalDrugComponent” and “EEGPattern.”
Figure 2:
EpSO class hierarchies for “Etiology,” “ClinicalDrugComponent,” “EEGPattern”
EpiDEA leverages EpSO to support a number of functionalities, including:
Term disambiguation: commonly used synonyms and acronyms of a term are modeled using the OWL annotation properties in EpSO and are used to reconcile variations to the correct ontology class.
Term normalization: Syntactic variations of a term, such as singular/plural and acronyms are normalized using EpSO classes together with customized rules.
Subsumption reasoning: The EpSO class hierarchy allows EpiDEA to correctly classify terms according to their broader semantic type. For example, polyspike and sharp wave EEG signal patterns are types of epileptiform patterns, which are classified as abnormal patterns in EpSO. This generalization-specialization information allows investigators to flexibly use either specific EEG patterns or broader semantic types for cohort identification. This functionality is leveraged by EpiDEA to address variability in usage of epilepsy terminology within and across EMUs in the PRISM project without affecting the quality of results for cohort identification.
With the help of two clinicians, we identified the following seven constraints that are used for cohort identification: sex, age, epileptogenic zone, etiology, EEG pattern, current antiepileptic medication, and past antiepileptic medication.
Among these constraints, sex, age, epileptogenic zone, and etiology values occur in semi-structured section while the rest of the constraints occur in the unstructured section of the discharge summary. As we discussed earlier, the relevant terms for deciding these constraints are modeled in EpSO.
Pre-processing
The discharge summary reports are stored as either portable document format (PDF) document or as image files. We use the Adobe Acrobat Optical Character Recognition (OCR) tool for text recognition in the image files and the output is stored as a PDF file. As the next phase the pdftotext utility from the Xpdf suite is used for converting PDF files to plain text files while retaining the original structure of the reports. The resulting set of text files are curated to address minor formatting issues due to the variability of the two tools.
As we discussed earlier, the discharge summary reports used in this work have two distinct interleaved formats that are processed separately. Branch I uses the components available in cTAKES to analyze free text, whereas Branch II uses specialized rules to extract information from the semi-structured attribute-value sections of the reports (Figure 1).
Branch I
This branch uses the first stages of the cTAKES pipeline followed by three specialized components to process free text from unstructured sections in the discharge summary (Figure 3).
Step 1. Sentence Detection. EpiDEA re-uses the cTAKES sentence boundary detector module that extends the OpenNLP maximum entropy sentence detector tool. The module identifies the use of punctuation marks, including period, question mark, and exclamation mark, to mark the end of a sentence[8].
Step 2. Tokenization. The cTAKES tokenizer used in EpiDEA extracts tokens using space and punctuation mark. Special cases corresponding to dates, acronyms, etc. are accounted for by merging back the tokens[8].
Step 3. Part-of-speech (POS) tagging. EpiDEA uses normalization rules to reconcile variations in lexical properties of the tokens as part of the preprocessing step. Hence, the output of the tokenization step can be directly used by the cTAKES POS tagger, which is a wrapper around the OpenNLP POS module.
Step 4. Shallow parsing. The cTAKES shallow parser is re-used in EpiDEA to tag noun phrases.
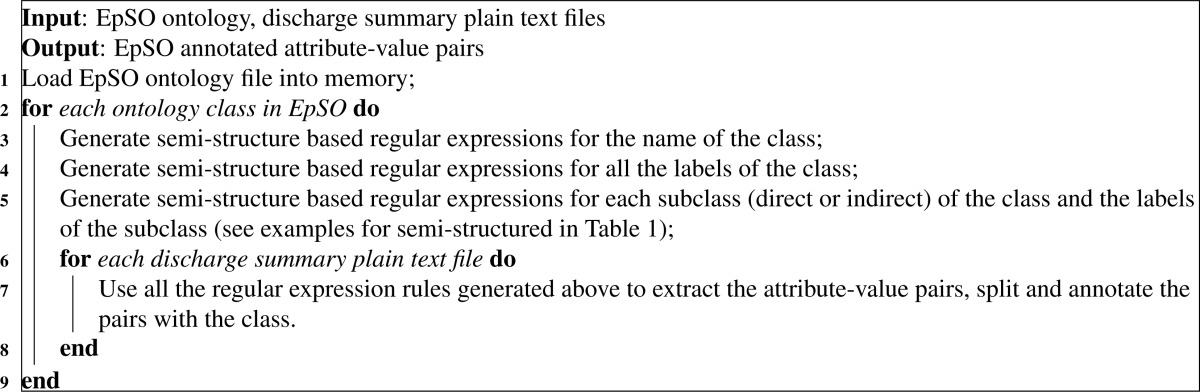
- Step 5. Epilepsy Named Entity Recognition (Ep-NER). The output of the shallow parsing are further filtered and annotated in terms of EpSO (see Algorithm 1). We are only interested in the noun phrases extracted by cTAKES. For each ontology class in EpSO, unstructured text based regular expression rules are created and applied to the noun phrases. Related noun phrases are annotated with the ontology class.
Algorithm 1:
Pseudocode for ontology and rule based annotator for unstructured text Step 6. Negation detection. To detect negation and uncertainty, we adapt the NegEx algorithm, which identifies negation for clinical text, developed by Chapman et al.[18] The trigger lists for negation and uncertainty detection are based on the existing negation triggers and the observation of the discharge summaries (see Table 2). For each EpSO annotated noun phrase, the adapted NegEx algorithm is applied to the sentence where the noun phrase locates and determines if the EpSO related term is negated or uncertain.
- Step 7. Post-processing.
- EEG pattern. One patient may have multiple EEG patterns appearing in different places. The EpSO class “EEGPattern” and its subclasses are used to extract related patterns, which are then concatenated by comma with duplicates removed.
- Current or past antiepileptic medications. Without considering the dose and time information, medications about drug brand names or ingredient names are extracted in terms of all the subclasses of the EpSO class “DrugBrandName” and “DrugIngredient”. To determine whether a medication is current or past, the beginning and ending indexes of the medication being annotated are compared to that of the headers “Past Antiepileptic Medication” and “Current Antiepileptic Medication.”
Figure 3:
An example of free text from unstructured sections
Table 2:
Examples of the trigger words for negation and uncertainty detection
| Category | Negation | Uncertainty |
|---|---|---|
| Pre trigger terms | no, no evidence of, no abnormal, etc. | probable, possible, likely, might have, etc. |
| Post trigger terms | is ruled out, unlikely, etc. | suspected, suspicious, etc. |
| Pseudo trigger terms | no change, no significant change, etc. | NA |
| Conjunction trigger terms | but, reason for, etc. | suffers from, suffered, etc. |
Branch II
The semi-structured sections of the discharge summaries have primarily an “attribute-value” structure (Figure 4), hence they are easier to process as compared to unstructured sections. Hence, EpiDEA does not perform the complete NLP pipeline in this branch, but uses a rule-based approach for extracting relevant information.
Step 1. Rule-based semi-structured text parser. The terms are extracted using EpSO based parsing rules to identify relevant terms. The extraction is performed using UIMA framework with EpSO ontology and rule based annotator (see Algorithm 2). After loading EpSO ontology, semi-structured text based regular expression rules are generated in terms of the name, labels, subclasses (direct or indirect), and labels of the subclasses for each class. The generated rules capture variations of the class, which are applied to each plain text file to extract, split and annotate the attribute-value pairs. For instance, the rules generated for “SemiologicalZone” take into account variant expressions for “SemiologicalZone” and its subclasses including “EpileptogenicZone” and “SeizureOnsetZone.”
Step 2. Post-processing. The values of sex, age, epileptogenic zone and etiology are extracted from the corresponding “attribute-value” pairs. The extracted information about sex includes “Female,” “F,” “Male” and “M,” which are normalized as “Female” and “Male.” Age information appears as only an integer or an integer followed by one of the following strings: “years old,” “year-old,” “years old.,” “years-old,” “year old,” “years,” “y.o.,” where only integers are extracted and integrated.
Figure 4:
An example of semi-structured sections
Algorithm 2:
Pseudocode for ontology and rule based annotator for semi-structured text
The PRISM Patient Knowledge Base
The result from both Branch I and II are integrated and stored in the PRISM knowledge base implemented using a MySQL relational database.
3. Results
Gold standard
There is no community gold standard for epilepsy named entities, hence we built our own gold standard using manual annotations created by two clinicians at the UH CMC EMU. The two clinicians annotated the 104 discharge summary reports processed by EpiDEA according to the cohort identification template. The template was implemented as a table, which was filled by each of the clinicians after reviewing each report. For “Current Antiepileptic Medications” and “Past Antiepileptic Medications,” only drug brand names or drug ingredients were recorded. To measure the quality of annotations, we calculated the inter-annotator agreement by manually comparing the two sets of annotations. Except partial variation annotating an EEG signal pattern, all the other annotations were found to be in agreement. The partial variation in the EEG signal pattern was due to use of terms at different levels of specialization, namely clinician 1 annotated a signal pattern as “Normal,” whereas clinician 2 annotated the same signal pattern as “Sharp transients” (a sub category of “Normal”). This difference was resolved after discussion among the clinicians and an additional epileptologist.
A total of 17,614 sentences were detected in the discharge summaries, including 118,786 word tokens and 27,899 noun phrases, which were mapped to appropriate EpSO classes. Table 3 lists the top 8 EpSO ontology classes occuring in the discharge summaries ordered by the number of extractions.
Table 3:
Top 8 EpSO ontology class extraction
| Ontology Class | Number of Extractions |
|---|---|
| PhysicalPathologicalProcess | 3219 |
| Seizure | 3195 |
| Organism | 968 |
| ClinicalDrugComponent | 957 |
| EpilepticSeizure | 713 |
| EEGPattern | 658 |
| AbnormalEEGPattern | 625 |
| EpileptiformPattern | 611 |
Evaluation
Our evaluation was done separately for Branch I, which processed only free text in the unstructured sections, and for Branch II, which processed the attribute-value in the semi-structured sections. The result of Branch II was almost completely accurate, hence we report the result for Branch I only (Table 4). The evaluations were performed on a MacBook Pro running Mac OS X Snow Leopard with 4 GB of main memory and an Intel Core i5 processor.
Table 4:
Evaluation of the extraction for EEG Pattern and Medications
| Topics | Precision | Recall | F-measure |
|---|---|---|---|
| EEG Pattern | 92.99% | 85.38% | 89.02% |
| Current Antiepileptic Medications | 96.21% | 86.41% | 91.05% |
| Past Antiepileptic Medications | 91.59% | 80.23% | 85.53% |
| Average | 93.59% | 84.01% | 88.53% |
The overall precision was 93.59% and the recall was 84.01% with a F-measure of 88.53% (Table 4) for the processing of free text in the discharge summaries.
The EpiDEA visual query interface for cohort identification
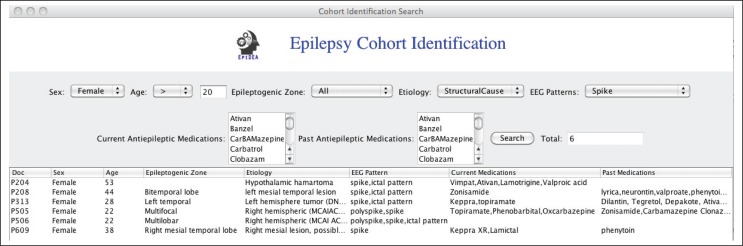
Figure 5 is an example of a typical cohort identification query used by investigators in the PRISM project.
Figure 5:
A typical cohort identification query
To support these patient cohort identification queries over data extracted from the patient discharge summaries, we implemented a visual query interface to allow investigators to directly query the PRISM patient knowledge base (Figure 6). The query interface consists of a set of drop-down menus that are directly populated with the EpSO classes. This allows users to flexibly construct queries, for example they can use either the drug ingredients or drug brand. For example, “CarBAMazepine” has three trade names “Carbatrol,” “TEGretol,” and “TEGretolXR,” so in the case a user selects “CarBAMazepine,” the query interface identifies all discharge summaries that mention the drug ingredient or its tradenames. The results of a query include the patient discharge summaries and the actual values in the discharge summaries, which allow investigators to manually verify the result values. The query interface has been implemented using Java 1.6 and is integrated with EpiDEA system, which provides an integrated environment for investigators in the PRISM project for cohort identification.
Figure 6:
Cohort Identification Interface
4. Discussion
Error Analysis
We manually reviewed the discharge summaries and analyzed the causes of the errors in Branch I, which processed the free text in unstructured sections. The following cases for false negatives were identified after manual review:
Spelling mistakes. The major source of spelling mistakes was the section recording anti-epileptic drug. For example, “Phenobarbital,” “Ethosuximide” and “Oxcarbazepine” were misspelled as “Phenobarbitol,” “Ethosuxsimide” and “Oxycarbazepine” respectively. This issue can be addressed through improving the Ep-NER rule base to support partial matches.
Errors introduced in the preprocessing step. The use of the Adobe OCR and Xpdf introduced errors during their parsing step. For example, “Polyspike-and-wave and spike-and-wave” was converted to “Polyspik an wave and spik an wave” in the plain text file. We are working with the UH CMC EMU staff to encourage them to save reports as PDF files, which will lead to reduced number of such conversion errors.
Errors in POS tagging. “Vimpat,” a drug brand, was sometimes tagged as prepositional phrase instead of the noun phrase, and thus it was missed during the noun phrase extraction. We expect to address this issue by training the cTAKES POS tagger module on epilepsy-related clinical notes (the downloadable version of cTAKES has been trained on generic clinical notes).
In addition to correcting the above issues for false negatives, the performance of EpiDEA is expected to keep improving both in terms of accuracy and coverage as a result of continued development of EpSO. At present, we are expanding the drug information modeled in EpSO with neuroleptic and anti-depressant drug information, both are often prescribed together with anti-epileptic drugs (already modeled in EpSO). In addition, we are using Formal Concept Analysis (FCA)[17] to dynamically classify epilepsy disease types along multiple dimensions. This is a critical step identified by the ILAE CTC to address the growing use of epilepsy classification system in communities beyond epilepsy and seizure clinical researchers, such as pharmaceutical companies[7]. The result of FCA-based classification will be incorporated in the EpSO class hierarchy, which will help improve EpiDEA in improving its classification functionality and visual query interface.
5. Conclusion
There is an urgent need for an automated clinical free text-processing tool to analyze epilepsy and seizure-related clinical notes such as patient discharge summaries in large clinical studies. As part of the PRISM project, involving four EMUs in the USA and UK, we have developed the EpiDEA system using a novel epilepsy ontology for extracting structured information for identifying patient cohorts for SUDEP research. EpiDEA achieves an overall precision of 93.59% and recall of 84.01% with a F-measure of 88.53%. The EpiDEA system also features a visual query interface that enable investigators to directly query the discharge summaries for cohort identification.
Table 1:
Regular expression examples generated in Algorithm 1 and Algorithm 2
| Term | Regular Expression (RE) | Comment |
|---|---|---|
| Semi-structured: | ||
| EpileptogenicZone (class) | \b[eE][pP][iI][lL][eE][pP][tT][oO][gG] [eE][nN][iI][cC]\s*([zZ][oO][nN][eE]| [zZ][oO][nN][eE][sS])\s*:\s*($|.*$) | Class names in CamelCase are split, and the RE for the last word contains plural and singular variants. |
| EZ (label of EpileptogenicZone) | \bEZ\s*:\s*($|.*$) | No other variants is included for the abbreviation EZ to avoid false positives. |
| Etiology (class) | \b([eE][tT][iI][oO][lL][oO][gG][yY]| [eE][tT][iI][oO][lL][oO][gG][iI][eE] [sS])\s*:\s*($|.*$) | Splitting is unnecessary. The RE for the last word contains plural and singular variants. |
| TraumaticBrainInjury (subclass of Etiology) | \b[tT][rR][aA][uU][mM][aA][tT][iI][cC] \s*[bB][rR][aA][iI][nN]\s*([iI][nN][jJ] [uU][rR][yY]|[iI][nN][jJ][uU][rR][iI] [eE][sS])\s*:\s*($|.*$) | Class names in CamelCase are split, and the RE for the last word contains plural and singular variants. |
| Unstructured: | ||
| SharpWave (class) | .*\b[sS][hH][aA][rR][pP]\s*([wW][aA] [vV][eE]|[wW][aA][vV][eE][sS]).* | Unlike the semi-structured section, \s*:\s*($|.*$) is replaced with .* to extract terms from noun phrases. |
| SW (label of SharpWave) | .*\\bSW\s{1,}.* | At least one space is required after the abbreviation SW. |
| EpileptiformPattern (class) | .*\b[eE][pP][iI][lL][eE][pP][tT][iI] ff[oO][rR][mM]\s*([pP][aA][tT][tT] [eE][rR][nN]|[pP][aA][tT][tT][eE][rR] [nN][sS]).* | Unlike the semi-structured section, \s*:\s*($|.*$) is replaced with .* to extract terms from noun phrases. |
| SpikeWaveComplex (subclass of EpileptiformPattern) | .*\b[sS][pP][iI][kK][eE]\s*[wW][aA] [vV][eE]\s*[nN][sS]).*([cC][oO][mM] [pP][lL][eE][xX]|[cC][oO][mM][pP][lL] [eE][xX][eE][sS]).* | Unlike the semi-structured section, \s*:\s*($|.*$) is replaced with .* to extract terms from noun phrases. |
| SWC (label of SpikeWaveComplex) | .*\\bSWC\s{1,}.* | At least one space is required after the abbreviation SWC. |
6 Acknowledgement
This research was supported by the PRISM (Prevention and Risk Identification of SUDEP Mortality) Project (1-P20-NS076965-01) and in part by the Case Western Reserve University CTSA Grant UL1 RR024989 and NIH/NCATS UL1TR000439.
References
- 1.Lhatoo SD, Faulkner HJ, Dembny K, Trippick K, Johnson C, Bird JM. An electroclinical case-control study of sudden unexpected death in epilepsy. Ann Neurol. 2010;68:787–796. doi: 10.1002/ana.22101. [DOI] [PubMed] [Google Scholar]
- 2.http://www.aesnet.org/files/dmfile/HirschSUDEPNIHAppendix_e-1_FULL_REPORTNeurology20114.pdf
- 3.Gold S, Elhadad N, Zhu X, et al. Extracting structured medication event information from discharge summaries. AMIA Annu Symp Proc; 2008. pp. 237–41. [PMC free article] [PubMed] [Google Scholar]
- 4.Turchin A, Pendergrass ML, Kohane IS. DITTO-a tool for identification of patient cohorts from the text of physician notes in the electronic medical record. AMIA Annu Symp Proc; 2005. pp. 744–8. [PMC free article] [PubMed] [Google Scholar]
- 5.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. IMIA Year book of Medical Informatics. 2008;47(Suppl 1):128–44. [PubMed] [Google Scholar]
- 6.Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of post-operative complications within an electronic medical record using natural language processing. JAMA. 2011;306(8):848–55. doi: 10.1001/jama.2011.1204. [DOI] [PubMed] [Google Scholar]
- 7.Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, Van Emde Boas W, Engel J, French J, Glauser TA, Mathern GW, Moshé SL, Nordli D, Plouin P, Scheffer IE. Revised terminology and concepts for organization of seizures and epilepsies: Report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2010;51:676–85. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- 8.Savova G, Masanz J, Ogren P, et al. Mayo Clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowley RS, Castine M, Mitchell KJ, Chavan G, McSherry T, Feldman M. caTIES-A Grid Based System for Coding and Retrieval of Surgical Pathology Reports and Tissue Specimens In Support Of Translational Research. J Am Med Inform Assoc. 2010;17(3):253–64. doi: 10.1136/jamia.2009.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic Detection of Acute Bacterial Pneumonia from Chest X-ray Reports. J Am Med Inform Assoc. 2000;7:593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain NL, Knirsch CA, Friedman C, Hripcsak G. Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. Proc AMIA Annu Fall Symp; 1996. pp. 542–6. [PMC free article] [PubMed] [Google Scholar]
- 12.Jain NL, Friedman VC. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp; 1997. pp. 829–33. [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp; 2000. pp. 270–4. [PMC free article] [PubMed] [Google Scholar]
- 14.Sneiderman CA, Rindflesch TC, Bean CA. Identification of anatomical terminology in medical text. Proc AMIA Fall Symp; 1998. pp. 428–32. [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10(3–4):327–48. [Google Scholar]
- 16.http://opennlp.apache.org/
- 17.Zhang GQ, Sahoo SS, Lhatoo SD. From Classification to Epilepsy Ontology and Informatics. Epilepsia. 2012 Jul;53(Suppl 2):28–32. doi: 10.1111/j.1528-1167.2012.03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]