Abstract
Syntactic parsers have made a leap in accuracy and speed in recent years. The high order structural information provided by dependency parsers is useful for a variety of NLP applications. We present a biomedical model for the EasyFirst parser, a fast and accurate parser for creating Stanford Dependencies. We evaluate the models trained in the biomedical domains of EasyFirst and Clear-Parser in a number of task oriented metrics. Both parsers provide stat of the art speed and accuracy in the Genia of over 89%. We show that Clear-Parser excels at tasks relating to negation identification while EasyFirst excels at tasks relating to Named Entities and is more robust to changes in domain.
Introduction
Syntactic parsing is a process assigning tree or graph structure to a free text sentence. These structures are useful for application such as information extraction[1], negation detection[2], entity disambiguation [3, 4]and other applications[5, 6]
Syntactic Dependency is a parsing scheme where we create edges between words in the sentence denoting different types of child➔parent relations (e.g. in “IL-8 activates CXCR1” the proper noun “IL-8” is the child of the verb “activates” with relation type of noun-subject). This scheme is very useful for many practical tasks: in protein-protein interaction (PPI) extraction[1] we may want to extract the subject and object of a verb such as “phosphorylates”, negation detection [7] can be achieved by finding the governor the negation word.
In the past parsing was a lengthy and error prone process, in a former review of parsing techniques for the medical domain [8] the parsing time ranged from 2–20 seconds per sentence with accuracy of less than 80%. Due to these drawbacks, syntactic parsing was not traditionally used in biomedical NLP pipelines such as: Medlee [9], MetaMap [10] or cTakes [11].
Recent advances in parsing change that reality with parsers which are both fast and accurate. These are complemented by new Gold standard training data in the medical domain[12] which allows training these parsers without the need for adaptation.
The biomedical NLP suit cTakes [13] recently integrated Clear-Parser[14], a dependency parser using the CONLL scheme. The accuracy of Clear-Parser on Genia is 89.6% with running time of 1.8 milliseconds per sentence.
In this paper we introduce a model of the Easy-First parser [15] trained on the Genia biomedical corpus1 for creating syntactic dependency trees in the Stanford Dependency scheme[16]. This parser produces parse trees with accuracy of 89.9% and with running time of 16 milliseconds per sentence.
The different parse tree schemes of the two parsers (Stanford and CONLL) are based on different linguistic representation choices. To provide a meaningful comparison of the parsers we use the task specific approach as suggested by[8, 17]. We apply metrics specific to down the line applications such as PPI extraction, negation detection, named entity recognition (NER) and disambiguation as well as the accuracy when applied in a different biomedical domain.
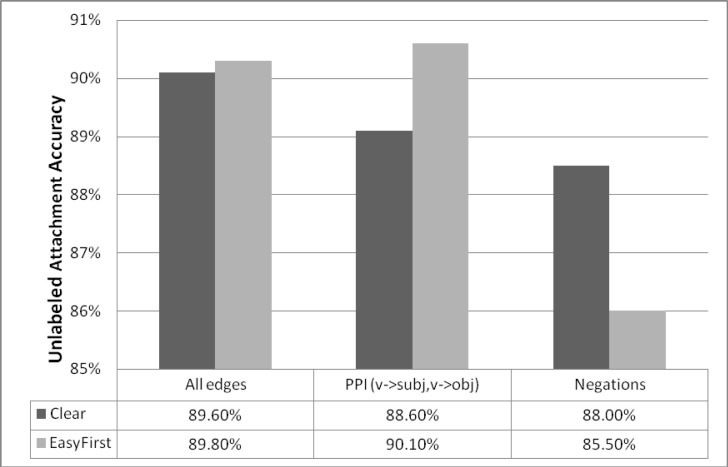
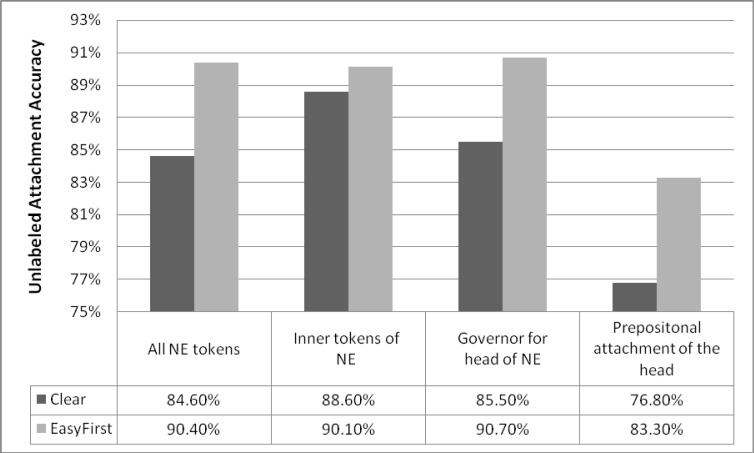
The overall results are similar with accuracy approaching 90% for both parsers. For the different tasks we found variation between the parsers: Clear-Parser is more accurate in predicting the object of a negation word (88% compared to 85.5%) while Easy-First is more accurate at predicting the structure of multi-word named entities (90% compared to 84%) and in predicting the governor of the named entity (90.7% compared to 85.5%). These differences suggest that the different parsers may be useful for different tasks or by ensemble combination, especially when extracting features for down the line classifiers.
Background
Syntactic dependency parsers have been extensively studied in recent years [14, 15, 18–20]. Various parsing methods (MST, MALT, BEAM, EasyFirst, etc.) achieve high accuracy, 90–93%, on the Wall Street Journal derived Penn Treebank. Comparison of parsers is based on micro averaged accuracy of edge attachment. However, this metric is dependent upon the parsing framework used in training and design of parser features and the gold standard trees of different representations are significantly different. The two commonly used parsing representations for dependency parsers are CONLL and Stanford Dependencies (SD) though other representations are in use in some state of the art parsers[21]. The different dependency representations make different linguistic assumptions about the correct structure of a dependency tree. This leads to very different trees with agreement on only ∼70% of the edges in the tree. Syntactic trees produced by using different representations can be compared using different metrics [22]or by using a task specific comparison [17].
We review two parsers. Clear-Parser [14] is based on the scheme of shift-reduce with beam search. In shift reduce parsing the sentence is processed left-to-right using a stack, each input word may be connected to the word on top the stack (either as child or as parent) or pushed into the stack. This is a very quick, O(n) complexity, greedy approach. The beam search version allows remembering key choices an enabling rollback for some of the greedy decisions. Easy-First [15] uses a greedy approach to parse the sentence, at each stage it connects to words with an edge by choosing the decision with minimal risk. This allows quick parsing, O(log(n)) complexity, combined with decision making in a broader context than the left-to-right of shift-reduce parsers. Both parsers yield state of the art results in the newswire domain.
Syntactic parsing accuracy is measured by comparing the produced syntactic tree to a gold standard. Accuracy is measured as the percentage of pair of parent-child words connected in the gold standard tree which are connected in parser output.
Practices for adapting syntactic parsers trained in the newswire domain to the biomedical domain have been studied in depth resulted in low accuracy compared to the newswire domain [19, 23, 24]. However, the amount of training data available for some biomedical domains make it possible to integrate syntactic parsers trained for these domains into biomedical NLP pipelines and practical tasks. Even without accurate domain specific models, the Stanford Dependency Parser is already in use for PPI extraction[1, 25], Noun Phrase identification[26], disease dictionary construction [27]and Negation Detection [28].
Clegg and Shepherd (2007) [8]compared syntactic parsers on a number of task oriented metrics as: accuracy of verb, object and subject edges (a measure on the accuracy of features used for PPI extraction if we concentrate only on verbs pertaining to protein interactions) and correct identification of the head of a negation (i.e. the negated term). They also explored the known Achilles heel of dependency parsers, prepositional attachments where the object is connected to its governor through a preposition (in “the effect of IL-2 in Jurkat Cells” the entity “Jurkat Cells” should be connected to “effect” through the preposition “in”, however, the parser may connect “in” to “IL-2” instead).
The representation scheme of a syntactic parser affects its usefulness for other applications. Miyao et al.[17]compared 3 parsing schemes (dependency, phrase structure and deep parsing) and 8 parsers on the task of PPI extraction. They concluded that the parser compared produced similar accuracy which was improved by training with in-domain data. Another observation made was that the speed advantage of dependency parsers is not offset by any decline in accuracy.
Methods
Experimental Settings
We split the Genia Treebank in two parts for training / testing, each part comprised of 9K sentences. For Clear-Parser, the treebank was transforms into CONLL dependency representation using the Clear Penn-to-Dependency converter [29]. For EasyFirst, we used the Stanford Parser[16] module for converting into Stanford Dependencies. See Table 1 for corpus statistics.
Table 1.
Corpus statistics for the corpora used in this paper.
| Genia | Genia-NER | WSJ | PennBioIE | |
|---|---|---|---|---|
| Trees | 18,419 | 5,954 | 41,532 | 3,320 |
| Tokens | 482,548 | 138,226 | 990,145 | 85,144 |
| Distinct Tokens | 22,354 | 10,737 | 44,389 | 7,945 |
The same conversion process is used for the PennBioIE Treebank[30]. See Table 1.
Each parser was trained on the aforementioned training data portion of Genia TB (both parsers were trained using 20 iterations). The model was then used to parse the test portion of the treebank.
For control we use EasyFirst and Clear-Parser models trained on sections 2–22 of the Penn Treebank (WSJ).
To assess the accuracy for Named Entity related tasks we used the Genia Entity Recognition corpus from the 2004 NER shared task [31]. We extracted 5,974 parse trees from the test portion of Genia that correlate to a sentence from the named entity challenge. This portion of the Treebank was used for NER evaluations. See Table 1.
Results
The accepted method for calculating parser accuracy is the number of edges predicted correctly by the parser compared to the gold standard. We first compare unlabeled accuracy (UAS) of both parsers over the Genia test set (9K sentences). See Figure 1.
Figure 1.
Attachment accuracy: for all edges (average accuracy), for edges relevant for PPI extraction and for negation words.
Evaluating the parsers using task specific metrics
For the task of PPI extraction we look at the accuracy of verb➔object and verb➔subject pairs for verbs pertaining to interaction, such as: “activate”, “modulate”, “phosphorylate”, “regulate”, “upregulate”, “downregulate”, “antagonize”, “suppress”, “stimulate”, “facilitate” and “induce”. These are common verbs in the Genia domain that pertain to biological activity. See Figure 1.
To estimate parser usefulness for negation detection we report the accuracy of predicting the head of a negation (the negated object) for negation words such as: “not”, “no”, “absent”, “none”, “negative”, “cannot”, “without”, “disprove”, “exclude” and “unlikely” (we use the heuristic rule list suggested by Clegg and Sheperd). See Figure 1.
Named entities play important role in understanding biomedical texts. We evaluate the following metrics using the Genia BioNLP NER data (See Figure 2).
Figure 2.
Accuracy of NER related tasks: accuracy over all NE tokens, accuracy of edges within the entity for multi-word entities, accuracy of predicting a correct governor for the head of the NE and accuracy for predicting the governor of the head when it is within a prepositional attachment.
Accuracy for predicting the correct parent for each token in the entity (See Table 2).
Table 2.
Accuracy on predicting the parent for all tokens which are a part of a named entity by entity type. There are 5 entity types in the Genia NER data.
| NE type | Clear-Parser | EasyFirst |
|---|---|---|
| DNA | 89.6% | 92.5% |
| RNA | 81.8% | 85.5% |
| Cell Line | 91.9% | 93.4% |
| Cell Type | 93.3% | 93.1% |
| Protein | 85.4% | 88.5% |
Accuracy in connecting all the words in a multi word entity (e.g. in “Jurkat Cells” we expect the two words to be connected by an edge), in this case we only examine the inner edges of the entity (i.e. we ignore the edge connecting head of the entity to its parent node).
Accuracy in predicting the governor of the head of the entity (e.g. in “IL-2 affects Jurkat Cells” the head of the entity “Cells” should be connected to “affects” as the object). This dependency edge was shown to be a useful feature for predicting the type of a named entity (Gene/Protein etc.) [4]
Accuracy in predicting the governor of the head of the entity in the more complex case of prepositional attachment (e.g. “the effect of IL-2 in Jurkat Cells”, “Cells” should be governed by the preposition “in” and through it to “IL-2”).
Evaluating the impact of training in-domain
We evaluate the parsers using models trained in the newswire domain for two reasons: parsers trained for the newswire domain in biomedical domains are already widely in use (see Background) and for assessing the portability of the two parsers inside the different biomedical domains.
To address the usefulness of newswire trained models for biomedical problems we evaluated both parsers on the same metrics described above. We note a dramatic difference in the accuracy with overall accuracy of EasyFirst dropping by over 10% and Clear-Parser accuracy by 16%. See table 3.
Table 3.
Accuracy of models trained in the newswire domain (Wall Street Journal) on the Genia domain.
| Clear-Genia | Clear-WSJ | Change | EasyFirst-Genia | EasyFirst-WSJ | Change | |
|---|---|---|---|---|---|---|
| All edges | 89.60% | 73.60% | −16.00% | 89.80% | 78.30% | −11.50% |
| PPI (v->subj,v->obj) | 88.60% | 78.70% | −9.90% | 90.10% | 80.90% | −9.20% |
| Negations | 88.00% | 77.90% | −10.10% | 85.50% | 78.30% | −7.20% |
| All NE tokens | 84.60% | 74.30% | −10.30% | 90.40% | 79.70% | −10.70% |
| Inner tokens of NE | 88.60% | 77.96% | −10.64% | 90.10% | 77.97% | −12.13% |
| Governor for head of NE | 85.50% | 70.80% | −14.70% | 90.70% | 81.30% | −9.40% |
| PP attachment of the head | 76.80% | 69.92% | −6.88% | 83.30% | 76.20% | −7.10% |
Due to the lexicalized nature of the parsers (i.e. the model incorporates knowledge on specific words during training) words appearing in the test corpus but missing from the training corpus (out of vocabulary or OOV) are a major source of errors[32]. The proportion of OOV tokens is very high with 24% when training on newswire and applying to Genia compared to only 4% when training on newswire and applying to the test set of newswire or 6.6% when training on the Genia test set and applying to the development set.
We examine the accuracy of the Genia trained models on a different biomedical domain, the PennBioIE corpus, a corpus composed of abstracts pertaining to cancer and enzyme inhibition (Genia corpus pertains to transcription factors in human blood cells). The proportion of OOV tokens in PennBioIE when training on Genia is 13%, markedly lower than the difference from the newswire corpus. Both parsers’ performance was degraded in the new domain, EasyFirst by 6% to 83% and Clear-Parser by 12% to 77% (see Table 4).
Table 4.
OOV expressions for different train/test sets. The smaller sized biomedical treebank PennBioIE, only 2,931 trees in total, leads to a lacking coverage of the vocabulary.
| WSJ-test | Genia-test | PennBioIE-test | |
|---|---|---|---|
| WSJ-Train | 4% | 24% | 23% |
| Genia-Train | 6% | 13% | |
| PennBioIE-Train | 8.5% |
Parsers Running Time
The training and running time of both parsers appear in Table 5, compared to state of the art MST parser, both parsers are faster by an order of magnitude in training and parsing (Clear-Parser is ∼100 times faster at parsing time).
Table 5.
Training and parsing speed for Clear-Parser, EasyFirst and MST parser.
| Clear-Parser | EasyFirst | MST | |
|---|---|---|---|
| Train (s/sentence) | 0.3 | 0.6 | 2.6 |
| Parse (ms/sentence) | 1.8 | 16 | 166 |
Discussion
We have shown that Clear-Parser and EasyFirst provide state of the art accuracy on biomedical text combined with parsing speed faster by orders of magnitude than that of previously used parsers.
The parsers use different schemes for representing syntactic dependency trees. We have shown that this leads to different accuracy in different tasks: Clear-Parser is more useful for negation detection while EasyFirst is more accurate in tasks concerning named entities. Both parsers performance is similarly high in the average accuracy and in accuracy in predicting edges relevant to PPI extraction.
Training in domain has a vast impact on the results of the two parsers with a sharper decline in accuracy for Clear-Parser when out of domain. EasyFirst shows greater robustness when migrating to another domain within the biomedical domain with a lesser reduction in accuracy.
Syntactic dependency parsers provide information useful for a variety of down the line applications. Integrating these parsers would be useful for improving many of these tasks, the choice of parser is task dependent and due to the different representation, some information may be gained from ensemble use of both parsers.
Creating an available resource for training syntactic parsers for the clinical domain would greatly improve the availability of syntactic parsing technology for extracting information from clinical notes. This is compounded by the need for annonimization, this problem should be addressed by future studies.
Footnotes
Available at http://www.cs.bgu.ac.il/~nlpproj/easymed.html
References
- 1.Fundel K, Küffner R, Zimmer R. RelEx—Relation extraction using dependency parse trees. Bioinformatics. 2007;23(3):365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 2.Wilson T, Wiebe J, Hoffmann P. Proceedings of the conference on Human Language Technology and Empirical Methods in Natural Language Processing. Association for Computational Linguistics; Vancouver, British Columbia, Canada: 2005. Recognizing contextual polarity in phrase-level sentiment analysis; pp. 347–354. [Google Scholar]
- 3.Finkel JR, Manning CD. Proceedings of Human Language Technologies: The 2009 Annual Conference of the North American Chapter of the Association for Computational Linguistics. Association for Computational Linguistics; Boulder, Colorado: 2009. Joint parsing and named entity recognition; pp. 326–334. [Google Scholar]
- 4.Finkel J, et al. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications. Association for Computational Linguistics; Geneva, Switzerland: 2004. Exploiting context for biomedical entity recognition: from syntax to the web; pp. 88–91. [Google Scholar]
- 5.Ping C, Barrera A, Rhodes C. Semantic analysis of free text and its application on automatically assigning ICD-9-CM codes to patient records. Cognitive Informatics (ICCI), 2010 9th IEEE International Conference; 2010. [Google Scholar]
- 6.Lewis N, Gruhl D, Hui Y. Dependency Parsing for Extracting Family History; Healthcare Informatics, Imaging and Systems Biology (HISB), 2011 First IEEE International Conference; 2011. [Google Scholar]
- 7.Chapman WW, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of biomedical informatics. 2001;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 8.Clegg A, Shepherd A. Benchmarking natural-language parsers for biological applications using dependency graphs. BMC bioinformatics. 2007;8(1):24. doi: 10.1186/1471-2105-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman A general natural - language text processor for clinical radiology. Jamia - Journal of the American Medical Informatics Association. 1994;1(2):161. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aronson A. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 11.Savova GK, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tateisi Y, et al. Syntax Annotation for the GENIA corpus. ACL. 2005 [Google Scholar]
- 13.Cairns BL, et al. The MiPACQ clinical question answering system. American Medical Informatics Association; 2011. [PMC free article] [PubMed] [Google Scholar]
- 14.Choi JD, Palmer M. Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies: short papers - Volume 2. Association for Computational Linguistics; Portland, Oregon: 2011. Getting the most out of transition-based dependency parsing; pp. 687–692. [Google Scholar]
- 15.Goldberg Y, Elhadad M. NAACL 2010. Association for Computational Linguistics; Los Angeles, California: 2010. An efficient algorithm for easy-first non-directional dependency parsing; pp. 742–750. [Google Scholar]
- 16.Marneffe M-Cd, Manning CD. Coling 2008. ACL; 2008. The Stanford typed dependencies representation; pp. 1–8. [Google Scholar]
- 17.Miyao Y, et al. Task-oriented evaluation of syntactic parsers and their representations. Proceedings of ACL-08: HLT. 2008:46–54. [Google Scholar]
- 18.McDonald R, et al. Non-projective dependency parsing using spanning tree algorithms. EMNLP. 2005:523–530. [Google Scholar]
- 19.Nivre J, et al. The CoNLL 2007 Shared Task on Dependency Parsing. 2007 EMNLP-CoNLL 2007. [Google Scholar]
- 20.Zhang R, et al. Evaluating Measures of Redundancy in Clinical Texts. Proc AMIA. 2011 [PMC free article] [PubMed] [Google Scholar]
- 21.Tratz S, Hovy E. Proceedings of the Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics; Edinburgh, United Kingdom: 2011. A fast, accurate, non-projective, semantically-enriched parser; pp. 1257–1268. [Google Scholar]
- 22.Tsarfaty R, Nivre J, Ndersson E. Proceedings of the Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics; Edinburgh, United Kingdom: 2011. Evaluating dependency parsing: robust and heuristics-free cross-nnotation evaluation; pp. 385–396. [Google Scholar]
- 23.Sagae K, Tsujii J-i. Dependency parsing and domain adaptation with LR models and parser ensembles. EMNLP-CoNLL 2007. 2007:1044–1050. [Google Scholar]
- 24.Dredze M, et al. Frustratingly hard domain adaptation for dependency parsing. CoNLL 2007. 2007 [Google Scholar]
- 25.Bui Q-C, Katrenko S, Sloot PMA. A hybrid approach to extract protein-protein interactions. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq620. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, et al. Improved Identification of Noun Phrases in Clinical Radiology Reports Using a High-Performance Statistical Natural Language Parser Augmented with the UMLS Specialist Lexicon. Journal of the American Medical Informatics Association. 2005;12(3):275–285. doi: 10.1197/jamia.M1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu R, et al. Unsupervised method for automatic construction of a disease dictionary from a large free text collection. American Medical Informatics Association; 2008. [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Lowe HJ. A Novel Hybrid Approach to Automated Negation Detection in Clinical Radiology Reports. Journal of the American Medical Informatics Association. 2007;14(3):304–311. doi: 10.1197/jamia.M2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi JD, Palmer M. Robust constituent-to-dependency conversion for English. Proceedings of the Ninth International Workshop on Treebanks and Linguistic Theories; 2010. [Google Scholar]
- 30.Liberman M, Mandel M. PennBioIE. Linguistic Data Consortium; Philadelphia: 2008. [Google Scholar]
- 31.Kim J-D, et al. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications. Association for Computational Linguistics; Geneva, Switzerland: 2004. Introduction to the bio-entity recognition task at JNLPBA; pp. 70–75. [Google Scholar]
- 32.Lease M, Charniak E. In: Parsing Biomedical LiteratureNatural Language Processing – IJCNLP 2005. Dale R, et al., editors. Springer Berlin; Heidelberg: 2005. pp. 58–69. [Google Scholar]