Abstract
Medical terminologies are large and complex. Frequently, errors are hidden in this complexity. Our objective is to find such errors, which can be aided by deriving abstraction networks from a large terminology. Abstraction networks preserve important features but eliminate many minor details, which are often not useful for identifying errors. Providing visualizations for such abstraction networks aids auditors by allowing them to quickly focus on elements of interest within a terminology. Previously we introduced area taxonomies and partial area taxonomies for SNOMED CT. In this paper, two advanced, novel kinds of abstraction networks, the relationship-constrained partial area subtaxonomy and the root-constrained partial area subtaxonomy are defined and their benefits are demonstrated. We also describe BLUSNO, an innovative software tool for quickly generating and visualizing these SNOMED CT abstraction networks. BLUSNO is a dynamic, interactive system that provides quick access to well organized information about SNOMED CT.
Introduction
Medical terminologies are often large and complex. Many times errors, omissions or inconsistencies are hidden in this complexity. Rector et al. point out numerous modeling problems and errors in SNOMED CT are discussed [1]. Héja et al. describe two kinds of errors in SNOMED CT, ontological problems and knowledge-engineering errors [2]. Our objective is to find such errors in large terminologies. Previous experience has shown that for a small terminology errors can often be recognized by visually inspecting the structure of the terminology represented as a diagram. However, in a large and complex terminology diagram this is typically impossible because an “interesting” (from the point of view of finding errors) structure is hidden in one of three possible ways. (1) The structure is so large that it is spread out over many screens that cannot be viewed simultaneously, hiding the relevant structural properties from view. (2) The diagram is laid out, due to space limitations, in a “compressed” way that does not make the structure comprehensible. Thus it is impossible to “see the forest for the trees” and recognize errors in structural irregularities. For example, “sibling concepts” may appear far apart and at different levels of the diagram. Sometimes the overwhelming amount of information makes it impossible to see problems with single concepts, in effect making it impossible to “see the trees for the forest.” (3) The structure is intermixed with elements from other structures that are irrelevant from the point of view of finding errors. For example, concepts of two unrelated hierarchies may appear “interleaved.” Sometimes a combination of (1), (2) and (3) may occur.
To overcome this problem, we have developed a theory of abstraction networks in previous research [3]. The goal of every abstraction network is to be substantially smaller than the medical terminology that it is summarizing, with visualizations of the abstraction network fitting on a single (or very few) screen(s), thus eliminating problem (1). Reducing the number of elements that are displayed and laying them out in a systematic way reduces problem (2). An abstraction method that cleanly separates elements from different hierarchies is needed to overcome problem (3). With all of these simplifications, an abstraction network needs to preserve enough of the original terminology to make it possible to recognize errors in it.
Reducing the size of a terminology of tens of thousands or even hundreds of thousands of concepts to an abstraction network of a size such that a visualization fits on a single or a few screens cannot be done indiscriminately, or too much of the structure will be lost that is necessary to recognize errors, omissions or inconsistencies. Thus an abstraction network needs to capture the most important elements of the large terminology and omit all the other elements. In previous research we have succeeded in defining two kinds of abstraction networks that were shown to achieve the contradictory goals of reducing the size of the abstraction network diagram while maintaining enough of the terminology structure to uncover errors in the original terminology from which the abstraction networks were derived. We will briefly review these two abstraction networks below, called the area taxonomy and the partial area taxonomy. Both were defined for SNOMED CT, the Systematized Nomenclature of Medicine – Clinical Terms, a very large terminology with nearly 400,000 concepts and over 1.5 million relationships [4].
One numeric parameter that can be used to characterize an abstraction network is its reduction factor. Informally, the reduction factor is the ratio of the “elements” in the reduced representation to the “elements” in the original representation, in percent. Thus, when generating an abstraction network for a terminology, the reduction factor describes the ratio of the number of elements in the abstraction network to the number of elements in the terminology. When generating a (smaller) new abstraction network from a given original abstraction network, the reduction factor describes the ratio of the number of elements in the new abstraction network to the number of elements in the old abstraction network. Defining the reduction factor at this point is necessary, because it used in expressing the motivation for this paper. However, an exposition of the exact nature of the “elements” will have to be delayed to the Background Section.
To review the reduction factors achieved by the partial area taxonomy for seven hierarchies of SNOMED CT see the last column in Table 1. (The other columns will be explained later.) As can be seen, the reduction factors vary widely, from 30.6% for the Specimen hierarchy down to 0.0738% for the Body Structure hierarchy of SNOMED CT. As the size of each hierarchy is given and constant (for one release of SNOMED CT) and the size of a screen imposes a limit of approximately 30 to 70 elements (such as boxes representing concepts in a diagram) on the size of an abstraction network, it would be desirable to gain control over the reduction factor. However, with the rigid definitions of the area taxonomy and the partial area taxonomy, this was previously not possible.
Table 1:
Seven SNOMED CT hierarchies and their reduction factors.
| Hierarchy | # Concepts (C) | #Areas (A) | # P-Areas (P) | C/A | C/P | Reduction Factor |
|---|---|---|---|---|---|---|
| Body Structure | 31155 | 2 | 23 | 15577.5 | 1354.57 | 0.0738% |
| Clinical Finding | 98414 | 347 | 10393 | 283.62 | 9.47 | 10.55% |
| Event | 3661 | 7 | 31 | 523 | 118.10 | 0.846% |
| Pharm/ Biologic Product | 17129 | 4 | 8540 | 4282 | 2.01 | 49.71% |
| Procedure | 52665 | 729 | 10731 | 72.24 | 4.91 | 20.32% |
| Situation | 3237 | 10 | 837 | 323.7 | 3.87 | 25.86% |
| Specimen | 1330 | 23 | 407 | 57.83 | 3.27 | 30.60% |
This paper advances the state of the art by introducing two new kinds of abstraction networks, with the goal of providing improved control over the reduction factors of abstraction networks, as follows: (1) The relationship-constrained partial area subtaxonomy can be derived from any area taxonomy of SNOMED CT. (2) The root-constrained partial area subtaxonomy can be derived from any partial area taxonomy of SNOMED CT. Both these new abstraction networks achieve a further reduction of size compared to the original SNOMED CT hierarchies. While there is only one area taxonomy and one partial area taxonomy for a SNOMED CT hierarchy, there are typically many relationship-constrained partial area subtaxonomies and many root-constrained partial area subtaxonomies of different sizes. Therefore, they give the user fine control over the reduction factor and ultimately the display size. This paper describes the methods for deriving the two new kinds of abstraction networks from existing abstraction networks.
Abstraction networks by themselves are not sufficient for finding problems in medical terminologies. Rather, software tools are needed to display these abstraction networks. This paper introduces such a tool, called BLUSNO (Biomedical Layout Utility for SNOmed CT). BLUSNO enables the generation, visualization and exploration of abstraction networks. It provides a user-friendly interface for deriving these new subtaxonomies, as well as the original area and partial area taxonomies. A detailed description of BLUSNO is given in this paper. We refer to the process of finding errors, omissions or inconsistencies in a (medical) terminology as “terminology auditing,” and BLUSNO is a tool that supports auditing.
Background: Abstraction Networks for SNOMED CT
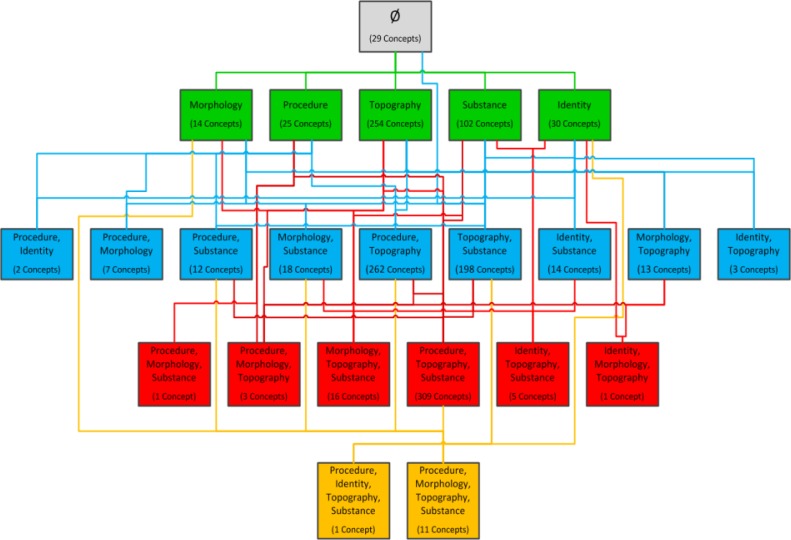
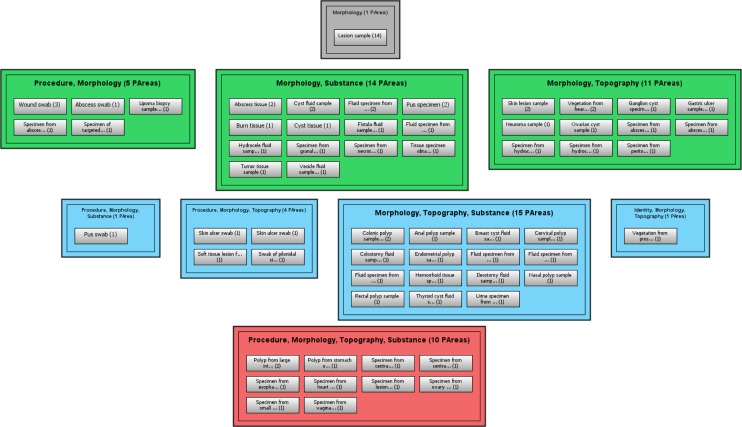
Definition: An abstraction network is a high-level, simplified, view of an otherwise complex structure. In our previous work [3], we developed two kinds of abstraction networks which supported the auditing of SNOMED CT. The first of these abstraction networks was the area taxonomy. Definition: An area is the set of all the concepts that share the same set of “attribute relationships” (the official SNOMED CT term for semantic relationships), regardless of the target values of the relationships. Definition: An area taxonomy is an abstraction network consisting of areas as nodes connected by CHILD OF links. In the diagram of an area taxonomy an area is represented by a single box. An area is named after the set of relationships that the concepts within it exhibit, i.e., it is a multi-part name. A CHILD OF link indicates that a specific concept in one area has a parent concept in a different area. In Figure 1, the green box labeled “Topography” is the area in which all concepts, 254 in total, have only the relationship “Topography.”
Figure 1.
The area taxonomy for SNOMED CT’s Specimen hierarchy. Concepts are not displayed.
In Figure 1, the concept Abscess swab in the area {Procedure, Morphology} (second box in the third row from the top) IS-A Specimen from abscess in the area {Morphology} and also IS-A Swab, which is in the area {Procedure} (first and second box in the second row from the top). In the area taxonomy, these IS-A links are reflected by the fact that the area {Procedure, Morphology} is a CHILD OF the areas {Morphology} and {Procedure}, as shown by the connecting lines. A concept can only exist within one area of a hierarchy. The null character in the first row (∅) indicates there are no relationships in this area’s relationship set, i.e. the included concepts have no relationships at all. According to the rules of SNOMED CT, the same attribute relationship may be introduced at more than one concept in a hierarchy. We only use those concepts introducing attribute relationships to construct area taxonomies.
Definition: A root of an area is a concept that is characterized by having at least one additional relationship compared to every one of its parents. The second form of abstraction network is the partial area taxonomy. Partial areas are the primary units of interest in our structural auditing methods. Definition: A partial area consists of a root and all of its descendants within the same area. Definition: A partial area taxonomy is an area taxonomy where every area is represented along with its partial areas. An area may contain one or more partial areas, as an area may have one or more root concepts. Naturally, all concepts in all partial areas of an area have exactly the same set of attribute relationships. While concepts always exist in only one area, a concept may exist in more than one partial area within the same area, because there may be paths from it to more than one root. In the diagram of a partial area taxonomy, a partial area appears as a white box within a colored area box, with the name of its root displayed in the white box. The name of the root is also used as the name of the partial area itself.
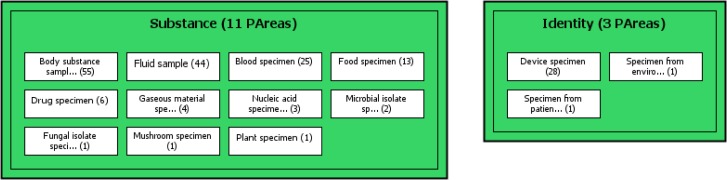
In Figure 2, the concepts Body substance sample and Fluid sample, in the Area {Substance}, introduce the {Substance} relationship for 54 descendants and 43 descendants, respectively. Similarly, the concept Device specimen introduces the {Identity} relationship for its 27 descendants. The numbers shown in parentheses after the partial area names describe the total numbers of concepts in the specific partial areas. A partial area contains a hierarchy reflecting the IS-A relationships of the original SNOMED CT hierarchy, however this hierarchy is never displayed.
Figure 2.
A small subset of the partial area taxonomy for SNOMED CT’s Specimen hierarchy.
With the description of the area taxonomy and the partial area taxonomy now in hand, we revisit Table 1, which contains, for seven SNOMED CT hierarchies, the exact numbers of concepts, partial areas and areas in each of the hierarchies. The fifth and sixth columns show the ratios of concepts to areas and concepts to partial areas, respectively. The last column shows the especially important reciprocal of the concept to partial area ratio, which is equal to the reduction factor for partial area taxonomies, given in percent.
Methods
We will now describe the two new kinds of abstraction networks, which allow a great degree of control over the reduction factor of a display of a SNOMED CT hierarchy.
The Relationship-Constrained Partial Area Subtaxonomy
As mentioned above, SNOMED CT refers to semantic “non-IS-A” relationships as “attribute relationships.” We will abbreviate this to “relationships.” Definition: A relationship-constrained partial area subtaxonomy is a partial area taxonomy generated using a subset of the outgoing attribute relationships in a terminology hierarchy. This subtaxonomy is based on the underlying structure of concepts in SNOMED CT. In previous research, an area taxonomy was generated using the set of all outgoing attribute relationships that exist within a given hierarchy. In extreme cases there can be over twenty-five different attribute relationship types in one SNOMED CT hierarchy. A large number of relationships leads to a comparatively large number of areas in a partial area taxonomy. For example, the Procedure hierarchy has twenty-seven different attribute relationship types, which results in a taxonomy of 729 different areas. This is in contrast to the Body Structure hierarchy, which has only a single outgoing relationship and two areas.
The relationship-constrained partial area subtaxonomy allows an auditor to generate areas with a chosen subset of relationships. Suppose a hierarchy has the set of outgoing attribute relationships R = {R1, R2, R3, … Rn}. For a relationship-constrained partial area subtaxonomy an auditor selects a strict subset from R to define the subtaxonomy. We denote this strict subset R′ = {Ri1, Ri2, Ri3, … Rm}, where the relationships in R′ are taken from R and m < n. R′ is then used to generate a partial area taxonomy for the hierarchy, using the same methods as for a “normal” partial area taxonomy. The result is a taxonomy where all of the areas (and therefore concepts) have a set of attribute relationships that is a subset of R. Figure 3 shows an abstract example where a given set of four relationships is reduced to three, resulting in a reduction of the number of areas from 13 to eight.
Figure 3:
Relationship-constrained area subtaxonomy derivation: Relationships on the right are a subset.
In theory, an area taxonomy for a hierarchy with n relationships could contain 2n different areas. However, an area corresponding to a specific subset of relationships is only created if there are actual concepts with this combination of relationships in SNOMED CT, which is not always the case. For example, on the left side in Figure 3 there is no area {R1, R3, R4}. However, on the right side, with three relationships defined, there are eight areas, which corresponds to 23, the maximum number of areas possible for three relationships. From this description it should be clear that there is only one possible area taxonomy, but there are many possible relationship-constrained area subtaxonomies, depending on the size of n. This brings us closer to our goal of giving fine control over the reduction factor to the user. The last step is to derive partial areas inside of all areas found by this method. The derivation method used for this purpose is unchanged from the method described in the Background Section. A SNOMED CT example of a relationship-constrained partial area subtaxonomy will be shown in the Result Section.
The Root-Constrained Partial Area Subtaxonomy
A second method for generating smaller taxonomies is based on what was called above a root-constrained partial area subtaxonomy. Definition: A root-constrained partial area subtaxonomy is a subset of a taxonomy based on the CHILD OF links of the partial area taxonomy. In previous research, partial area taxonomies were rooted at the partial area containing the root concept of the whole hierarchy, e.g. Specimen, and the taxonomy contained all of the descendant partial areas of the root, i.e., the entire taxonomy. (Because hierarchies of SNOMED CT have unique root concepts, there is only one partial area in each root area.) With a root-constrained partial area subtaxonomy, an auditor defines which partial area should be used as the root of the desired subtaxonomy. The resulting subtaxonomy consists of the selected root partial area along with all of its descendant partial areas. If a partial area is not a descendant of the chosen root, it is not included in the display. If a descendant partial-area has one or more parent partial areas that are outside of the root-constrained partial area subtaxonomy, the associated CHILD OF links are disregarded.
The root-constrained partial area subtaxonomy allows an auditor to analyze how the selected root concept’s descendants evolve throughout a SNOMED CT hierarchy. To generate a root-constrained partial area subtaxonomy, a breadth-first traversal of the hierarchy of CHILD OF links within the complete partial area taxonomy is performed, starting at the selected root partial area. Partial areas that can be reached during the breadth-first traversal are considered members of the root-constrained partial area subtaxonomy. Partial areas with identical sets of lateral relationships are then regrouped back into areas.
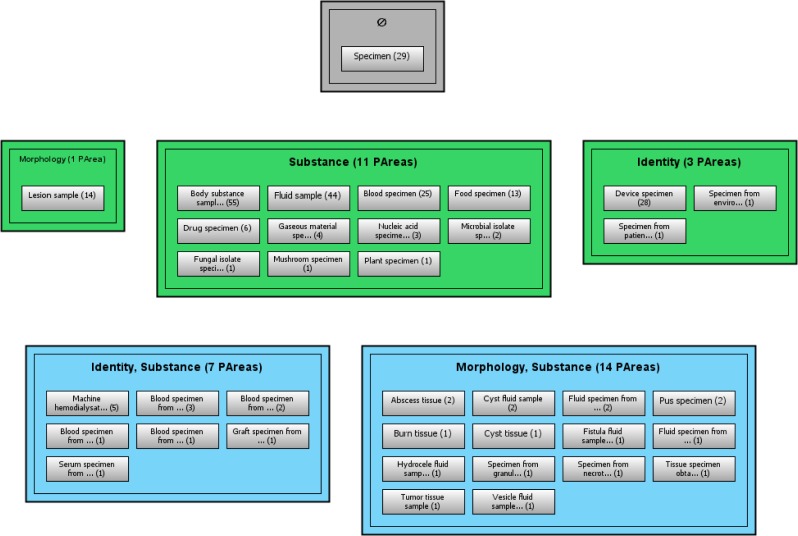
Using a root-constrained partial area subtaxonomy, an auditor can determine the amount of information a particular concept introduces into SNOMED CT. Figure 4 shows an example of a root-constrained partial area subtaxonomy. Reviewing Figure 4 shows that the number of partial areas is reduced from 11 to five. Naturally, there is a loss of information involved in this abstraction network, as the ancestors of the new root are all eliminated from the abstraction network. However, this might be exactly what is desired for a simplification of an abstraction network to enable structure-based auditing. Additionally, if information on ancestors is required, it can be obtained by traversing up the CHILD OF partial area hierarchy of the selected root, adding some or all of the ancestors. The tool for this is described below.
Figure 4:
A root-constrained partial area subtaxonomy. P2 is chosen by a user as the new root partial area.
As opposed to the relationship-constrained partial area subtaxonomy, there is no simple formula to describe the total number of possible partial areas and how they are reduced by selecting a specific partial area as a new root. However, the number of possible root-constrained partial area subtaxonomies is equal to the number of partial areas minus 1. (Selecting the original root partial area itself as a new root would not constrain the abstraction network and is therefore not a meaningful operation, which explains the “ – 1”.)
Biomedical Layout Utility for SNOMED CT
The Biomedical Layout Utility for SNOMED CT (BLUSNO) is a software tool that assists auditors by providing the ability to dynamically generate highly interactive visualizations of abstraction networks created by using the methods described in this paper. BLUSNO provides functionality to visualize, manipulate and modify all four kinds of taxonomies described above. Users may select any hierarchy of SNOMED CT with attribute relationships and have the option to choose from multiple SNOMED CT releases, going several years back.
Prior to BLUSNO, auditing with partial area taxonomies was accomplished using text reports generated by a variety of small, disconnected software utilities. Analyzing the data required extensive time and effort. Additionally, there was no way to visualize the abstraction networks, apart from drawing a diagram by hand with a graphics editor, which often required three or four days of work to accomplish. Auditing a hierarchy of SNOMED CT required the generation of a partial area taxonomy with one software tool, followed by analysis of the taxonomy with other tools, such as the CliniClue browser [5]. BLUSNO has overcome these limitations.
BLUSNO allows a user to view multiple taxonomies and subtaxonomies at once; this is accomplished through a windowing system where all of the information for a specific taxonomy is contained within a single window. Subtaxonomies can be requested for different hierarchies or for different releases of the same hierarchy. There are a number of preset layouts for taxonomy windows, provided to organize multiple views on the screen. The user has the ability to organize windows as s/he sees fit. Each window is a self-contained unit with options and functionality tied to the given taxonomy. Within each window the user can switch between interfaces for exploring and editing views of the taxonomy (not the original terminology). The main functionality of BLUSNO is defined by its graphical (diagram) interface, however, an auxiliary hybrid text-diagram interface was also implemented and is fully functional. Details follow below.
The Graphical Interface of BLUSNO
The graphical (diagrammatic) interface of BLUSNO produces highly interactive displays that are modeled after the static diagrams used in our previously published papers and presentations [3, 6, 7]. This representation provides high-level views of the previously described taxonomies. Users have the ability to move, pan and zoom throughout the hierarchy, quickly analyzing what partial areas exist within a specified area. To limit visual complexity, CHILD OF connections are only shown on request and typically limited to connections between a small number of partial areas. They are omitted in all figures below. All of the elements of the taxonomy (areas, partial areas, CHILD OF edges, etc.) are interactive in that they provide specific information when clicked on by a user.
The Hybrid Text-Diagram Interface of BLUSNO
In previous work [8] we have developed the Neighborhood Auditing Tool (NAT) for auditing the National Library of Medicine’s Unified Medical Language System (UMLS), a large and complex meta-terminology created by merging over 160 different source terminologies into a single structure. The NAT is a browser and auditing tool that provides the user with a hybrid text-diagram view that combines the best features of a graphical diagram display and a text-based representation of the UMLS or one of its source terminologies. For BLUSNO we have implemented a similar design, but instead of displaying information about a concept’s neighborhood, this paper introduces the new idea of a partial area neighborhood. A partial area neighborhood is anchored by a focus partial area, the partial area that an auditor is currently viewing. The neighborhood of this focus partial area includes the partial areas that are parent partial areas, child partial areas, and other partials areas that are within that same area, if any exist.
In a hybrid text-diagram view, textual information is displayed where it would be geometrically located in a diagram. For example, a list of parent partial areas is displayed strictly above the focus partial area. Similarly, the list of child partial areas is displayed strictly below the focus partial area. Detailed information about the selected partial area, as well as a history of viewed partial areas, is displayed at the center. Due to space limitations, examples had to be omitted.
Auditing Taxonomies through Time
New versions of SNOMED CT are incorporated into BLUSNO as they are released. BLUSNO includes a number of older versions of SNOMED CT, going back to July 2007, to aid in our auditing research efforts. Support for different releases of SNOMED CT enables an auditor to track the changes a given hierarchy has undergone, either because of auditing or because of “natural growth.” Different releases can be viewed side by side, so a direct comparison can be performed in seconds. Search functionality is provided to quickly find elements (partial areas, areas or concepts) that exist within multiple versions of a taxonomy.
Results
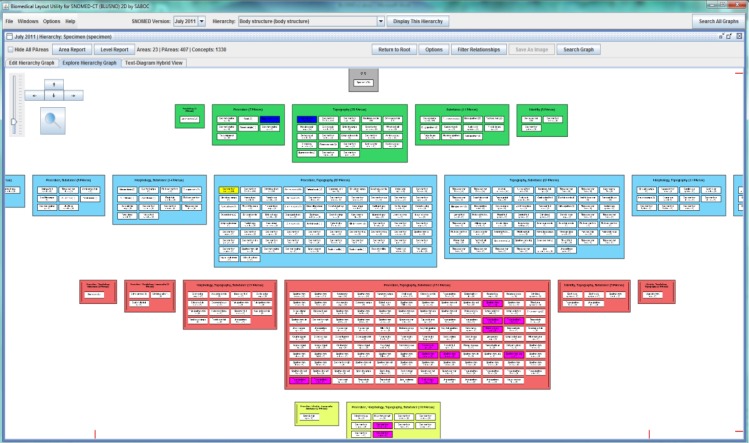
Figure 5 shows a layout of the partial area taxonomy for the SNOMED CT Specimen hierarchy, generated by BLUSNO. While this diagram is large and complex one must keep in mind that it summarizes 1330 concepts and the thousands of connections between them on a single screen. (A small amount of information is omitted at the left and right edges of Figure 5, however the visualization can be further zoomed out to view the whole taxonomy.) While we acknowledge that the details are not readable in this diagram, it still gives a good idea of the “gestalt” of the Specimen hierarchy. (Current desk top screens are also considerably larger than this figure, and the auditor can always zoom in on single areas.) For example, the row of boxes below the root area shows five areas, each defined by a single relationship. The displayed rows also indicate that no concept has all five possible relationships. Clicking on a partial area automatically highlights its parent partial areas and child partial areas in blue and purple respectively.
Figure 5.
The diagrammatic interface in BLUSNO. July 2011 Specimen hierarchy is shown.
Figure 5 makes the point that a partial area taxonomy may still be too large and complex, which has motivated the new abstraction networks introduced in this paper. Figure 6 demonstrates a relationship-constrained partial area subtaxonomy for the Specimen hierarchy, limited to three relationships. The comparison of Figure 5 and Figure 6 shows the dramatic reduction in size and complexity of the diagram that can be achieved by limiting the number of relationships used when constructing the relationship-constrained partial area subtaxonomy diagram. However, it should be noted that both Figure 5 and Figure 6 have a root area with no relationships and one single partial area Specimen with 29 concepts in it, namely the concept Specimen itself, and 28 of its descendants in SNOMED CT.
Figure 6.
A relationship-constrained partial area subtaxonomy for the Specimen hierarchy generated with R′ = {Morphology, Substance, Identity}.
Figure 7 shows a root-constrained partial area subtaxonomy from the Specimen hierarchy rooted at the partial area Lesion sample. A comparison of Figure 5 with Figure 7 again shows a considerable reduction in the size and complexity of the taxonomy. However, in this case the root of the taxonomy is “lost,” leading to a more severe loss of information. It is up to the user to determine which completeness/complexity tradeoff and which reduction factor is most acceptable to her/him. During an auditing session with BLUSNO this choice may be revised many times. A figure of the Hybrid Text-Diagram Interface of BLUSNO is omitted, as the detailed display is hard to render on paper. Interested readers are referred to related previous work of Morrey et al. [8].
Figure 7.
A root-constrained partial area subtaxonomy from the Specimen hierarchy rooted at the partial area Lesion sample.
Example Errors found in a SNOMED CT Audit
In 2007 we performed an audit of all partial areas in the Specimen hierarchy containing only one concept. Such partial areas are called “singletons.” The reason for concentrating on singletons is that according to Halper et al. [9], there is a higher percentage of erroneous concepts among singletons. This audit was severely impeded and decelerated by the lack of a tool such as BLUSNO. The necessary partial area diagrams had to be created by hand, and the data that the diagrams were based on had to be found by database queries. As can be seen in Figure 6, in BLUSNO the partial areas are displayed from left to right, ordered according to the numbers of concepts contained in them. Thus, the singletons are in the bottom row(s) of their areas, and BLUSNO makes it easy to find them. In 2007, 255 concepts out of the 1056 Specimen concepts were singletons. During the audit, errors of different kinds were found (Table 2). For example, Edema fluid sample in the area {morphology, substance} has a parent Fluid sample that is too general. The parent should instead be Body fluid sample. The audit was performed by JX, who is an MD with experience in using SNOMED CT. The errors were reviewed by Dr. Spackman of IHTSDO, the owner of SNOMED CT. Only errors confirmed by him are reported.
Table 2:
Sample of errors discovered in the Specimen hierarchy
| Concept Name | Area | Error Type | Correction |
|---|---|---|---|
| Edema fluid sample | morphology, substance | Incorrect parent: Fluid sample | Correct parent: Body fluid sample |
| Drainage fluid sample | morphology, procedure, substance | Incorrect morphology relationship to Discharge | Delete relationship |
| Tissue specimen obtained from anus by polypectomy | topography, procedure | Missing substance relationship | Add substance relationship to Body tissue material |
| Buccal smear sample | topography, procedure | Incorrect procedure target: Biopsy | Correct target: Smear |
| Tissue cell sample | topography, procedure | Ambiguous concept | Retire it |
| Cervical secretion sample | topography, substance | Duplicate concept with Cervical mucus specimen | Remove duplicate. Combine parents |
The audit found 88 erroneous concepts out of the 255 singletons. The use of BLUSNO reduces the described audit from a research project with a time horizon measured in weeks to a routine audit measured in hours. These errors were corrected in the 2008 release. In Table 3, we divided the 88 errors by their kinds. The majority of them, 69 in total, were incorrect or missing parents. In a study of SNOMED CT users’ preferences by Elhanan et al. [10], “incorrect parents” was the most troublesome of all errors. The use of the two new kinds of abstraction networks introduced in this paper makes this auditing paradigm practical even for the largest SNOMED CT hierarchies. Looking back at Table 1, the Procedure hierarchy has 729 areas and 10,731 partial areas, which would be hard to audit without the new abstraction networks.
Table 3:
Types of errors and their counts (auditing singletons)
| Type of Error | # | Type of Error | # | Type of Error | # | Total |
|---|---|---|---|---|---|---|
| Incorrect parent | 55 | Incorrect relationship | 4 | Duplicate concept | 1 | 60 |
| Missing parent | 14 | Incorrect target | 3 | Duplicate targets | 1 | 18 |
| Missing relationship | 8 | Ambiguous concept | 2 | 10 | ||
| Total: | 77 | 9 | 2 | 88 |
Discussion
We have demonstrated two new methods for generating abstraction networks, which define an advance towards the goal of letting terminology auditors generate abstraction networks with a reduction factor of their liking from any given hierarchy of a medical terminology. When going from a terminology to an abstraction network we used the ratio of partial areas to concepts as reduction factor. When going from a partial area taxonomy to a subtaxonomy, the reduction factor is best computed as ratio of partial areas in the subtaxonomy to partial areas in the partial area taxonomy. However, the user can rely on BLUSNO to manage the possible reduction factors for her/him. Naturally, the medical terminology needs to conform to certain structural requirements. Thus, there must be an “IS-A hierarchy” and a set of attribute relationships. SNOMED CT itself has attribute relationships in only seven of its 19 hierarchies. Therefore, the methods developed in the current paper cannot be applied to the twelve remaining hierarchies. BLUSNO is a fairly new tool, and its use for auditing has been limited to several small case studies.
Conclusions and Future Work
In this paper we introduced two new kinds of abstraction networks with the potential of giving auditors of medical terminologies a great degree of control over the displays of a SNOMED CT hierarchy. These two abstraction networks are called the relationship-constrained partial area subtaxonomy and the root-constrained partial area subtaxonomy. While there is only one partial area taxonomy for each hierarchy, there are typically many subtaxonomies of both kinds. We also discussed the BLUSNO tool for visualizing abstraction networks. BLUSNO provides a wide variety of features that support the auditing of SNOMED CT. It incorporates a graphical diagram interface and a hybrid text-diagram display. BLUSNO exists currently as a “beta release.” We plan to add support for other description logic-based terminologies, such as the National Cancer Institute thesaurus (NCIt) to BLUSNO. The opportunity for this is described in [11], where we discuss partial area taxonomies for NCIt. We are presently working on a new methodology for deriving abstraction networks for the twelve SNOMED CT hierarchies that lack attribute relationships.
Acknowledgments
Work on BLUSNO was partially supported by the NIH ARRA grant R01 LM008912-S2. We would like to thank David Daudelin, William Collins, Michael Gigliotti, Guemila Sawadogo, Fan Yin and Pavel Romanovski for their work on the BLUSNO project.
References
- 1.Rector AL, Brandt S, Schneider T. Getting the foot out of the pelvis: modeling problems affecting use of SNOMED CT hierarchies in practical applications. J Am Med Inform Assoc. 2011;18(4):432–40. doi: 10.1136/amiajnl-2010-000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gergely Héja GS, Péter Varga. Ontological analysis of SNOMED CT. BMC Med Inform Decis Mak. 2008;8(Supplement 1) doi: 10.1186/1472-6947-8-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Halper M, Min H, Perl Y, Chen Y, Spackman KA. Structural methodologies for auditing SNOMED. J Biomed Inform. 2007;40(5):561–81. doi: 10.1016/j.jbi.2006.12.003. Epub 2007/02/06. [DOI] [PubMed] [Google Scholar]
- 4.SNOMED CT User Guide
- 5.RECOMMENDATION ITU-R BT.601-5, 1982–1995
- 6.Wang Y, Halper M, Wei D, Gu H, Perl Y, Xu J, et al. Auditing complex concepts of SNOMED using a refined hierarchical abstraction network. J Biomed Inform. 2012;45(1):1–14. doi: 10.1016/j.jbi.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Halper M, Wei D, Perl Y, Geller J. Abstraction of complex concepts with a refined partial-area taxonomy of SNOMED. J Biomed Inform. 2012;45(1):15–29. doi: 10.1016/j.jbi.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrey CP, Geller J, Halper M, Perl Y. The Neighborhood Auditing Tool: a hybrid interface for auditing the UMLS. J Biomed Inform. 2009;42(3):468–89. doi: 10.1016/j.jbi.2009.01.006. Epub 2009/05/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halper M, Wang Y, Min H, Chen Y, Hripcsak G, Perl Y, et al. Analysis of error concentrations in SNOMED. AMIA Annu Symp Proc. 2007:314–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Elhanan G, Perl Y, Geller J. A survey of SNOMED CT direct users, 2010: impressions and preferences regarding content and quality. J Am Med Inform Assoc. 2011;18(Suppl 1):i36–44. doi: 10.1136/amiajnl-2011-000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min H, Cohen B, Halper M, Oren M, Perl Y. Detecting role errors in the gene hierarchy of the NCI Thesaurus. Cancer Inform. 2008;6:293–313. doi: 10.4137/cin.s440. Epub 2009/02/18. [DOI] [PMC free article] [PubMed] [Google Scholar]