Abstract
The widespread use of paper or document-based forms for capturing patient information in various clinical settings, for example in epilepsy centers, is a critical barrier for large-scale, multi-center research studies that require interoperable, consistent, and error-free data collection. This challenge can be addressed by a web-accessible and flexible patient data capture system that is supported by a common terminological system to facilitate data re-usability, sharing, and integration. We present OPIC, an Ontology-driven Patient Information Capture (OPIC) system that uses a domain-specific epilepsy and seizure ontology (EpSO) to (1) support structured entry of multi-modal epilepsy data, (2) proactively ensure quality of data through use of ontology terms in drop-down menus, and (3) identify and index clinically relevant ontology terms in free-text fields to improve accuracy of subsequent analytical queries (e.g. cohort identification). EpSO, modeled using the Web Ontology Language (OWL), conforms to the recommendations of the International League Against Epilepsy (ILAE) classification and terminological commission. OPIC has been developed using agile software engineering methodology for rapid development cycles in close collaboration with domain expert and end users. We report the result from the initial deployment of OPIC at the University Hospitals Case Medical Center (UH CMC) epilepsy monitoring unit (EMU) as part of the NIH-funded project on Sudden Unexpected Death in Epilepsy (SUDEP). Preliminary user evaluation shows that OPIC has achieved its design objectives to be an intuitive patient information capturing system that also reduces the potential for data entry errors and variability in use of epilepsy terms.
Introduction
Large scale multi-centers research studies require a consistent, flexible, and scalable informatics infrastructure to capture patient information that also supports real-time data access and analysis by clinical investigators. In contrast to free-text forms, which are either paper-based or word processor-based (e.g. Microsoft Word), web-based electronic forms ensure data quality through the use of built-in data validation features, use of common terminological systems to facilitate data sharing across participating study centers. Specifically, the use of common terminological systems in web-based electronic forms, modeled as a domain ontology, eliminates terminological heterogeneity across study centers[4,15,19]. Ontologies are formal knowledge models representing domain information in a logic-based language, such as the Web Ontology Language (OWL)[8]. Domain ontologies play a central role in data harmonization and integration in a number of biomedical domains, for example Gene Ontology (GO) for genome information[1], the Foundational Model of Anatomy (FMA) for human anatomy information[17], and SNOMED-CT for clinical terms[3]. The use of ontologies in clinical informatics systems also facilitate linking of patient data with related information. For example, epilepsy clinical data annotated with an epilepsy ontology can be cross-referenced with genotype information annotated with GO terms[1].
In this paper, we present the design, development, and preliminary user evaluation results of OPIC, an Ontology-driven Patient Information Capture System for Epilepsy, to support multi-center epilepsy clinical studies. The core knowledge resource of OPIC is the Epilepsy and Seizure ontology (EpSO), which is based on the new epilepsy and seizure classification system recommended by the International League Against Epilepsy (ILAE) classification and terminology committee[2]. Hence, the use of EpSO in OPIC allows it to be used for data management across different Epilepsy Monitoring Units (EMUs) that use the ILAE classification recommendations for reference thereby having a notable impact on collaborative studies across national and geographical regions. OPIC uses EpSO to implement a structured data entry interface, where the ontology terms are used to populate “multi-level drop-down” menu values and the EpSO terms identifiers are used to store the user-selected data values. In addition to data sharing and interoperability functions, OPIC also addresses an important logistical challenge faced by document-based forms in supporting a range of analytical queries.
Current document-based systems make it difficult for post-collection analysis of patient information, for example identifying patient cohorts, reporting adverse drug events, and uncovering disease associations[9,16]. These clinical research tasks require the use of complex natural language processing (NLP) approaches, and in some cases require use of optical character recognition (OCR) tools for analysis of document forms stored as images to retrieve and represent useful patient information in a machine understandable format. The NLP-based tools often have low accuracy and coverage, which reduces the utilization of patient information in clinical research. Further, EMU generate multi-modal patient information, including text and signal data corresponding to electroencephalograpy (EEG), electrocardiography (EKG), and magnetic resonance imaging (MRI) evaluations, which are difficult to be integrated with document-based forms and require tedious manual effort to copy and paste into the forms.
The OPIC system supports multiple types of data entry requirements, such as admission notes, progress notes for a patient, and discharge summary reports along with the functionality to support the input and storage of multi-modal patient data in form of EEG, EKG, and MRI reports. OPIC has been developed to support the NIH-funded Prevention and Risk Identification of SUDEP Mortality (PRISM) Project (P20-NS076965), which is a multi-institution study of the sudden unexpected death in epilepsy (SUDEP) funded by the National Institute on Neurological Disorders and Stroke (NINDS). The features of OPIC make it an ideal resource for capturing patient information and supporting multi-center epilepsy projects of similar magnitude nationwide.
1. Background
The goal of the PRISM project is to study SUDEP with or without evidence of seizure[14]. The precise mechanisms of death in SUDEP patients and effective prevention strategies are not yet known. Hence, following a 2008 NINDS-sponsored meeting, a new approach involving the use of multimodal parameters including cardiovascular, biochemical, and genetic factors has been adopted [12]. Further, due to the low rate of reported incidences (about 5000 patients deaths per year in the USA are SUDEP relating), multi-center studies are the key to collect sufficient SUDEP/near SUDEP events for statistical analysis. The PRISM project, funded as part of NINDS SUDEP Centers Without Walls initiative, will recruit patients from the EMU population, which has the highest number of potential SUDEP patients. We expect to enroll about 1200 patients at the participating EMUs at University Hospitals Case Medical Center (UH CMC Cleve-land), Ronald Reagan University of California Los Angeles (UCLA) Medical Center (RRUMC-Los Angeles), the National Hospital for Neurology and Neurosurgery (NHNN, London, UK), and the Northwestern Memorial Hospital (NMH Chicago).
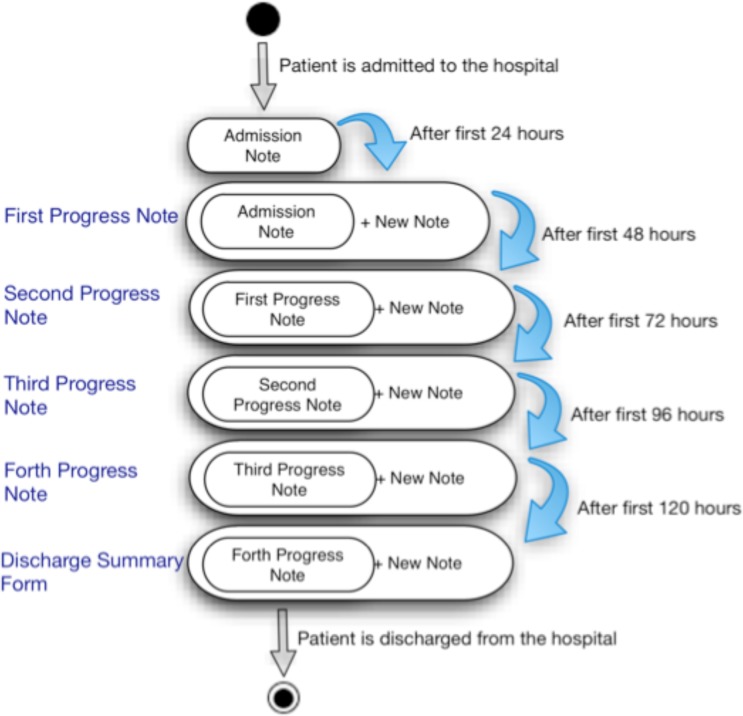
PRISM is the first use case for OPIC, as it requires the ability to uniformly capture patient information across the four EMUs and store the data in a common format with real-time access by researchers to support cohort identification. Patient information is captured at multiple stages during the patient’s stay in the EMU and three separate reports are generated: admission notes, progress notes, and discharge summary reports (Figure 1 illustrates the associated information flow in an EMU). The admission note is generated on the first day the patient is admitted to the hospital and records the demography, present history, past medical and surgical history, family history, psychosocial history, past and current antiepileptic medications, physical and neurological examinations, prior EEG, MRI, classification of paroxysmal episodes, impression and plan. On the second day of the patient’s stay, clinical fellows create a daily note, also known as progress note, for the patient. Information about history, examination, MRI, and medication information at time of admission are copied over from the admission note to the daily reports. Other sections, such as current EEG, classification of paroxysmal episodes, current AED, description of recorded seizures, are also added to the daily notes.
Figure 1:
Information flow in an EMU
A discharge summary report is created at the end of the patient’s stay in the EMU. The discharge summary includes some of the multi-modal patient data generated by the EMU, such as EEG and MRI reports. Hence, it is important for OPIC to support inclusion of images and additional multi-modal information in the patient discharge summaries.
2. Method
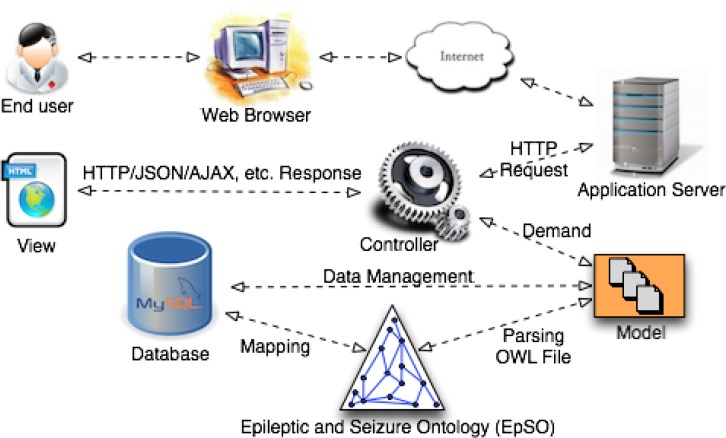
The OPIC system is built using agile methodology for rapid and iterative development in close consultation with the users. The Ruby on Rails (RoR) web development framework has been used to implement OPIC with a Model-View-Controller (MVC) architecture pattern (Figure 2). The MVC pattern involves a strict separation of the application logic from the user interface, which enable OPIC to seamlessly adapt to changing requirements of a clinical study (e.g. addition of blood oxygen level parameter for patient evaluation) or updates to the ILAE epilepsy and seizure classification system. This implementation approach ensures that OPIC is a flexible and user-centered system that can be used nationally as well as internationally to support effective capture of patient information. In the following subsections we describe the design and implementation of OPIC.
Figure 2:
Schematic overview of OPIC illustrating the different components and the associated process flow
2.1. The Epilepsy and Seizure Ontology
EpSO is the key knowledge resource for OPIC, driving many of its functionalities, including structured menu-driven data entry and data harmonization through use of uniform terminology across the participating study centers. Traditionally, the creation of a common terminology for epilepsy has been a difficult task due to the inherent complexity of epilepsy, its symptoms, the etiology, and the diverse community of users. Past ILAE classification systems, including the classification of seizures[10] and epilepsies[11], have been paper-based, which make it very difficult for these classification systems to be integrated with informatics applications. EpSO is a notable effort in creating a formal knowledge model corresponding to the 2010 ILAE epilepsy classification system through close collaboration between epileptologists, clinicians, members of the ILAE CTC, and computer scientists[20]. EpSO is modeled using the description logic-based Web Ontology Language (OWL)[8], which allows it to be easily integrated with OPIC.
The current version of EpSO has more than 590 classes and their associated properties to represent the ILAE classification system and additional terms needed to represent information generated during a patient’s stay in the EMU. These include classes modeling the etiology of epilepsy, which are categorized into four broad categories following the ILAE classification system[2]: “genetic” (e.g. chromosomal abnormality), “metabolic” (e.g. substance abuse), “structural” (e.g. cerebral palsy), and “unknown” (e.g. childhood febrile seizures). In addition, we have modeled the set of antiepileptic drugs using the RxNorm approach to capture drug ingredient, the associated brand names, the strength, and dosage types[13]. For example, EpSO models the information that drug ingredient “carbamazepine” is the constituent for “carbatrol”, “tegretol”, and “tegretolXR” brand name drugs. This mapping information between antiepileptic drug ingredients and brand name drugs allows clinicians and nursing staff to flexibly use either the drug ingredient name or the brand name in patient records, while OPIC automatically reconciles the terms to the same medication information. This important data harmonization step in OPIC is necessary for integration of patient data from multiple EMUs, which often have their conventions regarding terminology usage. Further, OPIC allows clinical researchers to use “carbamazepine” to identify relevant patient records, while OPIC automatically identifies and retrieves all patient records that mention one of the four drug names as medication (current or past). EpSO also models brain anatomy information by re-using FMA classes[17], and parameters associated with measurement of seizures (e.g. scalp electrodes versus intracranial electrodes for EEG recordings) by re-using classes defined in the Neural ElectroMagnetic Ontologies (NEMO)[7].
In addition to data harmonization and integration, the use of EpSO for annotating patient information enables linking of epilepsy phenotype information with closely related genotype information. Genetic information associated with epilepsy is increasingly important for better understanding of seizures, more accurate diagnosis, and patient care[6]. The use of EpSO in OPIC allows linking of patient information with genotype information through “schema mappings” between EpSO and widely used Gene Ontology (GO) annotations[18]. Similarly to genotype information, EpSO annotated epilepsy data can be linked to existing biological pathway data sources through mappings with the BioPAX ontology (for pathways)[5].
Finally, OPIC leverages the EpSO class hierarchy (Figure 3) to automatically reconcile variations in use of terms describing clinical findings also. For example, “fast ripples” is a special type of “high frequency oscillation” EEG feature that is typically analyzed in evaluating surgical outcomes for refractory patients. Similar to the drug information example, OPIC automatically identifies all terms linked to a clinical description by traversing the “parent-child” links in EpSO and retrieves the associated patient records that use either “fast ripples” or “ripples” in their description of intra-cranial EEG data.
Figure 3:

EpSO class hierarchy
2.2. Input Data Validation
Validation of input data is important to (a) ensure data quality before data is sent to the data repository, (b) reduce the need for expensive and labor-intensive “post-entry” data curation, and (c) improve the quality of query results in subsequent data analysis tasks (e.g. adverse event reporting). Data validation procedures, often integrated with data processing step in web-based forms, reduce typographical errors through a basic dictionary lookup, ensure “meaningful” data is added to the data fields through use of user-defined rules, and normalizes the data format before committing the data to the database. OPIC implements two kinds of validation resources: (1) a jQuery Validation Plugin, which is a standard library of jQuery-based validation solution that not only includes many of predefined validation rules (e.g. to make the field always required), but also allows users to create customized validation rules and, (2) a set of rules using regular expressions. We implement the OPIC data validation procedure as follows:
Presence check. The first check is implemented for “required” input data fields (e.g. patient name and epileptogenic zone) to ensure they are not empty and this validation procedure is triggered when the data field “loses” focus or the form is submitted. If a required field is empty, a warning message is displayed at both the data field and also at the top section of the data input form. In addition to the warning messages, the form “fails” to be submitted.
Data type check. Specific data fields are linked to pre-defined data types to ensure that the input data is of the correct data type. Currently five general data types are supported in OPIC, namely character, numeric, boolean, image (e.g. Portable Network Graphics, png), and date values.
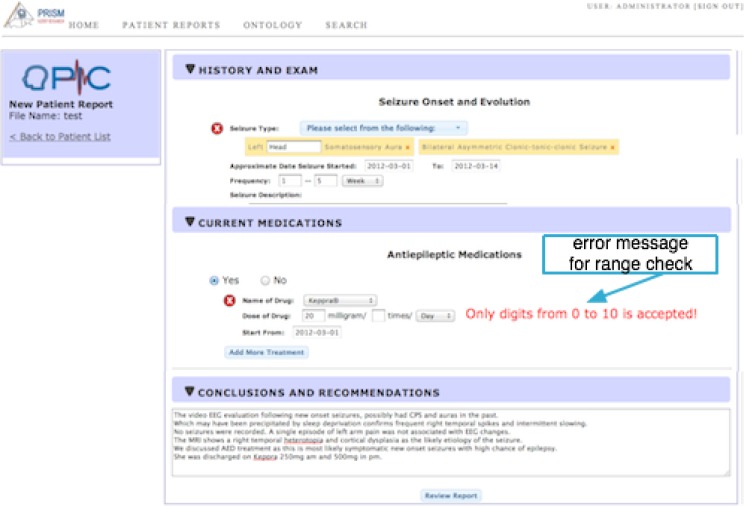
Range check. In many data fields, valid input values have to be within a pre-determined range. For example, numeric values for the “Age of the patient” data field have been defined to be within a specific range for the PRISM project.
Hence, all input values that violate the pre-defined validation rule are flagged and the user is presented with a warning message. To reduce the user effort in updating the incorrect values, the validated data values are retained and only the invalid data values are deleted during the data correction phase. Similar to the age data input field, OPIC implements a range validation procedure for the frequency of drug intake that allows entry of integer data values between the range of 0 to 10. Figure 4 illustrates an example warning message in case a data value outside of the appropriate data range is specified.
Figure 4:
“Range check” data validation for the dosage information
2.3. Logical Skip Patterns
In addition to input data validation, document-based forms cannot implement an important feature called “logical skip patterns”, which allows time constrained clinicians and nursing staff to input only relevant data items based on their preceding data entries. OPIC automatically “hides” sections of the data input forms based on the previous user selections. These skip patterns not only allow pre-defined sections of the form to be “hidden” from the users, but also enable the OPIC report generation module to remove these section from the published reports. This automated mechanism is especially helpful to reduce ambiguity in the data entry process and keep the context consistent during data entry.
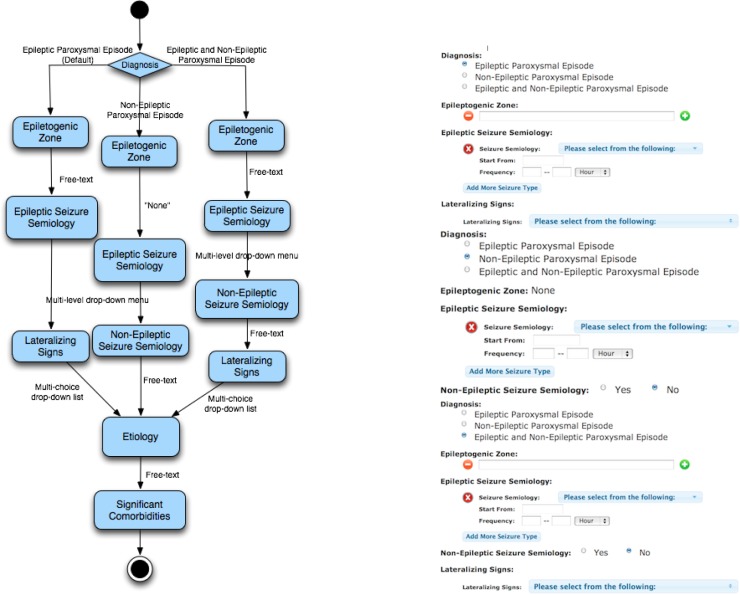
OPIC uses EpSO to define skip patterns, which creates a standardized representation format and facilitates re-use of skip patterns across projects. Figure 5 illustrates an example skip pattern implemented in the “Classification of Paroxysmal Episodes” section of a discharge summary form used in the UH CMC EMU. This skip pattern incorporates three possible scenarios and the corresponding “behavior” of the form in each of the three scenarios. In the first scenario, if the default option of Epileptic Paroxysmal Episode for the Diagnosis subsection is selected then the Epileptogenic Zone subsection is set as a free-text input field and the Non-Epileptic Seizure Semiology subsection is skipped. In the second scenario, if Non-epileptic Paroxysmal Episode option is selected for the Diagnosis subsection then a default value of “None” is assigned to the Epileptogenic Zone subsection and the Lateralizing Signs subsection is skipped. In the third scenario, if the Epileptic and Non-Epileptic Paroxysmal Episode option is selected for the Diagnosis subsection then no subsection is skipped.
Figure 5:
Conceptual view of “paroxysmal episode” skip pattern (left) and the corresponding sections (right)
The skip pattern implemented in the “Classification of Paroxysmal Episodes” is represented using the following template:
(subsection, value, skip-subsection, default-value).
Each skipped subsection will have an entry in the above template with an assigned default value (e.g. N/A for not applicable), such as
-
(Diagnosis, Non-Epileptic Paroxysmal Episode, Epileptogenic Zone, None),
(Diagnosis, Non-Epileptic Paroxysmal Episode, Lateralizing Signs, N/A).
Similarly, the skip pattern implemented in the “Evaluation” section is also useful in significantly reducing user effort to input only relevant data. Many skip patterns represent complex domain logic, hence the use of EpSO in standardizing their representation and streamlining their implementation enables OPIC to use skip patterns in variety of usage scenarios. Overall, OPIC implements skip patterns in 12 subsections of the patient report forms, which notably contributes to improving the turnaround time of patient data forms in an EMU.
2.4. Indexing Ontology Terms
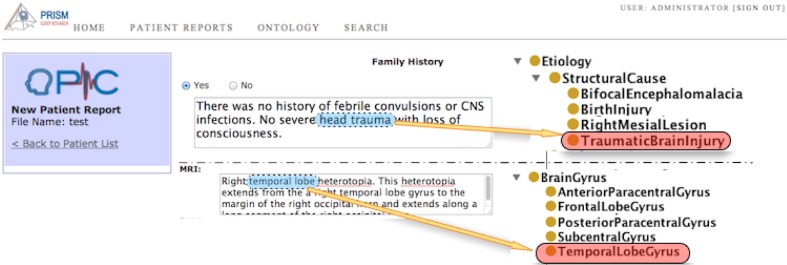
Though data entry fields in OPIC predominantly use drop-down menus (populated with values from EpSO), there are some free text fields and these data fields contain necessary information for supporting different investigator queries. OPIC implements a partial EpSO-driven indexing functionality that lexically parses data in the free-text fields and annotates terms corresponding to EpSO classes (Figure 6). These annotated terms are indexed by OPIC for subsequent use in query execution over both structured and free-text sections of the patient data entry forms.
Figure 6:
EpSO terms identified and indexed in free text fields
2.5. Dynamic Table Interface for Medication Information
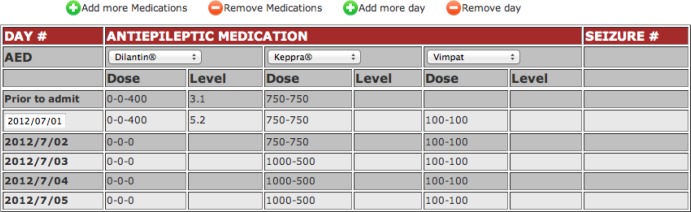
An important feature of OPIC is adaption of many existing data entry features from document-based systems, which have proven to be useful to the EMU staff, to a Web-based system. For example, the patient drug information table (Figure 7) allows systematic collection and representation of information about drug, dosage, and the level of antiepileptic medication in blood through a dynamic interface with built-in features for easy data entry. For example, the data validation mechanism ensures that the first date (Figure 7), corresponding to date of patient admission, is entered correctly. OPIC uses this date information to automatically generate subsequent dates corresponding to the daily updates of the patient and generation of the discharge summary.
Figure 7:
Table for input of medication information
2.6. Dynamic Report Generation and Database Support
The OPIC system supports generation of various types of reports that are required in EMUs, including generation of admission notes, progress notes and discharge summary reports. The user has the option to generate an editable HTML representation of the final version of a report or generate a PDF version that can be easily printed. In addition to support for report generation, OPIC has been designed as a flexible system that can be deployed in EMUs using multiple types of databases, including MySQL, Microsoft SQL server, and PostgresQL. OPIC uses the RoR ActiveRecord pattern to interface with the different types of databases, while providing an uniform and consistent user interface.
3. Results
OPIC was developed following an iterative and incremental development lifecycle conforming to agile software engineering methodology. This enabled OPIC to prioritize and implement “client-valued” functionality changes in a graduated manner interspersed with quick modifications following user feedback. The current version of OPIC is the result of three distinct phases over a period of 4 months. The result of the first phase was a prototype implementation featuring plain text input field, collapsible sections, the capability to review and edit the form as well as generation of a PDF version of the patient report. During the second phase, a series of user-friendly features, including check box and drop-down list were added, and logical skip pattern were implemented. The third phase included integration of controlled terminology system (modeled in EpSO) to implement single/multi-level drop-down menu with the ability to input data for repeated instances of a single type of event. For example multiple hospital visits or results of multiple evaluations (EEG or MRI reports) can also be captured flexibly by allowing the user to dynamically add or delete data field values. To validate the design principles and functionalities of OPIC, we conducted a systematic user evaluation after deployment at the UH CMC.
3.1. Comparative User Evaluation
The objective of our evaluation is to validate two key aspects of OPIC: (1) the ease of use and intuitiveness of the interface as compared to the previous document-based system used in UH CMC EMU, (2) potential to reduce errors through term suggestions. The evaluation was implemented in form of a questionnaire with 11 queries covering usability, effectiveness in data capture organization and validation of data. For example, Question 1 measured the ease of creating the discharge summary report using the paper-based system as compared to using OPIC. Questions 6–10 investigate the usability and customizability of OPIC as compared to the document-based system. Each query response was scored on a graduated scale of 1–10 points. The responses were categorized into two sets: From question 1 to question 5, 1 represented “not difficult at all” and 10 represented “very difficult”, while from question 6 to question 10, 1 represented “not at all” and 10 represented “very well”. Question 11 did not involve a score-based response but asked users to identify errors in the resulting report from both OPIC and the document-based system. This question was to evaluate the effectiveness of replacing most of the free-text data fields in the document-based system with multi-level ontology-driven drop down menus to reduce typographical and related errors.
The evaluation was conducted in two phases with the same group of users at the UH CMC EMU consisting of five clinical fellows on rotation in the department of neurology. In the first phase, the user group was asked to evaluate the document-based system without any exposure to the OPIC system, which elicited responses describing desired or missing features. In the second phase, the user group evaluated the OPIC system. The evaluation was conducted over a period of three weeks prior to and after deployment of the OPIC system.
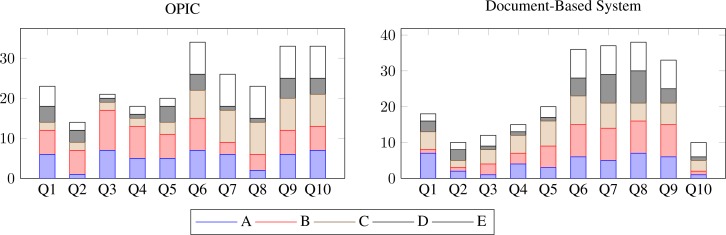
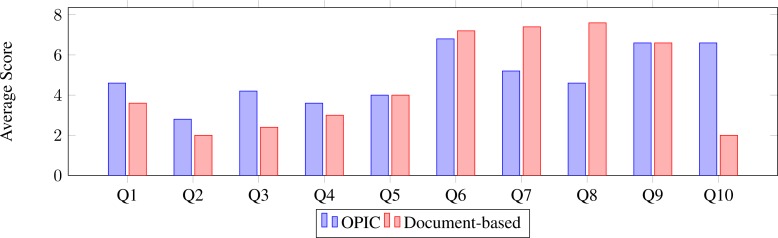
Figure 8 illustrates the variability in user responses to the questionnaire. It is interesting to note that there is greater variability in user response regarding use of OPIC, which can be explained in terms of the relative familiarity of users with computer-based systems (the UH CMC is currently in process of rolling out an electronic health record system). Figure 9 shows that the clinical fellows found the information presented in both OPIC and the document-based systems to be comparable, but unlike the document-based system, the potential for inadvertent modification of data is less in OPIC. This addresses the primary objective of OPIC, that is, to reduce errors during the patient information capture phase. Further, in response to question 10 regarding the importance of the “term suggestion feature” of OPIC (in form of drop-down menus), users ranked the original document-based system significantly lower as compared to OPIC.
Figure 8:
User responses to evaluation queries comparing OPIC (left) with the original document-based system (right)
Figure 9:
The average value of user responses comparing the original document-based system with OPIC
4. Discussion
At present, electronic health record systems have limited or no role for domain ontologies in capturing, storing, and retrieving patient information. Further, the incompatibility between vendor-specific EMR systems [21] makes it difficult to effectively conduct multi-center studies such as PRISM. The Research Electronic Data Capture (REDCap) system is a widely used system for online surveys and managing databases [22] but it is not geared towards management of discharge summary reports. In addition, REDCap features a library of data entry forms that can be re-used by its user community, but these forms have limited functionalities as compared to the domain ontology-driven features supported by OPIC. Hence, OPIC addresses an important requirement for a dedicated, ontology-driven patient information capture system that can be deployed across different centers to support large scale multi-center clinical studies.
OPIC is an evolving system and with the increasing coverage of the epilepsy domain in EpSO, including seizure features, EEG patterns, and semiological zones, it will be able to improve the quality of data capture and provide additional functionalities. For example, we are working to implement a EpSO-driven auto-complete feature in free-text input field, which is expected to help users avoid making typographical errors and reduce variations in use of terms to describe patient information. In addition, we are enhancing the report generation interface of OPIC to support a greater number of file formats and ability to generate multiple types of clinical reports from the same dataset. This is expected to significantly improve data re-use and prevent repeated data entry by users.
5. Conclusion
The use of document-based systems for patient data capture in EMUs is a significant challenge for large multi-center epilepsy studies. As part of the PRISM project, involving four study centers in the USA and UK, we report the development and deployment of an ontology-driven web-based patient data capture system called OPIC. Using EpSO, which formalizes the recommendations of the ILAE epilepsy and seizure classification system, OPIC supports flexible entry of multi-modal epilepsy data including EEG, ECG, and MRI reports. OPIC makes extensive use of EpSO driven multi-level drop-down menus to reduce user errors that are often introduced in free-text fields of document-based systems. OPIC has been deployed at the UH CMC EMU and initial user feedback shows that it has been successful in meeting its design objectives.
6 Acknowledgment
This research was supported by the PRISM (Prevention and Risk Identification of SUDEP Mortality) Project (1-P20-NS076965-01) and in part by the Case Western Reserve University CTSA Grant UL1 RR024989 and NIH/NCATS UL1TR000439.
References
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, Van Emde Boas W, Engel J, French J, Glauser TA, Mathern GW, MoshŽ SL, Nordli D, Plouin P, Scheffer IE. Revised terminology and concepts for organization of seizures and epilepsies: Report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2010;51:676–85. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- 3.Bodenreider O, Smith B, Kumar A, Burgun A. Investigating subsumption in SNOMED CT: An exploration into large Description Logic-based biomedical terminologies. Artificial Intelligence in Medicine. 2007;39(3):183–95. doi: 10.1016/j.artmed.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.BioPAX : Biological Pathways Exchange [cited 22 Jan 2008]; Available from: http://www.biopax.org/
- 6.Carranza Rojo D, Hamiwka L, McMahon JM, Dibbens LM, Arsov T, Suls A, Stšdberg T, Kelley K, Wirrell E, Appleton B, Mackay M, Freeman JL, Yendle SC, Berkovic SF, Bienvenu T, De Jonghe P, Thorburn DR, Mulley JC, Mefford HC, Scheffer IE. De novo SCN1A mutations in migrating partial seizures of infancy. Neurology. 2011 Jul 26;77(4):380–3. doi: 10.1212/WNL.0b013e318227046d. Epub 2011 Jul 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dou D, Frishkoff G, Rong J, Frank R, Malony A, Tucker D, editors. Development of NeuroElectroMagnetic Ontologies (NEMO): A framework for mining brain wave ontologies. Thirteenth International Conference on Knowledge Discovery and Data Mining (KDD2007); 2007; San Hose, CA. [Google Scholar]
- 8.Hitzler P, Krštzsch M, Parsia B, Patel-Schneider PF, Rudolph S. OWL 2 Web Ontology Language Primer: W3C; 2009 Contract No.: Document Number|.
- 9.Holmes AB, Hawson A, Liu F, Friedman C, Khiabanian H, Rabadan R. Discovering Disease Associations by Integrating Electronic Clinical Data and Medical Literature. PLoS ONE. 2011;6(6) doi: 10.1371/journal.pone.0021132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ILAE CTC Proposal for revised clinical and electrographic classification of epileptic seizures. Epilepsia. 1981;22:489–501. doi: 10.1111/j.1528-1157.1981.tb06159.x. [DOI] [PubMed] [Google Scholar]
- 11.ILAE CTC Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia. 1989;30:389–99. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- 12.http://www.aesnet.org/files/dmfile/HirschSUDEPNIHAppendix_e-1_FULL_REPORTNeurology20114.pdf
- 13.Liu S, Wei M, Moore R, Ganesan V, Nelson S. RxNorm: prescription for electronic drug information exchange. IT Professional. 2005;7(5):17–23. [Google Scholar]
- 14.Lhatoo SD, Faulkner HJ, Dembny K, Trippick K, Johnson C, Bird JM. An electroclinical case-control study of sudden unexpected death in epilepsy. Ann Neurol. 2010 Dec;68(6):787–96. doi: 10.1002/ana.22101. [DOI] [PubMed] [Google Scholar]
- 15.Murphy S, Mendis ME, Berkowitz DA, Kohane I, Chueh H, editors. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc; 2006. [PMC free article] [PubMed] [Google Scholar]
- 16.Murff HJ, Forster AJ, Peterson JF, Fiskio JM, Heiman HL, Bates DW. Electronically Screening Discharge Summaries for Adverse Medical Events. J Am Med Inform Assoc. 2003;10(4):339–50. doi: 10.1197/jamia.M1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosse C, Mejina JLV., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36(2003):478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Sahoo SS, Bodenreider O, Rutter JL, Skinner KJ, Sheth AP. An ontology-driven semantic mashup of gene and biological pathway information: Application to the domain of nicotine dependence. Journal of Biomedical Informatics. 2008;41(5 (Semantic Mashup of Biomedical Data)):752–65. doi: 10.1016/j.jbi.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahoo SS, Ogbuji C, Luo L, Dong X, Cui L, Redline SS, Zhang GQ. MiDas: automatic extraction of a common domain of discourse in sleep medicine for multi-center data integration. AMIA Annu Symp Proc; 2011. pp. 1196–205. Epub 2011 Oct 22. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang GQ, Sahoo SS, Lhatoo SD. From Classification to Epilepsy Ontology and Informatics. Epilepsia. 2012;53(Suppl. 2):28–32. doi: 10.1111/j.1528-1167.2012.03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FDA memo. H-IT Safety Issues, table 4, page 3, Appendix B, p. 7–8 (with examples), and p. 5, summary
- 22.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JS. Research electronic data capture (REDCap) - A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009 Apr;42(2):377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]