Abstract
Sepsis is a systemic inflammatory state due to an infection, and is associated with very high mortality and morbidity. Early diagnosis and prompt antibiotic and supportive therapy is associated with improved outcomes. Our objective was to detect the presence of sepsis soon after the patient visits the emergency department. We used Dynamic Bayesian Networks, a temporal probabilistic technique to model a system whose state changes over time. We built, trained and tested the model using data of 3,100 patients admitted to the emergency department, and measured the accuracy of detecting sepsis using data collected within the first 3 hours, 6 hours, 12 hours and 24 hours after admission. The area under the curve was 0.911, 0.915, 0.937 and 0.944 respectively. We describe the data, data preparation techniques, model, results, various statistical measures and the limitations of our experiments. We also briefly discuss techniques to improve accuracy, and the generalizability of our methods to other diseases.
INTRODUCTION
Sepsis is a heightened systemic immune response state due to an infection. It is defined as a combination of Systemic Inflammatory Response Syndrome (SIRS), and a confirmed or suspected infection, usually caused by bacteria. The infection may be localized to a part of the body such as lungs, skin, urinary tract, bone, or it may be generalized. Sepsis may occur whether the infection stays localized or spreads to other parts of the body. Bacteremia (presence of bacteria in blood) does not by itself denote sepsis in the absence of a systemic inflammatory response.
Untreated or inadequately treated cases of sepsis can lead to condition known as severe sepsis or septic shock, which are associated with high mortality and morbidity. Early diagnosis of sepsis is essential for successful treatment. Hence, we developed a Dynamic Bayesian Network (DBN) for the early detection of sepsis at the bedside in the emergency department. DBN is a generalization of Bayesian Networks (BN) and Hidden Markov Models (HMM), where the state transitions in the HMM are expressed using complex probabilistic interactions as in a BN[1]. DBN is a probabilistic technique where the input and output variables need not be predetermined and fixed, and algorithms are available to handle missing data, in addition to modeling temporal relationships using Markov properties. We included data from only the first 24 hours after admission for the test cases, and included only those variables that can be observed at the bedside or can be measured easily in the laboratory within this time. Our goal was to detect the presence or absence of sepsis within 24 hours after admission when the bacterial culture results are often unavailable. We have previously described a preliminary study about detecting sepsis using DBN[2]. Use of DBN to model organ failure in patients with sepsis has been described by Peelen et al.[3]
BACKGROUND
The high mortality of sepsis warrants early diagnosis and treatment. Sepsis is responsible for nearly 10% of the ICU admissions in the United States, totaling about 1 million cases nationwide every year[4]. The incidence rate of severe sepsis in the United States is about 300 per 100,000 persons per year, with a total of 750,000 cases nationwide per year. Direct costs per sepsis patient for ICU treatment in the United States have been estimated at more than $40,000. Gram negative bacteria have been implicated as the most common cause, followed by other bacteria and other pathogens.
The following definitions are from the American College of Chest Physicians (ACCP) and Society of Critical Care Medicine (SCCM) Consensus Conference held in 1991 to define common definitions for sepsis and related disorders, and were published in 1992[5]. Sepsis is defined as a systemic inflammatory reaction in response to an infection. In addition to SIRS, infection must be present or suspected to confirm a diagnosis of sepsis[5]. SIRS alone is not sufficient to confirm a diagnosis of sepsis, since SIRS can be caused due to non-infectious causes such as pulmonary embolism, adrenal insufficiency, anaphylaxis, pancreatitis, trauma, etc[5].
In adults, Systemic Inflammatory Response Syndrome (SIRS) is defined as the presence of two or more of the following[5]:
Body temperature below 36° C (degrees Celsius) or above 38° C.
Tachycardia, with heart rate above 90 beats per minute.
Tachypea (increased respiratory rate), with respiratory rate above 20 per minute, or arterial partial pressure of carbon dioxide (PaCO2) less than 4.3 kPa (kilo Pascals), equivalent to 32 mmHg (millimeters of mercury).
White blood cell (WBC) count less than 4,000/mm3(cubic millimeter) or above 12,000/mm3, or the presence of more than 10% immature neutrophils (band forms).
When sepsis causes Multiple Organ Dysfunction Syndrome (MODS), such as damage to vital organs, decreased perfusion, or hypotension, it is termed severe sepsis. Sepsis-induced hypotension is defined as a systolic pressure below 90 mmHg or a reduction in the baseline systolic blood pressure of more than 40 mmHg, in the absence of other causes of hypotension[5]. Sepsis can lead to a condition known as septic shock, which is indicated by hypotension (fall in blood pressure) that is not responsive to fluid replacement or vasopressor drugs[5].
Sepsis is a rapidly worsening clinical condition. Given the fast rate of change in the physiological parameters, the change in the clinical condition of sepsis patients lends itself well to a temporal probabilistic model such as a Dynamic Bayesian Network. Our objective was an early detection of sepsis even before many laboratory tests become available, ideally within the first few hours after admission.
MATERIALS AND METHODS
At LDS Hospital (LDSH) and Intermountain Medical Center (IMC), two tertiary care hospitals of Intermountain Healthcare in Salt Lake City, Utah, USA, the prevalence of sepsis in patients who directly present at the emergency department is between 1.7% and 2%. Clinical literature shows that patients with sepsis will have high mortality and morbidity if they are not treated immediately and aggressively. However, a confirmatory laboratory test for infections may take several hours to arrive, since culture and susceptibility tests cannot be performed immediately.
Many patients have atypical presentations, and may not have a clear picture of SIRS. To assist the clinicians in detection of sepsis, a clinical decision support system for early detection of sepsis is highly desirable. Sepsis presents a very good case for early detection using clinical decision support systems since the components of SIRS are easily measured at the bedside, or in the case of WBC and band counts, can be obtained in a short amount of time from the laboratory.
We wanted to use a temporal probabilistic model for the early detection of sepsis, and try to understand how the accuracy of the inference changes over time as more data become available. We used the Projeny toolkit[6] that we developed to build, train and test the Dynamic Bayesian Network models. We prepared our data using a sequence of steps and used the resulting data sets to train and test DBN models for the early detection of sepsis in the emergency department.
The Data Set
We obtained a data set of about 3,100 patients treated at Intermountain Healthcare, consisting of 20% cases (patients who had sepsis) and 80% controls (patients without sepsis). We used the anonymized data set for our sepsis detection modeling. The data elements available in the raw data set were the patients’ vital signs (heart rate, respiratory rate, body temperature, systolic blood pressure, diastolic blood pressure, and PaCO2); the patients’ lab test results (WBC count, bands percentage); and general encounter information (patient’s age, date of admission, date of discharge, etc). The data set also contained a variable named ‘Sepsis’, which was entered by a clinician during a retrospective review done for clinical research. Mean blood pressure or mean arterial pressure is the weighted average of the systolic and diastolic pressure. It is calculated using the formula
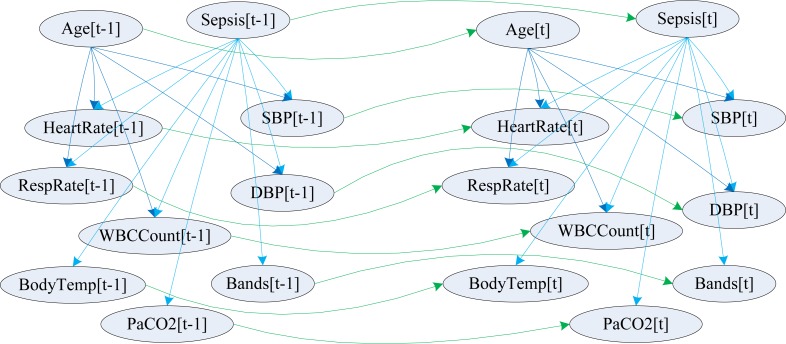
where MAP denotes mean arterial pressure, SP denotes systolic pressure and DP represents diastolic pressure. This formula is applicable to adult patients when the blood pressure is not extremely high or low. The variables included in our model and their probabilistic relationships within and across timeslices are shown in figure 1.
Figure 1.
Sepsis modeled as a two timeslice Dynamic Bayesian Network
The data set did not have information about blood (or other specimen) culture and sensitivity results from the microbiology laboratory, information signifying multi-organ dysfunction syndrome (MODS), or treatment information such as the administration of IV fluids and vasopressors, which help with a diagnosis of septic shock. Hence, we did not have the necessary clinical variables for diagnosing severe sepsis or septic shock.
We did not have clinical information denoting suspected or confirmed infection (culture and sensitivity results, clinical notes, etc.). However, we wanted to model a sepsis detection system with the currently available data. Our objective was to detect signs and symptoms to indicate impending or existing sepsis, which in turn can trigger treatment to prevent or treat sepsis, rather than confirming the presence of sepsis.
Vital signs were the most numerous type of data in our data set, and they were often measured between 1 to 2 intervals, even though some measurements were up to 20 hours apart. Hence, we used 1-hour as the width of the timeslices in our DBN models to help with early detection of sepsis within a few hours after admission. The lab tests were not measured at such frequent intervals. Not all vital signs were measured at 1-hour intervals. Hence, we had a large amount of missing data in our temporally aggregated and transformed data set. Our data preparation, data aggregation, temporal data abstraction and data discretization techniques are presented in detail in a separate publication[7], and a brief description is provided below.
Data Preparation
The accuracy of inferences performed by a machine learning system depends on the quality of data provided to the system. The input data determine the accuracy of the model and the technique, because many machine learning systems learn the structure of the model, the parameters or both from the training data. Machine learning systems such as Dynamic Bayesian Networks cannot directly use the raw data obtained from an electronic medical record system. The data need to be preprocessed and transformed into an appropriate format before they can be used by a Dynamic Bayesian Network-based model or system.
Probabilistic machine learning models require data in a continuous or discrete format. They cannot use unstructured or free-text data. The algorithms we used require discrete data. Clinical data from various sources need to be compiled together and transformed into a time-stamped, discretized format with a common structure for use by automated tools.
At Intermountain Healthcare’s LDS Hospital in Salt Lake City, the data are captured and stored in a well-structured form using an information model and an enterprise reference terminology. The original electronic medical record system, HELP (Health Evaluation through Logical Processing) encodes the data using a hierarchical data dictionary known as PTXT (Pointer to text), and stores the data in the HELP database mostly in encoded and partly in free-text forms[8]. Intermountain Healthcare also has a newer electronic medical record system known as HELP2, which uses a multihierarchical concept-based Healthcare Data Dictionary (HDD). The HELP2 data are stored in a Clinical Data Repository (CDR). HDD and CDR were developed in collaboration with 3M Health Information Systems, Inc.[9]
The data from HELP and HELP2 are highly structured and encoded using biomedical terminologies, and are usable for clinical documentation as well as decision support. However, all the data required for the temporal probabilistic models needed to be aggregated and abstracted before they could be used by the temporal reasoning tools.
Data Aggregation
The raw clinical data was available in an entity-time-attribute-value table format. Each row in the laboratory or vital signs table had columns identifying the patient and the date/time-stamp of the observation. The tables had additional columns that specified the data element and the value of the data element. The name, attribute or the value of the observation may be contained in a single column each, or spread across a group of columns. If the observation is a simple data element such as heart rate, it may be present in a single column. In cases where the data convey complex information such as blood pressure, which includes the systolic pressure, diastolic pressure, body site, posture of the patient, and the device used, this information would need to be post-coordinated from multiple pieces of data.
A denormalized table format was required to support the temporal reasoning tools used in our experiments. The denormalized table had two of its columns representing the patient identifier and the date/time identifier respectively. However, the remaining columns were not in the attribute-value format as in the source data tables. There were multiple additional columns instead, each representing a single, meaningful reconstituted clinical variable.
Different clinical variables were measured with different frequency and periodicity in the clinical setting. For example, vital signs were measured once every fifteen minutes to an hour. Lab tests were performed less often. Data elements that were measured together did not have the same date/time-stamp in some cases. They were often 1 to 15 minutes apart. Hence, storing them in the denormalized table produced several rows where only a handful of columns were populated. For each patient, we first loaded the timestamp and the values of the most numerous clinical variable into the denormalized table. We then selected the second most numerous clinical variable. If the patient identifier and timestamp of a given row of this second clinical variable existed in the denormalized table, we updated the row in the denormalized table to store the value of this second clinical variable in its own column. If the combination of the patient identifier and the timestamp did not exist, then a new row was inserted into the denormalized table with this value. This process was repeated for all clinical variables in the data set.
Temporal Data Abstraction
At the end of the data aggregation step, all the data reflecting the clinical variables in the model were stored in a single denormalized relational database table. The data present in this table were used to train and test the model. However, the data rows differed by a few minutes to a few hours, and produced a very sparse data table. A very sparse data table when used for training necessitates the use of the expectation maximization (EM) algorithm, which increases the computational expense of the model while reducing the accuracy of the learned parameters. However, the data can be temporally consolidated to pick one representative data point per time interval for the smallest time interval represented in the model, which will reduce the need for imputing missing values using the EM algorithm.
The smallest time interval to be supported by the model is based on both the nature of the model and the availability of data. In the case of the sepsis data set, which consisted of both cases and controls from the emergency department, the model consisted mostly of vital signs, which were available once every hour in most cases. So, a timeslice interval of 1-hour was chosen for the sepsis models, and a representative data point was chosen for each clinical variable during every 1-hour interval.
In the sepsis data set, we encountered both multiple instances and no instances of various clinical variables observed during each timeslice interval. Missing data can mean a variety of things: the data were not measured, measured and then lost, or they were uneventful and in line with the expected values given the prior measurements, and hence not recorded in this case. It may also mean that a value was measured on paper or was stored in a different part of the electronic medical record system, and hence unavailable at the time of data preparation. Several approaches have been discussed to define and overcome the missing data problem. Little and Rubin classify reasons for missing data as missing but completely at random (MCAR), missing at random (MAR), and not missing at random (NMAR)[10].
We did not apply special treatment for missing data in our experiments. Bayes Net Toolbox (BNT), which implemented all the algorithms we needed to train and test the models, supported parameter learning with missing values using the Expectation Maximization (EM) algorithm. Hence, we were able to leave missing values as null values in the database, and we designed our temporal modeling toolkit, Projeny, to support null values in the data set and to call the EM algorithm in BNT.
Approaches to choosing the representative data point for a variable if multiple data points are available in a temporal data set are discussed by various authors under the context of temporal data clustering[11,12][13]. Some simple approaches for temporal data sampling include selecting the average, or selecting the most abnormal measurement. We decided to select the average, since this process can be automatically applied for all numerical variables.
At the end of this process, we had a temporally abstracted denormalized table, with one data point or a null value for each variable per timeslice per patient. The numerical data were continuous in this data set. The data can be used in this form with models and algorithms that support continuous data. However, we designed our models with entirely discrete variables, since models with discrete variables are computationally less expensive and more tractable than models with continuous variables. Furthermore, only discrete variables were supported by our toolkits. Hence, we chose to discretize the continuous variables in our data set.
Data Discretization
Several data discretization methods are available to discretize continuous data for use with machine learning algorithms. It is helpful to evaluate the distribution of the continuous variables before we choose the discretization technique and any manually selected cut-off points. We used histograms and cluster analysis to find clusters and study the distribution of each continuous variable individually. Visually discernible clustering was not found for the continuous variables, and many continuous variables in our data set formed one large cluster each with few outliers.
We tried equal interval, domain knowledge based, k-means clustering and minimum description length (MDL) discretization techniques. MDL algorithm provided the most accurate results, and is described here. The lessons we learned from other discretization algorithms, and how choosing the correct algorithm can drastically affect the accuracy of a model are described in a separate publication[7].
The minimum description length (MDL) algorithm described by Fayyad and Irani (1993) finds the minimum number of clusters of the input variable required to describe the variation in the output variable[14]. All the relationships in our models were directional; hence describing the variation in one variable using variation in one or more variables is straightforward. The variations in the child nodes can be explained in terms of variation in parent nodes. Hence, the MDL algorithm seemed to be appropriate for discretizing the variables in our models. The model works by sorting and then splitting the distribution of input values at specific cut-off points that reduce class entropy of the resulting classes[14]. We used the MDL algorithm implemented in Weka[15] to discretize our data.
We found that MDL discretization produced a much smaller number of discrete states for most of the continuous variables in the sepsis model. Using the MDL algorithm also reduced the time taken for training and testing the model, and produced higher accuracy than other discretization techniques.
Model Structure, Training and Testing
A Dynamic Bayesian Network (DBN), also known as a Temporal Bayesian Network, allows complex causal relationships within and across time instances to be represented as a directed acyclic graph (DAG) with Bayesian probabilities[1]. A DBN may be considered as a generalization of a Hidden Markov Model (HMM), in which the probabilistic relationships are represented using complex interactions and dependencies between many variables as a Bayesian Network. The temporal processes in the model are designed as time invariant Markov processes. This reduces the computational complexity and the number of parameters required to describe the model, since the conditional probabilities learned from a two timeslice DBN can be used to infer the variables in test cases of arbitrary lengths. A detailed description of Hidden Markov Models is provided in [16].
We created a two timeslice Dynamic Bayesian Network using the nodes and edges shown in figure 1. We created several models, and the only latest and the most accurate model is described in this article. The model was built using the Projeny toolkit that we have created. Projeny is a front-end application written in Java, and it provides a user-friendly graphical environment to create, train and test Dynamic Bayesian Network models[6]. It also allows easy data binding between the nodes in the model and various columns in a relational database table. Projeny allows the user to call the Bayes Net Toolbox (BNT)[17], a DBN algorithm implementation running inside Matlab, for both training and testing. Projeny then saves the results from the inference algorithm in a relational database table. Projeny is based on the source code of Bayesian Network tools in Java (BNJ)[18], and it uses the JMatLink library[19] to communicate with Matlab. Projeny is released as open source software under the GNU GPL v2 license[20].
We first created the nodes of the two timeslice DBN model, and then we defined the states of the nodes as described earlier under the data discretization section. The variables included in our model (and their names used in figure 1) are clinicians’ diagnosis of sepsis (Sepsis), patient’s age (Age), systolic blood pressure (SBP), diastolic blood pressure (DBP), heart rate (HeartRate), respiratory rate (RespRate), body temperature (BodyTemp), WBC count (WBCCount), PaCO2 (PaCO2) and percentage of immature neutrophils (Bands). We then created the intra-slice (atemporal, shown in blue) and inter-slice (temporal, shown in green) edges (probabilistic relationships), as shown in figure 1. When we train and test the model, BNT automatically ‘unrolls’ (expands) the model to as many timeslices as are present in the data of each patient, and learns the probabilities or infers the marginal probability distributions. BNT uses the EM algorithm[21] to learn the conditional probability tables in spite of missing data.
In this model, age and sepsis are d-separated by the nodes that represent systolic blood pressure, diastolic blood pressure, heart rate, respiratory rate and WBC count. Age and sepsis together explain the variation in these physiological parameters in the model. If none of these five physiological parameters in the model are known, then age and sepsis are mathematically conditionally independent. [1]
The discretized data set was divided into training data set and test data set. Two-thirds of anonymized patients were allocated to the training data set at random, and the remaining one-third of the patients were allocated to the test data set. Training was performed using an EM-based parameter learning algorithm implemented in BNT. Training was performed with a data set having a maximum of 168 timeslices (approximately 7 days since admission) for each patient. Training took about 9 hours, on the same computer with two quad-core 2.25GHz Intel Xeon processors, 24GB of RAM and 32GB of swap space. Training completed successfully in less than 9 hours, with less than 7GB of memory utilization by Matlab.
The value of sepsis was replaced with null values for all the timeslices for all the patients in the test data set. Our goal was to repeat testing with data sets having different number of timeslices, so that we can simulate testing after the patient has been in the hospital for increasing durations of time. The test data set was then divided into four separate data sets having up to 3, up to 6, up to 12 and up to 24 timeslices for each patient. Testing was performed using a junction-tree algorithm[22], with test data sets having a maximum of 3, 6, 12 and 24 timeslices, since our goal was to detect sepsis within 24 hours after admission. These timeslices correspond to approximately 2, 5, 11and 23 hours after admission respectively, since the first timeslice was measured when the patient arrived at the emergency department at time t = 0 hours after admission.
RESULTS
Sepsis is a binary variable in our model, with values of true and false. The goal of our DBN model was to detect signs and symptoms indicating impending or existing sepsis, to trigger interventions to prevent or treat sepsis. Hence, the presence of sepsis as entered by the clinician provided a reference standard against which our DBN model’s inferences were compared. This makes our model similar to laboratory tests that are performed to detect specific diseases.
Given the similarity of our sepsis detection models to laboratory tests that detect a disease, our DBN models were evaluated using the same evaluation techniques applied to laboratory tests. We decided to perform statistical analysis of our model’s inferences in terms of sensitivity, specificity, positive predictive value, negative predictive value, F-value and area under the ROC (receiver operator characteristic) curve.
The clinician-entered values of sepsis, also considered as the reference standard or ‘disease‘, was left intact in the training data set. The clinician-entered values of sepsis in the test data set were hidden from the inference algorithm during the test iteration, and were later used to evaluate the performance of the predicted values. All other variables were left intact in both the training and test data sets. The state of sepsis estimated by the DBN model was considered analogous to a lab test finding. If the probability of sepsis estimated by the DBN model was equal to or above 0.5, it was considered as a positive test. If the estimated probability of sepsis was below 0.5, it was considered a negative test.
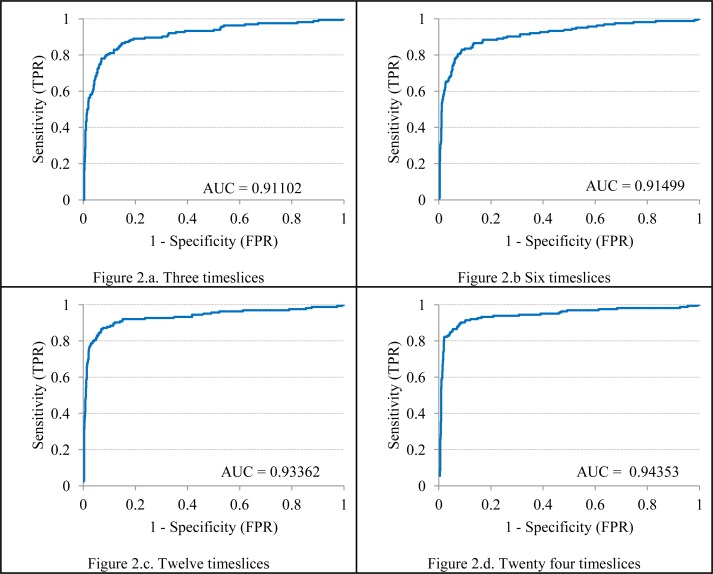
A 2x2 confusion matrix and standard epidemiologic techniques could then be applied to calculate the sensitivity, specificity, positive predictive value, negative predictive value, and the F-measure. The ROC curve was constructed and the area under the ROC curve were calculated using a procedure described by Morrison using Microsoft Excel[23]. A confusion matrix was plotted with the actual values of sepsis considered as the ‘disease’ and the estimated values of sepsis considered as the ‘test‘. The confusion matrices are shown in the table 1 (sub-tables 1.a. through 1.d.). The area under the ROC curve (AUC) obtained from the model using test data sets with 3, 6, 12 and 24 timeslices is shown in figure 2 (sub-figures 2.a. through 2.d.).
Table 1.
Confusion matrices for detecting sepsis using test data sets with 3, 6, 12 and 24 timeslices.
| Table 1.a. Three timeslices | Table 1.b. Six timeslices | ||||||
| Sepsis | Actual Yes | Actual No | Total | Sepsis | Actual Yes | Actual No | Total |
| Estimated Yes | 113 | 45 | 158 | Estimated Yes | 116 | 44 | 160 |
| Estimated No | 51 | 834 | 885 | Estimated No | 48 | 835 | 883 |
| Total | 164 | 879 | 1043 | Total | 164 | 879 | 1043 |
| Table 1.c. Twelve timeslices | Table 1.d. Twenty four timeslices | ||||||
| Sepsis | Actual Yes | Actual No | Total | Sepsis | Actual Yes | Actual No | Total |
| Estimated Yes | 134 | 45 | 179 | Estimated Yes | 141 | 48 | 189 |
| Estimated No | 30 | 834 | 864 | Estimated No | 23 | 831 | 854 |
| Total | 164 | 879 | 1043 | Total | 164 | 879 | 1043 |
Table 2.
Comparison of statistical measures of models using test data sets with 3, 6, 12 and 24 timeslices.
| 3 timeslices | 6 timeslices | 12 timeslices | 24 timeslices | |
|---|---|---|---|---|
| Sensitivity (recall) | 0.68902 | 0.70732 | 0.81707 | 0.85976 |
| Specificity | 0.94881 | 0.94994 | 0.94881 | 0.94539 |
| PPV (precision) | 0.71519 | 0.72500 | 0.74860 | 0.74603 |
| NPV | 0.94237 | 0.94564 | 0.96528 | 0.97307 |
| F-measure | 0.70186 | 0.71605 | 0.78134 | 0.79887 |
| AUC | 0.91102 | 0.91499 | 0.93362 | 0.94353 |
Figure 2.
Area under the ROC curve (AUC) obtained using test data sets with 3, 6, 12 and 24 timeslices
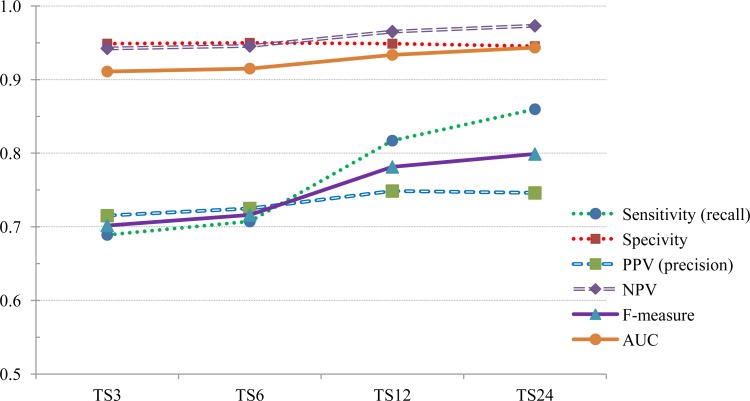
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), F-measure and the area under the ROC curve (AUC) are given in table 2.
Figure 3 shows how the statistical measures of sensitivity, specificity, PPV, NPV, F-measure and AUC increase as the number of timeslices in the test data set increases.
Figure 3.
Comparison of statistical measures using test data sets with 3, 6, 12 and 24 timeslices.
Discussion
The shortest number of timeslices supported by our algorithms is three. A three timeslice model includes data collected at approximately 0, 1 and 2 hours after admission in the emergency department. Hence, we tried to detect the presence of sepsis in less than 3 hours (approximately two hours, plus or minus a few minutes) in the first experiment. The three timeslice model shows a sensitivity of 0.69, a specificity of 0.95, positive predictive value of 0.72, negative predictive value of 0.94 and an area under the curve of 0.91 while testing with three timeslices.
From these results, we can demonstrate that DBN methods can be used to successfully detect sepsis in patients in the emergency department within two hours of admission. The model can detect sepsis with variables that are mostly collected at the bedside, and WBC count and bands percentage, which are easily obtained from the lab in a short duration of time. These measures again show that the model is more specific than it is sensitive.
However, the sensitivity increases further as more data become available, as can be seen from the 6, 12 and 24 timeslice models. Other statistical measures such as area under the ROC curve, specificity, PPV, NPV and F-measure also increase as more data become available for a patient.
It must be noted that the prior probability of sepsis in the emergency departments of Intermountain Healthcare was about 2%. But our data set was enriched to have a prior probability of 20% and our data set consisted of 20% cases and 80% controls. The real world prior probability of sepsis (0.02) is only 1/10 of the prior probability in our data set (0.2). Hence, the PPV and NPV of the model will change in the real world. Hence, further testing and validation needs to be done before this model may be used in an emergency department to detect sepsis.
A majority of probabilistic temporal reasoning experiments in the biomedical domain use Hidden Markov Models which in turn use equal-interval and equal-frequency data preparation techniques. We have shown that by using information content based methods of data preparation such as the MDL algorithm which analyzes the variation in the dependent variable to discretize the independent variable, we can obtain smaller but more meaningful state-spaces. This is a novel application and finding in the field of medicine, where probabilistic temporal reasoning methods have not been used extensively.
Our experiments showed that both data preparation and model structure affect the accuracy as well as the computational complexity of the model. We also found that simple models that are designed to be intuitive for human experts’ understanding such as the SIRS model may not be computationally efficient or accurate for probabilistic modeling[7]. Models that take into account the complex conditional inter-dependencies and reflect them accurately, while defining the state-space in a meaningful way, prove to be more accurate in probabilistic learning and inference[7].
CONCLUSIONS AND FURTHER RESEARCH
An overview of temporal reasoning techniques, detailed descriptions of the data preparation and discretization techniques, and detailed descriptions of multiple sepsis modeling experiments and glucose homeostasis (insulin dosing) models are described in the author’s doctoral dissertation, along with a description of factors influencing the computational complexity and accuracy of these models[7].
Our Dynamic Bayesian Network methods and the Projeny toolkit were built to be generalizable. These methods were successfully tested in estimating serum glucose and recommending insulin drip rates for patients in the intensive care unit. The results were validated by comparing them to the estimations of the computerized rule-based protocol currently in use. These results were described in a previous publication[24].
We intend to perform further experiments with data sets with real-world prior probability, variables that denote a suspected or confirmed infection, and differential diagnoses of sepsis or SIRS. We also intend to include clinical variables that are necessary to estimate the presence of severe sepsis or septic shock. We also hope to build and test models that predict future probability of sepsis rather than estimating the current probability. Such prognostic methods can help to provide treatment that can reduce or avert severe mortality and morbidity.
Acknowledgments
The authors thank Dr. Jason Jones at Intermountain Healthcare, Salt Lake City, UT for his help with the sepsis data set.
References
- [1].Murphy KP. U.C.Berkeley; 2002. Dynamic Bayesian Networks: representation, inference and learning. PhD thesis. [Google Scholar]
- [2].Wong A, Nachimuthu SK, Haug PJ. Predicting Sepsis in the ICU using Dynamic Bayesian Networks. AMIA Annu Symp Proc; 2009. [Google Scholar]
- [3].Peelen L, de Keizer NF, Jonge Ed, Bosman RJ, Abu-Hanna A, Peek N. Using hierarchical Dynamic Bayesian Networks to investigate dynamics of organ failure in patients in the Intensive Care Unit. J Biomed Inform. 2010 Apr;43(2):273–286. doi: 10.1016/j.jbi.2009.10.002. [DOI] [PubMed] [Google Scholar]
- [4].Dremsizov TT, Kellum JA, Angus DC. Incidence and definition of sepsis and associated organ dysfunction. Int J Artif Organs. 2004 May;27(5):352–359. doi: 10.1177/039139880402700503. [DOI] [PubMed] [Google Scholar]
- [5].Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992 Jun;101(6):1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- [6].Nachimuthu SK. Projeny: An Open Source Toolkit for Probabilistic Temporal Reasoning. AMIA Annu Symp Proc; 2009. [Google Scholar]
- [7].Nachimuthu SK. 2012. Temporal Reasoning in Medicine using Dynamic Bayesian Networks. PhD thesis.University of Utah. [Google Scholar]
- [8].Pryor TA, Gardner RM, Clayton PD, Warner HR. The HELP system. J Med Syst. 1983 Apr;7(2):87–102. doi: 10.1007/BF00995116. [DOI] [PubMed] [Google Scholar]
- [9].Clayton PD, Narus SP, Huff SM, Pryor TA, Haug PJ, Larkin T, et al. Building a comprehensive clinical information system from components. The approach at Intermountain Health Care. Methods Inf Med. 2003;42(1):1–7. [PubMed] [Google Scholar]
- [10].Little RJA, Rubin DB. Statistical analysis with missing data. Wiley; New York: 1987. [Google Scholar]
- [11].Mitsa T. Chapman & Hall/CRC data mining and knowledge discovery series. Chapman & Hall/CRC; 2009. Temporal Data Mining. [Google Scholar]
- [12].Warren Liao T. Clustering of time series data–a survey. Pattern Recognition. 2005;38(11):1857–1874. [Google Scholar]
- [13].Bar-Joseph Z. Analyzing time series gene expression data. Bioinformatics. 2004;20(16):2493. doi: 10.1093/bioinformatics/bth283. [DOI] [PubMed] [Google Scholar]
- [14].Fayyad UM, Irani KB. Multi-interval discretization of continuous-valued attributes for classification learning. Proceedings of the 13th International Joint Conference on Artificial Intelligence; Morgan-Kaufmann; 1993. pp. 1022–1027. [Google Scholar]
- [15].Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. ACM SIGKDD Explorations Newsletter. 2009;11(1):10–18. [Google Scholar]
- [16].Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Readings in speech recognition. 1990;53(3):267–296. [Google Scholar]
- [17].Murphy KP. BNT - Bayes Net Toolbox. Cited on March 13, 2009. Available from: http://people.cs.ubc.ca/~murphyk/Software/BNT/bnt.html.
- [18].Hsu, et al. BNJ - Bayesian Network Tools in Java. Cited on March 13, 2009. Available from: http://bnj.sourceforge.net.
- [19].Müller S. JMatLink. Cited on March 13, 2009. Available from: http://jmatlink.sourceforge.net.
- [20].Nachimuthu SK. Projeny website. Cited on March 10, 2012. Available from: http://projeny.sourceforge.net.
- [21].Lauritzen SL. The EM algorithm for graphical association models with missing data. Computational Statistics & Data Analysis. 1995;19(2):191–201. [Google Scholar]
- [22].Lauritzen S, Spiegelhalter D. Local computations with probabilities on graphical structures and their application to expert systems. Journal of the Royal Statistical Society Series B (Methodological) 1988;50(2):157–224. [Google Scholar]
- [23].Morrison AM. Receiver Operating Characteristic (ROC) Curve Preparation: A Tutorial. 5 Vol. 20. Boston: Massachusetts Water Resources Authority Report ENQUAD; 2005. [Google Scholar]
- [24].Nachimuthu SK, Wong A, Haug PJ. Modeling Glucose Homeostasis and Insulin Dosing in an Intensive Care Unit using Dynamic Bayesian Networks. AMIA Annu Symp Proc. 2010;2010:532–536. [PMC free article] [PubMed] [Google Scholar]