Abstract
During the past decade, molecularly targeted drugs have had a transformative impact on the treatment of several cancer types. Although the clinical benefits of these drugs are impressive, their effects are generally short-lived due to the acquisition of resistance. Unlike most cytotoxic agents, for which resistance mechanisms have remained obscure despite decades of clinical use, an understanding of the molecular basis of resistance to most targeted agents has emerged quickly. This rapid progress has been possible due to advances in molecular technologies that allow genome-wide profiling of patient samples. One important and consistent theme is that resistance is almost invariably associated with restoration of the signaling pathway inhibited by the targeted agent. Here I review examples from three diseases — chronic myeloid leukemia, prostate cancer, and lung cancer — that illustrate these points and reveal how insights into resistance mechanisms can rapidly accelerate the development of second-generation targeted therapies or combination regimens to improve patient outcome.
INTRODUCTION
Cancer drug development in 2012 is almost exclusively focused on therapies that target cancer-relevant proteins, largely identified from studies of cancer genomes. This is a striking difference from prior decades when cytotoxic agents with broad antitumor activity dominated the pipelines of most pharmaceutical companies. Currently these drugs are developed as single agents and, when evaluated in the right patient population, can have spectacular clinical benefit. However, even in best-case scenarios, responses tend to be short-lived and resistance develops. There is no doubt that appropriate drug combinations are needed, but the number of possibilities is too daunting to envision how an empiric approach, as was used for cytotoxics, might be successful. Here I review three examples in which studies of resistance to targeted agents in chronic myeloid leukemia, prostate cancer, and lung cancer have provided insight into the development of next-generation inhibitors and rational combinations. These examples might serve as a blueprint for other cancers and cancer targets.
ABL KINASE INHIBITORS IN CHRONIC MYELOID LEUKEMIA
During the past 10 years, the ABL kinase inhibitor imatinib has transformed the treatment of chronic myeloid leukemia (CML) from a disease with a 5- to 6-year life expectancy to a chronic condition that, if treated early, can be managed for decades with relatively non-toxic oral therapy. Imatinib induces cytogenetic and molecular remissions in most patients, but therapy must be continued indefinitely because stem and early progenitor cells derived from the leukemic clone are generally spared. In addition, roughly 20% of patients relapse on imatinib during the first 5 years of therapy (∼4 percent per year) (1). The mechanism of relapse, first shown in patients with blast crisis CML and Philadelphia chromosome–positive acute lymphoid leukemia that extended to chronic phase CML, is most commonly caused by point mutations in BCR-ABL that impair binding of imatinib to the kinase domain (2). Initially it was suspected that only a limited number of mutations were capable of causing resistance; however, further investigations have revealed more than 50 different amino acid substitutions in imatinib-resistant CML patients (3).
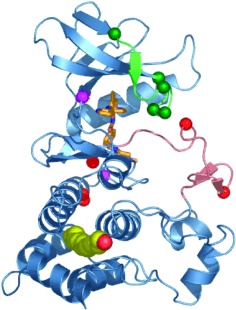
Such a diversity of resistance mutations would typically preclude an effective strategy to overcome resistance through inhibition of the same target with a second-generation compound due to the multiple mechanisms of potential escape. However, structural studies of imatinib bound to the ABL kinase domain revealed conformational requirements that explain why so many different mutations are capable of conferring drug resistance. Specifically, imatinib binds ABL when the activation loop of the kinase domain is in the closed, inactive conformation. Although some of the resistance mutations map to amino acids that make direct contact with imatinib (such as T315), most occur in residues quite distant from the sites of drug binding (Figure 1). Modeling studies show that the primary consequence of mutation at these distant residues is to restrict the conformational flexibility of the kinase domain such that it is incapable of achieving the shape required for optimal imatinib binding. Indeed, most mutations are predicted to generate a kinase domain that favors a conformation in which the activation loop is open rather than closed (2–4).
Fig. 1.
Structure of the BCR-ABL kinase domain and the location of several mutations that confer imatinib resistance in patients. (Modified from Ref. 3, with permission.)
An important corollary of this prediction is that inhibitors that bind the open conformation of the ABL kinase domain should retain activity against many of these “conformational” resistance mutants. A test of this hypothesis was quickly realized through studies of the ABL inhibitor dasatinib. At the time, the crystal structure of dasatinib bound to ABL had not been solved, but the different activity profile of dasatinib versus imatinib against other kinases (particularly SRC family kinases) suggested that the two compounds bound differently. Preclinical studies revealed that dasatinib remained active against all imatinib-resistant BCR-ABL mutants with the exception of the T315I gatekeeper mutation, which disrupts a critical hydrogen bond made with both drugs and also introduces a bulkier amino acid (isoleucine for threonine) into the ATP binding pocket (5). The clinical activity of dasatinib in imatinib-resistant CML patients was quickly shown in a phase I clinical trial and, as predicted from the preclinical work, efficacy was tightly linked to BCR-ABL mutation status (6). Initially approved for imatinib-resistant CML (with the exception of patients with the T315I mutation), dasatinib was subsequently found to be superior to imatinib as up-front therapy for CML (7, 8).
ANTIANDROGENS IN PROSTATE CANCER
For decades the standard of care for treatment of metastatic prostate cancer has been hormone therapy using drugs that decrease serum testosterone levels by blocking secretion of androgens by the testes (e.g., leuprolide) or competitive antagonists that block the binding of androgens to the androgen receptor (e.g., bicalutamide and flutamide). Both modalities, given alone or in combination, confer considerable clinical benefit in most patients but are not curative. The duration of treatment response is highly variable, ranging from months to years, and is inevitably followed by the growth of drug-resistant tumor cells, a phase of the disease that is now called castration-resistant prostate cancer (CRPC).
Although initially considered to be hormone independent, it is now clear that CRPC remains dependent on androgen receptor (AR) signaling (9). The clearest evidence for this first emerged from preclinical xenograft models which revealed that increased levels of androgen receptor expression are both necessary and sufficient to confer castration resistance (10). Furthermore, increased AR expression altered the response of prostate cancer cells to the AR antagonist bicalutamide, converting its activity from an antagonist to an agonist. Increased AR expression was also commonly observed in CRPC patient samples, often through AR gene amplification, and provided rationale to search for more potent inhibitors of AR pathway signaling.
Our group, in collaboration with synthetic chemist Michael Jung at UCLA, screened for such compounds using a cell-based assay in which a prostate tumor line was engineered to express increased levels of AR, thereby mimicking the situation in tumors from CRPC patients. As expected, these cells were resistant to current antiandrogens such as bicalutamide. Among the most compelling hits that emerged from this screen were derivatives of a previously reported compound called RU-59063 that has ∼100-fold higher binding affinity for the ligand binding domain of AR than bicalutamide. Although RU-59063 is a potent agonist, certain derivatives now had potent antagonist activity in AR overexpressing cells. Further chemical modifications resulted in a few orally bioavailable compounds with favorable pharmacokinetic properties that caused substantial tumor regressions in a xenograft model of CRPC (11, 12). A compound known as RD162' was selected for clinical development and renamed MDV3100.
MDV3100 was first evaluated in patients in a phase I clinical trial conducted at Memorial Sloan Kettering Cancer Center led by Howard Scher. Eligibility was restricted to men with metastatic CRPC who had failed multiple prior hormonal therapies. Remarkably, the first six patients all had substantial decreases in their serum prostate-specific antigen (PSA) levels, indicative of AR inhibition and potential tumor response. On the basis of this unprecedented early result, the trial was amended to allow rapid accrual of an additional 24 patients at each dose level once the safety of that dose was established in the first three patients. Consequently, 140 men had received MDV3100 within 18 months at doses ranging from 30 to 600 mg. More than half of the patients had reductions in serum PSA by greater than 50% that were sustained for at least 12 weeks, regardless of prior treatment with chemotherapy (13). On the basis of these encouraging findings as well as radiographic evidence of tumor responses, a phase 3 randomized trial was initiated in 2009 in 1199 men with chemotherapy refractory CRPC. In 2011, the trial was terminated early based on an interim analysis that revealed a significant survival advantage for MDV3100 treated patients, with a 37% reduction in risk of death (hazard ratio 0.63). US Food and Drug Administration (FDA) approval of MDV3100 is anticipated in 2012.
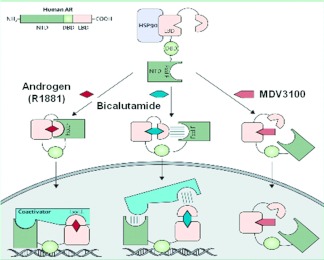
Although MDV3100 does bind to AR protein with greater affinity than bicalutamide, molecular studies of AR signaling in prostate cancer cells exposed to MDV3100 have revealed additional effects that may explain its superior antitumor activity (Figure 2) (14). In cells treated with bicalutamide, AR protein translocates to the nucleus and binds DNA at many of the same AR consensus binding sites where AR normally binds after stimulation with androgens. In contrast, approximately 70% of the pool of AR protein in MDV3100-treated cells remains in the cytoplasm. Furthermore, genome-wide analyses of AR DNA binding using ChIP-Seq technology reveal near-complete absence of AR protein bound to DNA. This striking property, which almost certainly disables signaling by AR, could serve as the basis for developing even more potent AR inhibitors.
Fig. 2.
Cartoon showing the effects of androgen, bicalutamide, or MDV3100 on androgen receptor localization and DNA binding. (Modified from Ref. 14, with permission.)
Although MDV3100 and a second drug called abiraterone that blocks residual testosterone production by the adrenal gland through inhibition of the enzyme CYP17 (15) are both important new therapies for prostate cancer, neither is curative. Initial studies were conducted in men with end-stage CRPC who had failed chemotherapy, but both drugs are likely to be used primarily in the pre-chemotherapy setting where their impact on survival may be even greater. It is also anticipated that these drugs could be used even earlier in prostate cancer treatment, before progression to CRPC. In this scenario, MDV3100 may be preferable to abiraterone because it does not require co-administration of prednisone to prevent endocrine side effects. Another next-generation antiandrogen ARN-509, currently in phase 2 clinical trials, has also shown favorable safety and efficacy (16).
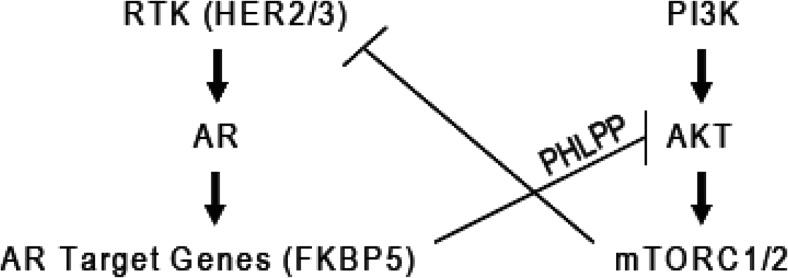
In parallel to efforts to move these new drugs earlier in treatment, various AR pathway inhibitor combination strategies are under consideration, based on preclinical and clinical studies of resistance to single-agent therapy. Data from xenografts and patients treated with abiraterone have shown that increased AR activity is observed in drug-resistant tumors, as measured by AR mRNA levels and nuclear AR protein localization (17, 18). These findings provide a strong rationale for combined CYP17 and AR inhibition. Another compelling combination regimen is based on preclinical studies of MDV3100 that have revealed relative resistance in prostate tumors driven by loss of the PTEN tumor suppressor gene (19). PTEN regulates the PI3K signaling pathway; therefore, PI3K pathway inhibitors using drugs targeting PI3K, AKT, or mTOR are under intense investigation. PTEN-negative prostate cancers were found to have reduced AR pathway signaling as a consequence of negative feedback of sustained PI3K activation on HER family kinases that normally enhance AKT signaling. Consequently, PI3K inhibitors “activate” AR signaling due to relief of negative feedback. Similarly, AR inhibition in PTEN-negative tumors “activates” PI3K signaling through reciprocal feedback (Figure 3). However, combined AR plus PI3K inhibition produced near-complete responses in PTEN-negative tumors, whereas each single-agent treatment had only modest effects. Clinical trials evaluating both combination regimens (CYP17/AR and PI3K/AR) are just getting underway.
Fig. 3.
Reciprocal negative feedback between PI3K and androgen receptor signaling pathways in prostate cancer cells with PTEN mutation. (Reprinted from Ref. 19, with permission.)
EGFR INHIBITORS IN LUNG CANCER
The discovery of next-generation inhibitors of BCR-ABL in CML or AR in prostate cancer was driven by a precise understanding of acquired resistance to earlier generation compounds. Similar investigations are underway in lung cancers with EGFR mutations that respond to EGFR inhibitors such as erlotinib but subsequently relapse. Acquired resistance to EGFR inhibitors occurs through secondary mutations in EGFR analogous to the T315I gatekeeper mutation in CML, as well as other mechanisms (20–22). However, a closer examination of initial response to EGFR inhibitors reveals considerable heterogeneity among patients with identical EGFR mutations, suggesting that additional factors beyond the defining driver mutation impact treatment response. (This phenomenon is not unique to EGFR-mutant lung cancer. A similar argument applies to other kinase-driven cancers such as ALK fusion positive lung cancer, BRAF mutant melanoma and KIT mutant gastrointestinal stromal tumors.) One hypothesis to explain this heterogeneity of response is that genetic modifiers regulate the degree to which tumor cells are EGFR dependent. To search for such modifiers, we conducted an RNA interference screen to identify genes that enhance cell death induced by EGFR inhibition when knocked down (23).
In considering which cell line models to evaluate in the screen, we intentionally selected lung cancer lines with EGFR mutation but suboptimal response to EGFR inhibitors, mimicking the clinical situation of incomplete tumor response in a patient whose tumor otherwise seemed “poised” to respond. After screening shRNAs representing ∼2500 genes involved in cancer and signaling, we identified 36 screening hits, 18 of which targeted genes in the NFkB signaling pathway. Validation studies confirmed that activation of NFkB signaling conferred resistance to EGFR inhibitors in EGFR dependent tumor models and, conversely, that NFkB inhibition enhanced sensitivity to EGFR inhibitors. The clinical relevance of these data is suggested by evidence that patients with EGFR mutant lung cancer whose tumors have elevated IkB expression (which correlates with low levels of NFkB activity) have superior clinical outcomes when treated with EGFR inhibitors. Collectively, these findings establish that NFkB pathway activation can promote resistant to EGFR kinase inhibitors, that IkB expression may be a biomarker of treatment response to EGFR inhibitors, and that combined EGFR/NFkB therapy could enhance response rates and extend response duration.
CONCLUSION
Targeted therapies are rapidly changing the face of cancer therapy. Although the current practice of developing these drugs as single agents results in meaningful responses in many patients, these responses are usually temporary. There is widespread consensus that combination therapies are needed, but there is no clear direction as to how to get there. The fact that targeted therapies are specifically designed to target a cancer-relevant protein provides an opportunity to unravel the molecular basis of response and resistance in a very precise, mechanistic manner. Here I have showcased three examples from three different cancer types in which studies of resistance have yielded insights that resulted in new drug development and combination therapy regimens. There is no reason to assume that these examples are in any way unique. Broader application of these principles to all targeted cancer therapies should hasten the pace of new drug development and optimal deployment of rational, effective combinations.
Footnotes
Potential Conflicts of Interest: The author is a co-inventor of MDV 3100 and may be entitled to royalty payments.
DISCUSSION
Sawyers, New York: I'm happy to answer any questions if there are any. Hey Marty.
Blaser, New York: Charles, really wonderful presentation. As you know, I am a microbiologist, and sometimes we treat infections with agents that are bacteriostatic. They are not bacteriocidal and the patients recover because we are buying time, we are decreasing the replication rate of the agent and we are buying time until the host response sets in. So the question is, in a way, what's the analogy there with cancer? And I'll just make the possibility that maybe drugs that are working on NF-kB aren't necessarily working just on the tumor but the inflammatory cells, which are part of the substrate. Some tumors are suppressed by inflammation and some tumors are facilitated by it.
Sawyers, New York: Absolutely. So the analogy would be, as you know, the large cancer immunotherapy community. The enthusiasm for this has been resurrected at a very high level in the last year with the approval of ipilimumab, an antibody that enhances the recognition of the tumor cell by the immune system by blocking a target called CTLA-4 on T cells. The thinking is that a kinase inhibitor shrinks tumor volume down to a smaller level, then the small population that remains could be eliminated by the patient's immune system. The place where this experiment is going to play out first is in melanoma where there is a driver kinase called BRAF. The kinase inhibitor has just been approved. Ipilimumab has clinical activity in that same population so the question is, what is the activity of the combination? As far as the NF-kB inhibitors and their activity perhaps being through the immune system, I think it's too early to know. We saw in-vitro activity where there was no immune system in our model system as well as in-vivo. I don't know if the in-vivo activity was accentuated by a host effect. I think you could also make the argument that that might be a problem with NF-kB therapy where you may prevent the immune system from clearing remaining tumor cells if the drugs are too immune suppressive. So we will have to see.
Rosenblatt, New Jersey: I wanted to go back to the Gleevec story. So you said in the beginning the chemists were reluctant to go for kinase targets because they were afraid they would hit other kinases and that Gleevec exquisitely matched just a limited number of conformations of the enzyme and then when resistance emerged, you had to find a compound that actually reacted with more conformations, dasatinib. I am wondering if, as a result of that, did the new compound lose specificity and spill onto other kinases?
Sawyers, New York: Absolutely, great point. Dasatinib was looked at a bit askance originally because it has “one of the dirtier profiles” when you look at its activity across a panel of kinases. It inhibits essentially all Src family kinases. Of course the concern going into the phase I trial was that it would be a much more toxic compound. But, in fact, it is quite well-tolerated. There is now a marketing battle between two separate pharmaceutical companies competing with each other as to which drug is safer. Both compounds, in my view, are quite, quite safe and certainly are far superior to the chemotherapeutic drugs or interferon that have been used previously in this setting. There are tweaks to the schedule to give dasatinib as an intermittent pulse rather than continuous exposure that have improved its safety without compromising efficacy.
Nathan, Boston: Well first of all I want to just congratulate and thank you for the terrific work you've done over the years. I've watched it very, very carefully and it's really outstanding. I just want to call attention to this group to a wonderful book by Mel Greaves on cancer as an evolutionary problem and this particularly relates to prostate and lung cancer. Mel has very carefully studied childhood leukemia, and the chromosomes in childhood leukemia look great most of the time. There may be one translocation that's driving the system but it doesn't look anything like the bomb that goes off in the nucleus in prostate and lung cancer. These cells are a mess and they are mutating like mad and there is no such thing as a cancer stem cell. Every cell is a stem cell. So to me, the problem with targeted therapy is that every clone is a different one and to get them all, as you point out, is going to require a lot of combination chemotherapy and we may not be able to even do that. I mean that's the gloomy side of this, that the cell is smarter than we are and it keeps evading these drugs and I wonder whether we are going to see, in this wonderful example you gave with prostate cancer, that there will be an escape from a clone that never really was sensitive to the drug in the first place.
Sawyers, New York: I have no doubt tumors will escape eventually but David, in prostate cancer, because the age of onset is late in most men, the goal could be to extend life just 10 more years and you've really had a huge public health impact. That said, you know that I don't like the compromise of living with cancer versus killing it. I am more optimistic than you that we could eliminate more clones if we give potent combination therapy up front and more aggressively. Time will tell. Perhaps the most optimistic thing I can tell you is that the current molecular profiling technologies allow us to track how the tumor responds to treatment. If we get biopsies, we can anticipate much sooner than we have in the past.
Alexander, Atlanta: I also congratulate you and thank you for a brilliant series of studies. I also have a question that's related, I think, to the one that Dr. Nathan was asking, about the bomb and the mess that the nucleus is at the time. This disease, or most of these diseases start at some point with a mutation and that probably is almost always in a preclinical setting. Then as you showed, multiple mutations in the various pathways accumulate. It's as if mutation begets mutation and, if you are in an environment at the beginning that permitted or enabled the mutation, then it just keeps getting worse. My question is, what is known about the characteristics about that general environment that are enabling the whole process?
Sawyers, New York: The mutation rate in cancer is dependent upon two things, the number of cell doublings and the efficiency of DNA repair. Just by probability, the error rate of DNA polymerase in normal cells leads to more mutations if there are more cell doublings. But one of the key insights came more than a decade ago that cancer cells can have mutations in the repair machinery, particularly in colon cancer. Significant fractions of colon cancers have mutations in DNA repair machinery and therefore if you've got one of those mutations, you're going to accumulate more mutations at a much faster rate.
Benz, Boston: First of all Charles, I didn't become editor of the New England Journal until 2002.
Sawyers, New York: I didn't mean to imply that you delayed publication of our first paper.
Benz, Boston: Your 2006 paper got in real fast.
Sawyers, New York: It certainly did.
Benz, Boston: I just want to pick up on a couple comments. Something I'm pretty sure you didn't mean to imply about Gleevec is its specificity for just BCR ABL kinase. Gleevec also targets a range of kinases and that's actually been put to good effect for the treatment of things like sarcoma, which is driven by a related kinase in that family. And making that point, another reason is that one of the things that makes development of these drugs difficult sometimes is seemingly small population of patients, only 10% of lung cancer patients instead of all of them, that can diminish enthusiasm for developing the drug because the market might not be big enough and, in fact, many of these drugs, the BRAF inhibitor being another, is finding uses in other forms of cancer that either are driven by the same family of mutations or are driven by related kinases. So this actually is changing the way to look at classifications of tumor from a therapeutic point of view and that is some cause for optimism in using these drugs. Now the other comment. David Nathan is usually trying to cheer me up. I am going to try to cheer him up for a change. I think you pointed out that many of these drugs that are in development are for patients with advanced cancers who have tumor burdens of literally trillions of cells and many of those with resistant mutations. The movement of these medications has to be in earlier tumors with better markers of who should get them. And you reduce the chances, first of having as many of those mutations and second, taking a lot longer for resistant clone to grow out.
Oates, Nashville: Charles I have one question. In the lung adenocarcinoma where the emergence of the NF-kB pathway is a second promoter of the tumor, do you think it's entirely mutations that drive the development of that traditional pathway or are there epigenetic changes that might occur in the context of the original mutation?
Sawyers, New York: I don't think it is only mutational events. Mutations in NF-kB family members have been looked for in lung cancer, maybe not to completion, but they haven't been found. But they have emerged in other tumor types such as myeloma. So I think it will be different mechanisms of NF-kB activation but yet the same final dependence on the pathway.
REFERENCES
- 1.Druker BJ, Guilhot F, O'Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–17. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 2.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 3.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–25. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 4.Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci U S A. 2005;102:3395–400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 6.Talpaz M, Shah NP, Kantarjian H, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–41. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 7.Kantarjian H, Shah NP, Hochhaus A, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362:2260–70. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- 8.Kantarjian HM, Shah NP, Cortes JE, et al. Dasatinib or imatinib in newly diagnosed chronic-phase chronic myeloid leukemia: 2-year follow-up from a randomized phase 3 trial (DASISION) Blood. 2012;119:1123–9. doi: 10.1182/blood-2011-08-376087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–61. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 10.Chen CD, Welsbie DS, Tran C, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–9. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 11.Tran C, Ouk S, Clegg NJ, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung ME, Ouk S, Yoo D, et al. Structure-activity relationship for thiohydantoin androgen receptor antagonists for castration-resistant prostate cancer (CRPC) J Med Chem. 2010;53:2779–96. doi: 10.1021/jm901488g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scher HI, Beer TM, Higano CS, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1-2 study. Lancet. 2010;375:1437–46. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Clegg NJ, Scher HI. Anti-androgens and androgen-depleting therapies in prostate cancer: new agents for an established target. Lancet Oncol. 2009;10:981–91. doi: 10.1016/S1470-2045(09)70229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clegg NJ, Wongvipat J, Tran C, et al. ARN-509: a novel anti-androgen for prostate cancer treatment. Cancer Res. 2012;72:1494–1503. doi: 10.1158/0008-5472.CAN-11-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mostaghel EA, Marck BT, Plymate SR, et al. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clin Cancer Res. 2011;17:5913–25. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Efstathiou E, Titus M, Tsavachidou D, et al. Effects of abiraterone acetate on androgen signaling in castrate-resistant prostate cancer in bone. J Clin Oncol. 2012;30:637–43. doi: 10.1200/JCO.2010.33.7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver BS, Chapinski C, Wongvipat J, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 22.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bivona TG, Hieronymus H, Parker J, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–6. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]