Abstract
This study investigated the use of direct gradient analysis of bacterial 16S pyrosequencing surveys to identify relevant bacterial community signals in the midst of a "noisy" background, and to facilitate hypothesis-testing both within and beyond the realm of ecological surveys. The results, utilizing 3 different real world data sets, demonstrate the utility of adding direct gradient analysis to any analysis that draws conclusions from indirect methods such as Principal Component Analysis (PCA) and Principal Coordinates Analysis (PCoA). Direct gradient analysis produces testable models, and can identify significant patterns in the midst of noisy data. Additionally, we demonstrate that direct gradient analysis can be used with other kinds of multivariate data sets, such as flow cytometric data, to identify differentially expressed populations. The results of this study demonstrate the utility of direct gradient analysis in microbial ecology and in other areas of research where large multivariate data sets are involved.
The study of the bacterial microbiome focuses on the bacteria that normally live on and within other organisms. For many years, microbial ecology was limited to the study of those organisms that could be easily grown in the laboratory; however, the bacterial 16S ribosomal RNA has been identified as a surrogate marker that enables the study of both culturable and unculturable microorganisms1,2. In recent years, the combination of 16S surveys with high-throughput, next-generation 454-pyrosequencing has revolutionized the field of microbial ecology3. 16S surveys are uniquely positioned to take advantage of many well-established ecological analysis techniques for assessing differences between communities and for determining what “environmental” effects might explain those differences1,4,5,6,7. Many recent studies have guided how one can analyze these data, both through a new application of existing ecological methods5,8, as well as via the invention of new methods9,10,11,12,13. However, the majority of publications involving 16S surveys do not extend beyond exploratory methods of analysis5. The common practice is to create a low dimensional representation of the bacterial community data (commonly referred to as an ordination) and to colorize data points based on a hypothesis or some other grouping variable.
Ordination is a method of displaying large multivariate data sets through dimensional reduction. The reduction falls into two classes: distance-based and eigenvector-based. In either case, the visual clustering of points is the primary method by which data of this type are usually displayed and interpreted. Of the distance-based methods, Non-Metric Multi-Dimensional Scaling (NMDS) is considered to be the most robust14. In practice, NMDS tries to maintain the distance between each point while minimizing the stress on the system. The only drawback of NMDS plots is that the individual points tend to spread out, making the visual clustering more difficult. More commonly used are eigenvector-based methods, such as principal component analysis (PCA); a variation of PCA that utilize non-Euclidean distances (frequently, ecologically-relevant distances) called principal coordinates analysis (PCoA); and correspondence analysis (CA), which performs a single-value decomposition (svd) on the fitted values of the chi-square transformed data matrix15. Irrespective of the method employed, the components of variation are ordered such that, when graphed, the component that contributed most to the variation will be graphed along the first axis, the next most along the second axis and so on for n-1 axes (where n = number of samples). The practical result is that huge numbers of variables can rapidly be reduced to a plot with 2–3 informative axes permitting ready visualization of the complexity of an ecological community.
The difficulty with this methodology is that the noise inherent in large biological data sets frequently limits the ability to distinguish real differences. Frequently, biology is inferred from ordinations by colorizing the points on the plot (e.g. by treatment), even though the hypothesis in question was not involved in the calculation of the ordination. This approach is known as indirect gradient analysis15. Its advantage is that all the data is considered, but indirect gradient analysis carries the disadvantage that the environment is rarely so controlled that all of the variation present in the system can be explained by the experimental question being asked. To get around this problem, it is very common to try multiple different metrics (for PCoA or NMDS) and methods of ordination and to choose what looks best for the experimental question being asked. By contrast, direct gradient analysis directly examines the hypothesis by finding correlations between the data and the hypothesis prior to variance partitioning. In direct gradient analysis, the orthogonal vectors of the eigen analysis are required to be linear combinations of the explanatory variable (in this case, the biological question being asked). Therefore, the variance displayed in the ordination is linearly related to the explanatory variables15. In classical ecology, explanatory variables usually include environmental factors such as soil pH or moisture content. In many areas of microbial ecology relevant to human health, the “environment” is the host; and perhaps for this reason, direct gradient analysis has rarely been used. In the current study, we detail how direct gradient analysis can be used, in combination with high-resolution 16S survey data or indeed other large multivariate data sets, to uncover specific responses that correlate with biological responses.
Results
The effect of data resolution on population detection
The first objective of this study was to determine whether analysis of data at higher effective taxonomic resolution provides greater insight into sample grouping, or alternatively if the increase in variation renders sample groups indistinguishable. To investigate this question, we used a 16S data set obtained from clinical lung tissue samples removed at the time of lung transplantation of five patients with severe emphysema16. Data were binned into phylotyped OTUs (limited to a genus level of taxonomic classification), or into OTUs binned at a 3% cutoff (roughly a species level of taxonomic classification). The number of OTUs associated with phylotyped data or 3% cutoff data were 109 and 143, respectively, after OTU selection criteria were applied (see Methods and Materials). Next, ordinations were generated as described in “Methods and Materials,” with individual data points colored according to the experimental question under consideration.
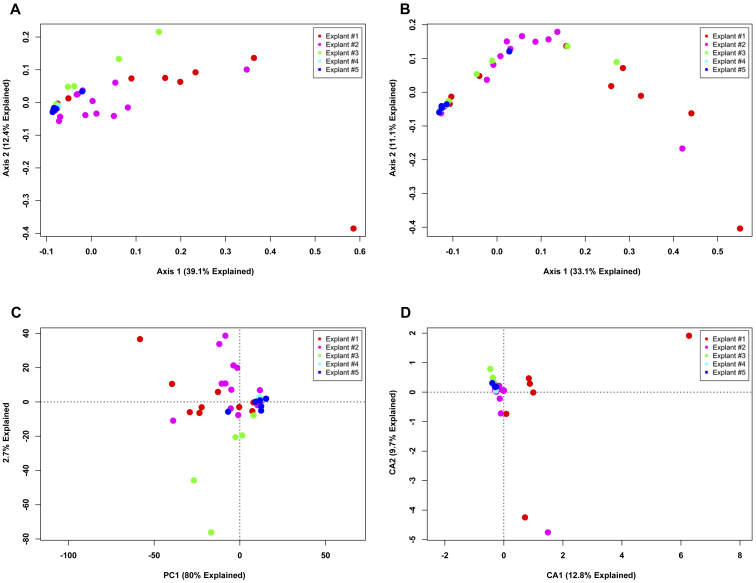
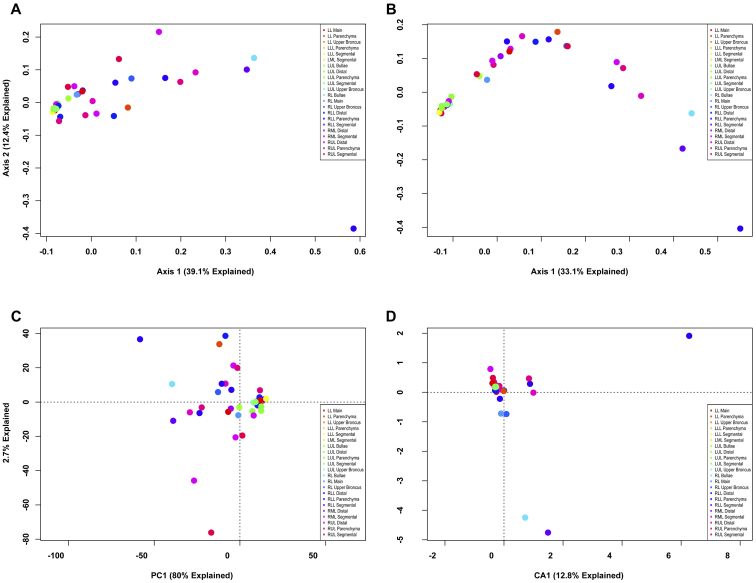
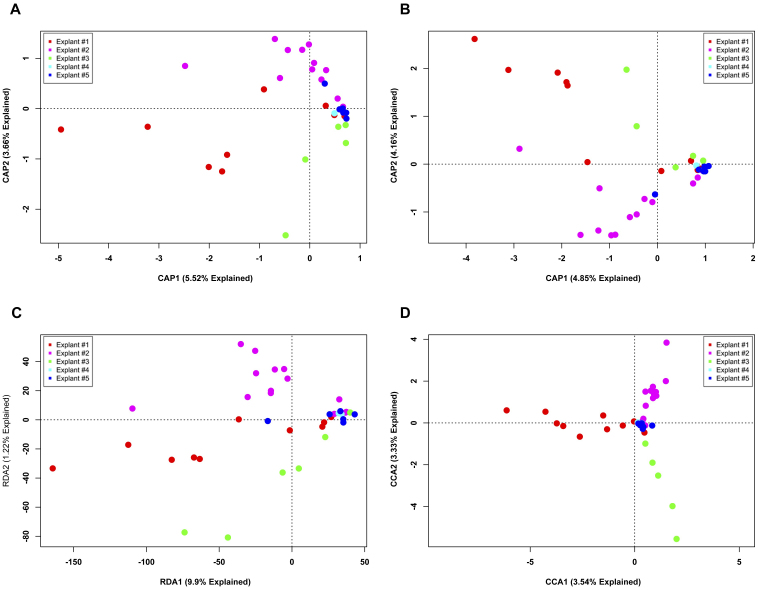
We carried out ordination of phylotype data using four separate analytical metrics (Bray-Curtis, Jaccard, PCA and CA) to investigate two experimental questions. First, are there differences between explants (Fig. 1)? Second, are there bacterial communities that are associated with different regions of the lungs (Fig. 2)? The ordinations demonstrated that, regardless of analytical method, a large proportion of the variation could be explained in the first two axes of each PCoA or PCA ordination. However, despite explaining a large portion of the variation between samples, there is no visible clustering either by explant (Fig. 1) or by anatomic location (Fig. 2), regardless of the method employed. The extensive overlap of samples utilizing the Jaccard metric (Figs. 1B, 2B) indicates that there is extensive shared membership between individual samples. However, these membership differences do not cluster by explant (Fig. 1B) or by location within the lung (Fig. 2B). Some samples do stand out as distinct from the other samples, some based on differences in membership (Figs. 1B and 2B), others based on the differences in membership and abundance (Figs. 1A, 2A, 1C and 2C); but, no identifiable pattern exists. Collectively, these data indicate that at a phylotype resolution there are no bacterial community differences between individual subjects with end-stage emphysema and that no bacterial communities are specific to particular airways, even within a given individual.
Figure 1. Unconstrained ordination of lung explant OTUs at phylotype resolution by subject.
The bacterial communities of human lung explants were binned at phylotype resolution and were ordinated using PCoA based on (A) Bray-Curtis distance, (B) Jaccard distance, (C) PCA and (D) CA. Individual samples were colored based on the explant from which they were obtained, regardless of anatomic location.
Figure 2. Unconstrained ordination of lung explant OTUs at phylotype resolution by anatomic location.
The bacterial communities of human lung explants were binned at phylotype resolution and ordinated using PCoA based on (A) Bray-Curtis distance, (B) Jaccard distance, (C) PCA and (D) CA. Individual samples were colored based on the location within the lung from which the sample was obtained, regardless of subject.
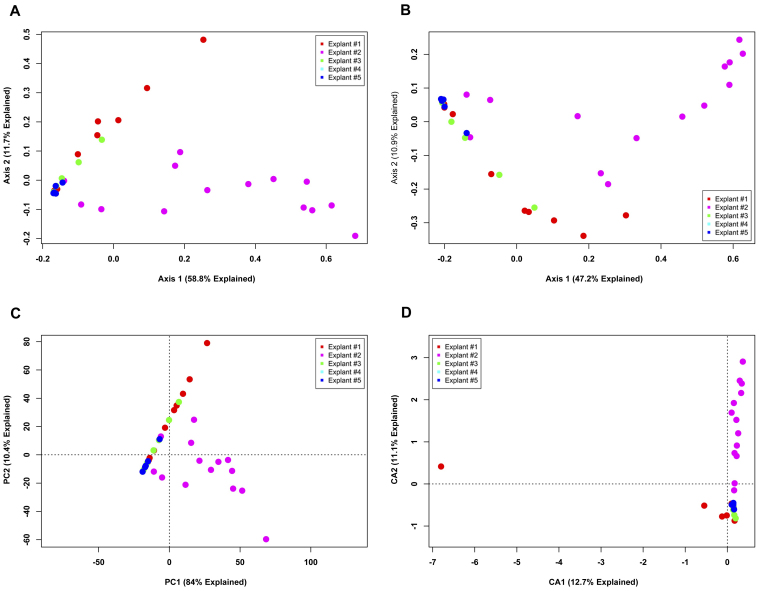
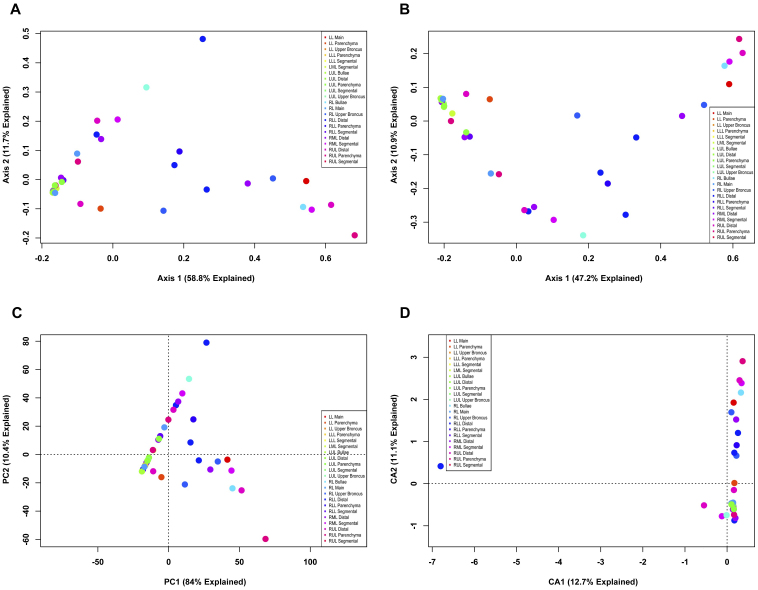
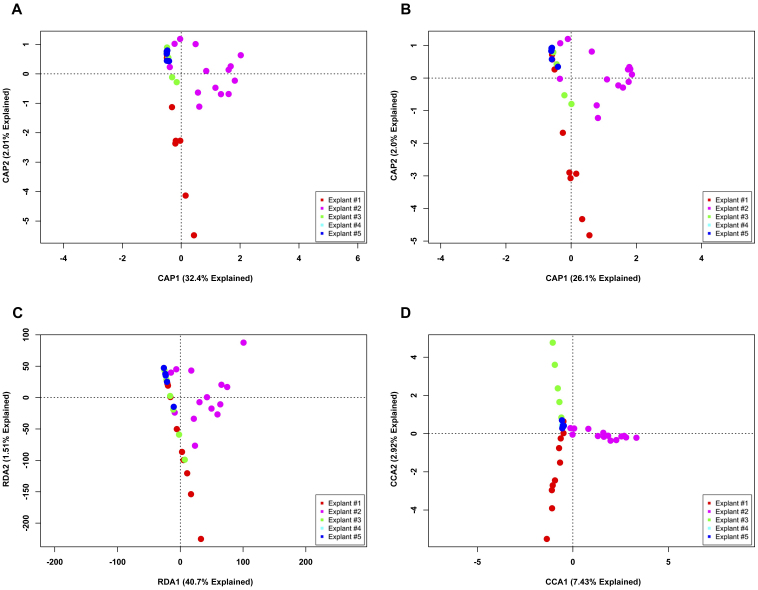
Next, data were binned using a species level (3% OTU) cutoff and analyzed to assess the same two questions (Figs. 3 and 4, respectively). As was seen in the phylotype ordinations, a large proportion of variation is explainable in the first two axes. However, unlike the phylotype data, there are readily apparent differences in sample clustering. Ordinations investigating differences between explants (Fig. 3) demonstrate differences between explant #2 and the other explants. The Bray-Curtis and Jaccard PCoA plots (Figs. 3A and B) show a strong shift along the first axis for explant #2. These metrics are complementary, in that the shift in the Jaccard PCoA indicates that the membership is different for explant #2 and the other explants, whereas a similar shift in the Bray-Curtis PCoA indicates that the OTU that is different is significantly abundant. The PCA plot (Fig. 3C) most clearly separates explant #2 from the other explants, but merges the other explants as indistinguishable. The CA plot (Fig. 3D) provides the most compact groupings with the least intra-sample dispersion; however, as in the PCA plot, the majority of the remaining samples are compacted into a small area in ordination space. An examination of the OTU tables reveals that the dominant OTU of explant #2, using a 3% OTU cutoff, is all but unique to that human subject. However, the sequences that define this OTU belong to the dominant OTU found in the other samples in the data table from the phylotype cutoff (data not shown).
Figure 3. Unconstrained ordination of lung explant OTUs at a 3% OTU cutoff by subject.
The bacterial communities of human lung explants were binned at a 3% OTU cutoff to produce high-resolution data. The data were ordinated using PCoA based on (A) Bray-Curtis distance, (B) Jaccard distance, (C) PCA and (D) CA. Individual samples were colored based on the explant from which they were obtained, regardless of anatomic location.
Figure 4. Unconstrained ordination of lung explant OTUs at 3% OTU cutoff by anatomic location.
The bacterial communities of human lung explants were binned at a 3% OTU cutoff to produce high-resolution data. The data were ordinated using PCoA based on (A) Bray-Curtis distance, (B) Jaccard distance, (C) PCA and (D) CA. Individual samples were colored based on the anatomic location within the lung from which the sample was obtained, regardless of subject.
When data points are colored based on their anatomic location (Fig. 4), there is no difference in point clustering compared to what was seen in figure 2. These data indicate that differences in the microbial communities of the lung can be seen between emphysema patients at a species-level resolution. However, no evidence was found that the microbial communities of the lung are specific to particular anatomic regions of the lung.
Together, these data demonstrate that increasing the effective taxonomic resolution of the data (and the total variation of the system) does not cause the sample groups to become indistinguishable, but rather increases the ability to differentiate between groups.
The effect of constraints on population detection
The next objective was to test directly the hypotheses investigated in the previous section using the method known as direct gradient analysis. The procedures performed in figures 1 through 4 are known as indirect, or unconstrained, analysis because all of the variance present in the system is displayed. Oftentimes the causes of the variance are not known; however, one can hypothesize what they might be and attempt to constrain the variation within those variables that are highly correlated with the hypothesis. If the hypotheses, also called explanatory variables, are good, then populations should visibly cluster, or, if the explanatory variable is continuous, populations should demonstrate a clear gradient along which a linear relationship exists. This method is known as constrained or direct gradient analysis17.
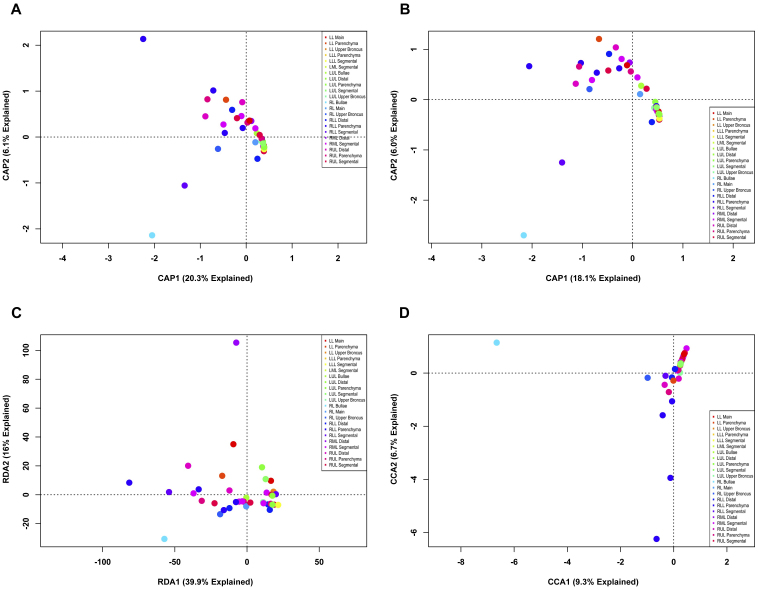
To test whether direct gradient analysis provides better ability to detect differences in microbial populations, new ordinations were generated for both the phylotype and 3% OTU explant data sets by constraining the data to the explant from which the samples originated. This approach directly addresses the question of whether similar bacterial communities inhabit the explanted lungs of patients with end-stage emphysema (and hence, whether those same communities were present before surgery). At the phylotype level, neither ecological metrics nor redundancy analysis (RDA) (Figs. 5A–C) were effective at separating the explants, although each method produced a similar distribution of points. Interestingly, canonical correspondence analysis (CCA) ordination (Fig. 5D) produced three distinct groupings which, by the standard method of visual assessment, would have been considered different; however, these differences were not found to be significant (p > 0.05) when using an ANOVA-like permutation test for testing the significance of constraints (anova.cca())18.
Figure 5. Direct gradient analysis of lung explant OTUs at phylotype resolution constrained by subject.
The bacterial communities of human lung explants were binned at a phylotype resolution and constrained ordinations were constructed using the explant from which the sample originated as the constraint. Ordinations were CAP (constrained analysis of principle coordinates) based on (A) Bray-Curtis distance, or (B) Jaccard distance; (C) RDA (the constrained form of PCA); (D) or CCA (the constrained form of CA). Individual samples were colored based on the explant from which they were obtained.
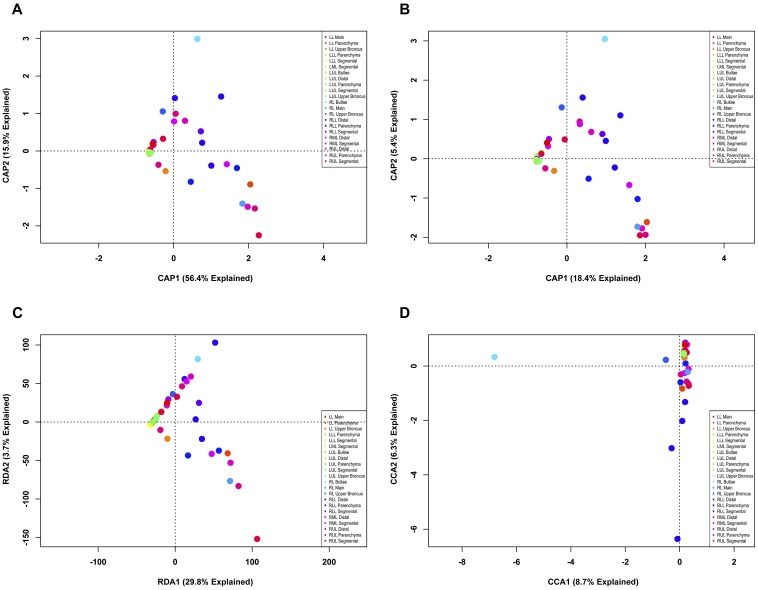
Next, we analyzed the high-resolution (OTU) explant data set in a similar fashion, which demonstrated that when the data are constrained by explant, sample clustering becomes apparent. Using the ecological metrics and RDA (Figs. 6A–C), explant #2 separates clearly from the other explants. The CCA ordination (Fig. 6D) again separated explants 1, 2, and 3 from 4 and 5 in a manner that is similar to what was seen in figure 5D. However, using higher resolution data sets results in a model that is significantly different (p < 0.005) from the null hypothesis that all end-stage lungs from humans with emphysema contain similar bacterial communities; significant differences were present along the first two axes {CCA1 (p < 0.005); CCA2 (p = 0.0485)}. Similar tests performed on the models from figures 6A–6C found them all to be significant (p < 0.005). However, under these models, significant differences could only be found along the first axis. This indicates that the bacterial community of explant #2 was significantly different from that of the other explants, and was so under all the analytic models. The CCA model also finds significant differences between explant #1 and explant #3 (p < 0.05; One-way ANOVA of CCA2 site scores, Tukey’s Multiple Comparisons Test). Together, these data yield two conclusions. First, consistent with our previous analyses of these samples16, the lung bacterial communities of these five subjects did not differ significantly at a phylotype level. Second, by contrast, constrained analysis of the lung bacterial communities at the operational definition of a bacterial species discloses significant differences between human subjects.
Figure 6. Direct gradient analysis of lung explant OTUs at 3% OTU cutoff constrained by subject.
The bacterial communities of COPD lung explants were binned at high-resolution, and constrained ordinations were constructed using the explant from which the sample originated at the constraint. Ordinations were CAP ordinations based on (A) Bray-Curtis distance, or (B) Jaccard distance; (C) RDA; (D) CCA. Individual samples were colored based on the explant from which they were obtained.
We have previously shown that there are marked differences in bacterial communities within a single lung explant16. We next examined both phylotype and 3% OTU data, constrained by anatomic location, to examine directly in a larger data set whether different bacterial communities consistently reside in particular regions of the lung. This possibility is plausible, given the known predilection of aspirated secretions to deposit in certain lung lobes in a gravity-dependent fashion, and is supported at the level of total variation by the current data set (Figs. 2 & 4). Similar to what was seen before (Fig. 2), however, constraining either the phylotype data (Fig. 7) or the 3% OTU data (Fig. 8) by anatomic location failed to identify any significant difference (p > 0.05) by any of the methods. Thus, within these five lung explants, there is no correlation between the region of the lung and the bacterial community present.
Figure 7. Direct gradient analysis of lung explant OTUs at phylotype resolution constrained by anatomic location.
The bacterial communities of COPD lung explants were binned at a phylotype resolution and constrained ordinations were constructed based on the anatomic lung location from which the sample originated. Ordinations were CAP ordinations based on (A) Bray-Curtis distance, or (B) Jaccard distance; (C) RDA; (D) CCA. Individual samples were colored based on the location in the lung from which they were obtained.
Figure 8. Direct gradient analysis of human lung explant OTUs at high-resolution constrained by anatomic location.
The bacterial communities of COPD lung explants were binned at high-resolution, and constrained ordinations were constructed using the location in the lung from which the sample originated at the constraint. Ordinations were CAP ordinations based on (A) Bray-Curtis distance, or (B) Jaccard distance; (C) RDA; (D) CCA. Individual samples were colored based on the anatomic location in the lung from which they were obtained.
It can be seen that by constraining data variation based on a specific hypothesis, direct gradient analysis provides an effective method of identifying bacterial communities that correlate with that hypothesis. Similar to the unconstrained analysis, higher-resolution data produced better results. However, relative to unconstrained analyses, constrained analyses enjoys two important advantages: (1) they are much less sensitive than unconstrained ordinations to the metric used; and (2) they permit apparent differences between communities to be tested for statistical significance.
Using constraints to detect biological differences
One useful aspect of a constrained methodology for analyzing bacterial community data is that, by focusing on bacterial communities that correlate with biological responses, it should be possible to detect correlated community changes that might be hidden by systemic noise. To test this possibility, we chose a more controlled data set with known outcomes. Samples were taken from a model of allergic airway responses resulting from the co-administration of cefoperazone and Candida albicans, that is dependent on changes in the gut microbiome. It has been well established that mice treated with both cefoperazone and C. albicans will develop allergic-type airway responses, whereas mice treated with either cefoperazone alone or C. albicans alone, will not. Quantitative plating of cecal contents has previously indicated that while the microbiome of cefoperazone treated mice is different from untreated, it is very different from the dually treated. Data for this analysis was a high-resolution 16S data set (3% OTU cutoff) of cecum samples from day 7 mice that were either untreated, treated with cefoperazone, given a gavage of C. albicans, or co-administered cefoperazone and C. albicans. Over the course of a year, three separate experiments were performed. One confounding factor was that during that time, the microbiota of the mice within the animal care facility drifted. This drift resulted in “noisy” data where differences in the microbiome did not seem to match the biologic readout, i.e. allergic airway responses.
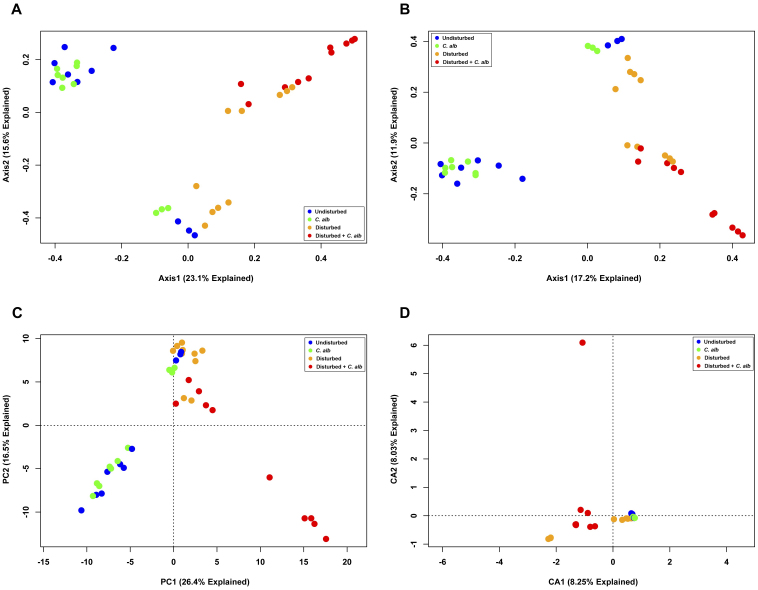
We first created unconstrained ordinations to examine the total variation present in the system and to determine the extent of the drift in the bacterial community, displayed by treatment group (Fig. 9). Clear drift in the total bacterial population was seen over the course of a year, in that even the bacterial community of the untreated mice displayed demonstrable differences over time (Fig. 9A–9C). The Jaccard PCoA (Fig. 9B) indicates that certain OTUs were present in some experiments but not others. The CA ordination again demonstrated itself to be vulnerable to outliers (Fig. 9D), making differentiation of treatment groups difficult. Visual assessment of these figures demonstrates extensive overlap between the bacterial communities of the different treatment regimes. Were this finding accepted without further analysis, this set of experiments would argue against published correlations4,19,20.
Figure 9. Indirect gradient analysis of cecal bacterial communities of mice undergoing gut microbiome modification.
Bacterial OTUs were binned at a 3% OTU cutoff and ordinations were plotted based on (A, B) PCoA using (A) Bray-Curtis distance or (B) Jaccard distance; (C) PCA ; and (D) CA. Individual samples were colored based on the treatment regime the mice received.
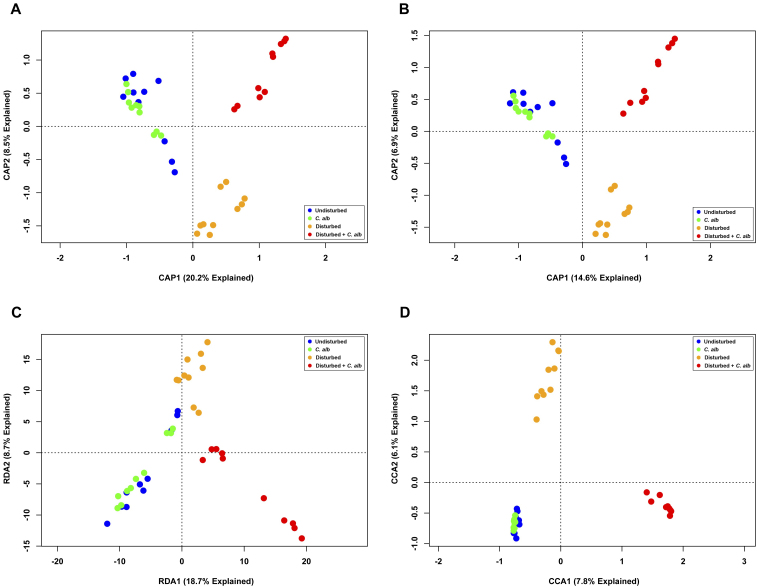
Accordingly, we tested the hypothesis that not all of the differences in the microbiota were associated with treatments. To that end, a direct gradient analysis was performed that constrained the data to that which correlated with treatments (Fig. 10). The CAP plots based on the ecological metrics (Figs. 10A–B) demonstrate clear separation between the bacterial communities of mice in three of the treatment groups (untreated, treated with cefoperazone, or co-administered cefoperazone and C. albicans), which perfectly matches previous observations. The bacterial communities of mice receiving C. albicans alone were indistinguishable from untreated, also matching earlier results. The RDA model (Fig. 10C) displayed similar trends; however, because of sample spread, this method appears less optimal. The CCA model (Fig. 10D) again provided very clear visual clustering with the maximal distance between samples. Regardless of these visual differences, each model proved to be statistically significant (p < 0.005), with significant differences along both the first and second axes (p < 0.005). These results demonstrate how constraining data can uncover significant correlations between bacterial communities and hypotheses of interest that would otherwise be masked by systemic noise.
Figure 10. Direct gradient analysis of the cecal bacterial communities of mice constrained by treatment.
Bacterial OTUs were binned at a 3% OTU cutoff and the data were constrained by treatment regime. Constrained ordinations were plotted based on (A, B) PCoA using (A) Bray-Curtis distance or (B) Jaccard distance; (C) PCA; and (D) CA Individual samples were colored based on the treatment regime the mice received.
Using constraints to differentially detect leukocyte populations
The final objective was to investigate the application of the methodology of direct gradient analysis outside the realm of ecology. In theory, direct gradient analysis should be possible on any large multivariate data set that is structured like a community matrix. We chose flow cytometric data to achieve this end. Flow cytometry is a method by which the expression of proteins on, or in, immune cells (leukocytes) can be assessed. The patterns of expression of various cell surface markers on a cell allow the identification of particular cell subsets and further the understanding of the immune response taking place. Such basic immunophenotyping can be extended by identifying leukocyte subsets that are expressed under one condition, but not another. Flow cytometric data sets are large multivariate data sets that make them ideal candidates for both indirect and direct gradient analysis.
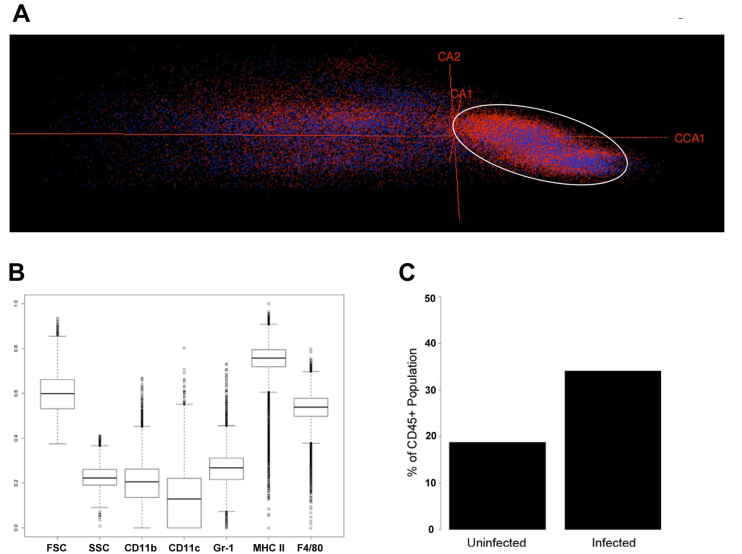
To determine if direct gradient analysis can differentially identify leukocyte populations in flow cytometric data, we generated a data set containing multi-color stained interstitial epithelial leukocytes (IEL) from the ceca of mice that had either been left untreated or had been infected with C. difficile for 42 hours (Fig. 11). A constrained correspondence analysis constraining the combined data set by infection status (Fig. 11A) resulted in a single large population that correlated with infection, whereas extensive overlap was seen throughout the rest of the ordination. This population was characterized (Fig. 11B) and the expression of surface markers was found to be consistent with a MHC Class II+ leukocyte that was not a macrophage or a dendritic cell, but rather unexpectedly, a B-cell. Traditional flow cytometric analysis of the leukocyte populations also confirmed this conclusion (data not shown). Furthermore it could rapidly be demonstrated that infected IEL had much higher levels of this cell than uninfected IEL (Fig. 11C). Together these data demonstrate a proof of principle that direct gradient analysis can be used with flow cytometric data, and more generally, outside the realm of ecology.
Figure 11. Direct gradient analysis of CD45+ interstitial epithelial leukocytes during C. difficile infection.
A. CD45+ IEL from untreated mice and infected mice were constrained by infection status using CCA and plotted using rgl. Blue, cells from untreated mice; red, cells from infected mice. B. Boxplots characterizing relative fluorescent intensities of the cells in the region indicated in panel A by the white ellipse. The vertical axis depicts cell frequency, relative to all CD45+ events; FSC, forward scatter; SSC, side scatter. Figure 11C depicts the frequency of cells in both the uninfected and infected state (as a % of CD45+ cells ) within the selected region.
Discussion
The current study details methodology by which analysis of complex data sets, such as those arising from 16S surveys of microbial communities, can be improved to detect underlying meaningful biological variables (frequently termed “gradients” in ecology), even in the midst of “noisy” data. The two crucial steps to this process are (1) the use of data with a fine level of differentiation between samples (in this case, a cutoff of 3% OTU rather than phylotypes); and (2) the application of constraints based on explicit hypotheses. We have applied these methods to two 16S studies: one involving samples from human lung explants, and the other a murine study in which drift in the bacterial community over the course of a year made correlation of microbial data with biologic outcomes difficult. In each case, these methods uncovered patterns relevant to our hypotheses that had been hidden by systemic noise. Finally, we demonstrate the general applicability of this approach, by extending it “beyond ecology” in an example of flow cytometric data analysis, to which this approach has not, to our knowledge, previously been applied.
Increasing the resolution of a data set as a means of uncovering underlying patterns may seem counter-intuitive, because the process magnifies rather than reduces variation between samples. However, fundamental to ordinations that are rooted in eigen analysis is the rank-order organization of data based on the elements responsible for the greatest amount of variation. Thus, the greater variation present in high-resolution data, relative to a less refined analysis of the same population, will tangibly affect ordination only if the new source of variation is significant. Contrasting each pair of phylotypic analysis (Figs. 1, 2, 5, 7) with its respective 3% OTU analysis (Figs 3, 4, 6, 8) provides excellent examples of the utility of this principle. For example, when analyzed at a phylotype resolution, the human lung explant data set possessed 109 OTUs present at > 1% of the total population, versus 143 OTUs at the same frequency in the high-resolution version of the same data set. Of those additional 34 OTUs, only one contributed to the large shifts that separated explant #2 from the others. This single OTU (“species”) existed almost exclusively in explant #2 but it merged into the dominant OTUs present in the other explants at the phylotype level. Thus, despite adding 34 additional sources of variation by a finer parsing of the same data, the process uncovered significant changes brought about by a single OTU and attained a deeper understanding of the microbial communities.
Our second crucial step, the use of constraints to address directly the effects of environmental factors, has been employed for many years by ecologists5,15,17, but is almost unknown in the remainder of biology. This methodology depends on the underlying reasoning that a major source of variation in any given community data set is differences in the environment (e.g., soil pH, abundance of food or water, etc.). In theory, if one completely understood the environment of the community in question, then a direct gradient analysis encompassing all of its environmental variables would produce the same result as the indirect gradient analysis. In most cases relevant to human health or experimental animal models, too little is known about the bacterial microenvironment to apply constraints such as differences in local pH. By contrast, often much is known about the macroenvironment (e.g., the origin of the sample, a particular treatment group, etc.). We demonstrate that such “macro” constraints can identify effects and correlations that would otherwise be hidden by systemic noise. Other methods for hypothesis testing of ecological models exist and are well-established5,15,21,22,23,24. However, the ability to isolate the data of interest, to display these data in a visually appealing manner, and to test whether significant differences exist, make direct gradient analysis ideal for asking biological questions of 16S data. The success of ordinations among microbial ecologists5 and flexibility of this method with regard to categorical data suggest that it is likely to be easily adapted and well-received.
Importantly, however, for the exploding field of microbiome analysis by 16S surveys, constrained ordination has largely been unused, with only 8.7% of 1141 publications using any form of hypothesis testing5. Instead, emphasis has been placed on various exploratory metrics to detect large or subtle effects within the data set as a whole25. This approach has the advantage of examining all of the data, but is limited by the production of radically different results depending upon the specific metric chosen. We hypothesized that applying constraints to “noisy” data would allow the detection of underlying effects in a manner that is much less dependent upon the metric, but more dependent upon the biological question. This was what was seen in our analyses of microbial communities in the lungs of subjects undergoing transplantation for end-stage emphysema. Constrained analysis allowed us to demonstrate that microbial communities differed significantly between human subject, but not in a reproducible manner between similar anatomic sites between or within subjects. Thus, the approach also has the key feature of being able to reject incorrect hypotheses in a statistically rigorous fashion.
Another important feature of the current study is the demonstration that direct gradient analysis is generally applicable to datasets farther removed from microbial ecology. For this purpose, we chose flow cytometric data not only because it was readily available to us but more cogently, because the technique is broadly used in contemporary immunology and cell biology. Structurally, flow cytometric data is both similar to ecological survey data in involving a very large numbers of events, but also differs because in flow cytometry, there are always many more “sites” (individual cells) than “species” (fluorescent channels). Nevertheless, these differences did not adversely affect direct gradient analysis in our study. From a practical viewpoint, the calculation of distance matrix becomes computationally more intensive with very large flow cytometry datasets; however, this has not prevented the previous use of unconstrained ordination methods such as PCA which also rely on a distance calculation26,27,28. To the best of our knowledge, the usage of CCA in conjunction with flow cytometric data has not been reported previously. As was shown in figure 11, a constrained model can be used effectively to identify important populations.
We consistently observed that CCA performed better than the other metrics tested. Even when analyzing the phylotyped explant data, although the differences proved not to be significant, CCA correctly identified the groups proven to be significant using the high-resolution data. Whereas each of the constrained metrics performed similarly in identifying statistically significant differences, CCA consistently produced ordinations that were easier to interpret visually. CCA is rooted in CA and, although not based on an ecological metric, has a long history of use in ecology. The strength of CA is that it tries when calculating site scores to maximize the correlation between sites and species. This feature differs from PCA and PCoA, which calculate site scores based on distances calculated from species abundances. One drawback of CA, as was observed in both data sets, is that it can be prone to the compression of the ordination due to a single outlier in normalized data sets. However, no such problem was encountered with CCA. In general, whichever method of direct gradient analysis is chosen, the indirect form of the same analysis should be run in parallel as a check on the quality of the constraint. Based on the data presented here, we believe that CCA is the constrained method of choice when a strong a priori hypothesis is available. We have demonstrated that this method is well-suited for finding correlations between 16S data and biological readouts.
In summary, we demonstrate the generalized utility of direct gradient analysis in large multivariate datasets derived from both translational and basic experiments. We are confident that this approach will provide a robust means of analyzing even larger biological datasets, including the exponentially more complex examples emerging from metabolomic surveys. Keeping biostatistical tools for data visualization and statistical analysis abreast of advances in sequencing techniques is an important goal that merits continued research interest.
Methods
16S survey data sets
Two 16S datasets that spanned V1–V3 were used in this study. Data set #1 included 281,822 sequences from 48 independent samples taken from various airways or lung parenchyma of five lung explants removed from patients undergoing lung transplantation for advanced emphysema. The number of samples per explant ranged from 4–15. Clinical data associated with these samples has been previously published16. Data set #2 included 130,796 sequences from 40 samples of tissue taken from the cecal tip of mice that had either been treated with antibiotics, given a gavage of the fungus Candida albicans, treated with both antibiotics and C. albicans, or left untreated. This treatment regime has previously been demonstrated to alter the systemic allergic-type responses in a microbiota-dependent manner4,19,29.
Flow cytometric data sets
Flow cytometric data were obtained from an experiment analyzing the cecum of mice that had been either left untreated or infected for two days with Clostridium difficile. Enrichment of intestinal leukocytes was performed as previously described30. The cells were stained for the cell surface markers CD45 (clone 30-F11), CD11b (clone M1/70), CD11c (clone HL3), Gr-1 (clone RB6-8C5), MHC II(clone M5/114.15.2), and F4/80 (clone BM8). Data on the samples were acquired on a 3-laser Canto II flow cytometer using FACSDiVa software (BD Biosciences).
16S survey data analysis
The data sets were processed and analyzed separately using the program mothur11 to bin the sequences into operational taxonomic units (OTUs). This process created .shared output files, based on a phylotype-level of sequence differentiation; i.e., OTUs binned into classifiable phylotypes, where genus is the lowest taxonomic level (phylotype() followed by make.shared()), or an average-neighbor clustered 3% OTU. These .shared output files – were then imported into and analyzed in the program R (http://cran.r-project.org). These files follow the format of a community data matrix, i.e., sites (samples) define the rows and species (OTUs) define the columns. Before further analysis, a 1% cutoff was applied to the data (i.e., only OTU populations at a frequency of >1% of the total population were considered for further analysis). The data were then normalized to # sequences/10,000 sequences. Further analysis was performed, heavily leveraging a number of the functions found in the R-package vegan18. Principle Coordinates Analysis was performed using the R base function Classical Multidimensional Scaling (cmdscale()) together with distance matrices created using the Jaccard (Jac) or Bray-Curtis (BC) ecological distance metrics (from the function vegdist()). Bray-Curtis is a metric frequently used in community ecology that balances both membership and abundance, whereas the Jaccard metric considers membership to be more important than abundance. PCA and CA were performed using the rda() and cca() functions, respectively. PCA is a general method for assessing differences between samples based on their variance (similar to PCoA utilizing a Euclidean distance metric). CA is a method that is conceptually similar to PCA, except that the chi-squared statistic of the data frame is used instead of the data frame itself. This latter method is often used in ecology because ordinations maximize the correlation between the sites where the data were taken and the species found at that site. It is worth noting that in CA what is explained is the contribution to the mean squared of the contingency coefficient, as opposed to overall variance, but their meaning is analogous for the purposes of these analyses. Constrained ordinations were generated using the Constrained Analysis of Principle Coordinates (capscale()), Redundancy Analysis (rda()), or Canonical Correspondence Analysis (cca()) functions which follow the method laid out by Legendre and Legendre15. When not available otherwise, axis loading (inertia) values were obtained by taking the eigenvalue of the axis and dividing by the sum of the positive eigenvalues.
Flow cytometry data analysis
Listmode data files were loaded into R using the flowCore package31, and the fluorescence channel information was extracted. CD45+ cells were selected by gating on CD45-high, SSC-high and FSC-mid populations using select3d() and the rgl package32. The resulting data matrix containing the CD45+ cells was combined into a new data matrix containing the CD45+ cells from both in uninfected and infected treatments. A vector containing the explanatory variable of "Treatment" was created for each row of the data frame. Fluorescent channels were log-transformed after adjusting for zeros. Population densities were adjusted so that they fell between the first and second decade. Finally, the combined CD45+ cell data frame was standardized using method = "max" from the decostand() function in the package vegan. The CD45+ data were constrained by Treatment to generate a CCA and the resulting ordination plotted using the ordirgl() function. To maximize visualization of overlapping populations, transparency was added (alpha = 0.5). Selection of unique populations was performed using select3d() function and the populations characterized using the function boxplot().
Author Contributions
Conceived and designed the experiments: JED, GBH, JLC, MRG, BH, NS, and JW. Performed the experiments: JED. Analyzed the data: JED, AASA. Contributed reagents/materials/analysis tools: AASA, FJM. Wrote the manuscript: JED, GBH, JLC.
Acknowledgments
Supported by: Joint Initiative for Translational and Clinical Research at the University of Michigan and Peking University Health Sciences; R01 HL082480, U19 AI090871 (GBH) and U01 HL098961 from the USPHS; and a Research Enhancement Award Program from the Biomedical Laboratory Research & Development Service, Department of Veterans Affairs.
References
- Liu Z., DeSantis T. Z., Andersen G. L. & Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36, e120 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. M. & Relman D. A. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Methods Enzymol 235, 205–222 (1994). [DOI] [PubMed] [Google Scholar]
- Rothberg J. M. & Leamon J. H. The development and impact of 454 sequencing. Nat Biotechnol 26, 1117–1124 (2008). [DOI] [PubMed] [Google Scholar]
- Mason K. L., Erb Downward J. R., Falkowski N. R., Young V. B., Kao J. Y. et al. Interplay between the Gastric Bacterial Microbiota and Candida albicans during Postantibiotic Recolonization and Gastritis. Infect Immun 80, 150–158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62, 142–160 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J. & Gordon J. I. The core gut microbiome, energy balance and obesity. J Physiol 587, 4153–4158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- Muegge B. D., Kuczynski J., Knights D., Clemente J. C., Gonzalez A. et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. & Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71, 8228–8235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Cai Y., Mai V., Farmerie W., Yu F. et al. Advanced computational algorithms for microbial community analysis using massive 16S rRNA sequence data. Nucleic Acids Res 38, e205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin P. R. An evaluation of relative robustness of techniques for ecological ordinations. Vegetatio 69, 89–107 (1987). [Google Scholar]
- Legendre P. & Legendre L. Numerical Ecology. 853. (1998).
- Erb-Downward J. R., Thompson D. L., Han M. K., Freeman C. M., McCloskey L. et al. Analysis of the lung microbiome in the "healthy" smoker and in COPD. PLoS One 6, e16384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Braak C. J. F. Canonical Correspondence Analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67, 1167–1179 (1986). [Google Scholar]
- Jari Oksanen F., Guillaume Blanchet, Roeland Kindt, Pierre Legendre, R. B. O'Hara, et al.vegan: Community Ecology Package. R package version 1.17-3. 1.17-3 ed. (2010).
- Noverr M. C., Noggle R. M., Toews G. B. & Huffnagle G. B. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun 72, 4996–5003 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason K. L., Erb Downward J. R., Mason K. D., Falkowski N. R., Eaton K. A. et al. Candida albicans and Bacterial Microbiota Interactions in the Cecum during Recolonization following Broad-Spectrum Antibiotic Therapy. Infect Immun 80, 3371–3380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R. Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology 18, 117–143 (1993). [Google Scholar]
- Peres-Neto P. & Jackson D. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129, 169–178 (2001). [DOI] [PubMed] [Google Scholar]
- Mardia K. V., Kent J. T. & Bibby J. M. Multivariate analysis: Academic Press. (1979).
- Zapala M. A. & Schork N. J. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc Natl Acad Sci USA 103, 19430–19435 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J., Liu Z., Lozupone C., McDonald D., Fierer N. et al. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods 7, 813–819 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi Y., Sato R., Genka S., Shitara N. & Takakura K. An interactive multivariate analysis of FCM data. Cytometry 9, 405–408 (1988). [DOI] [PubMed] [Google Scholar]
- De Zen L., Bicciato S., te Kronnie G. & Basso G. Computational analysis of flow-cytometry antigen expression profiles in childhood acute lymphoblastic leukemia: an MLL/AF4 identification. Leukemia 17, 1557–1565 (2003). [DOI] [PubMed] [Google Scholar]
- Lugli E., Pinti M., Nasi M., Troiano L., Ferraresi R. et al. Subject classification obtained by cluster analysis and principal component analysis applied to flow cytometric data. Cytometry Part A 71A, 334–344 (2007). [DOI] [PubMed] [Google Scholar]
- Noverr M. C., Falkowski N. R., McDonald R. A., McKenzie A. N. & Huffnagle G. B. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun 73, 30–38 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Yamazaki T., Kamada N., Tawaratsumida K., Kim Y. G. et al. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol 186, 4872–4880 (2011). [DOI] [PubMed] [Google Scholar]
- Hahne F., LeMeur N., Brinkman R. R., Ellis B., Haaland P. et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics 10, 106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch D. A. a. D. rgl: 3D visualization device system (OpenGL). R package version 0.92.798 ed (2011). [Google Scholar]