Abstract
Tyrosinase is a ubiquitous enzyme with diverse physiologic roles related to pigment production. Tyrosinase inhibition has been well studied for cosmetic, medicinal, and agricultural purposes. We simulated the docking of tyrosinase and D-(−)-arabinose and found a binding energy of −4.5 kcal/mol for theup-formof D-(−)-arabinose and −4.4 kcal/mol for thedown-form of D-(−)-arabinose. The results of molecular dynamics simulation suggested that D-(−)-arabinose interacts mostly with HIS85, HIS259, and HIS263, which are believed to be in the active site. Our kinetic study showed that D-(−)-arabinose is a reversible, mixed-type inhibitor of tyrosinase (α-value = 6.11 ± 0.98, K i = 0.21 ± 0.19 M). Measurements of intrinsic fluorescence showed that D-(−)-arabinose induced obvious tertiary changes to tyrosinase (binding constant K = 1.58 ± 0.02 M−1, binding number n = 1.49 ± 0.06). This strategy of predicting tyrosinase inhibition based on specific interactions of aldehyde and hydroxyl groups with the enzyme may prove useful for screening potential tyrosinase inhibitors.
1. Introduction
Tyrosinase (EC 1.14.18.1) is a ubiquitous enzyme with diverse physiologic roles related to pigment production. It plays a central role in melanin synthesis in skin [1, 2], the browning of vegetables [3, 4], wound healing [5], and cuticle formation in insects [6, 7]. Structurally, tyrosinase belongs to the type 3 copper protein family [8, 9], which consists of two copper ions individually coordinated with three histidine residues at the active site. Tyrosinases are directly involved in several reactions and carry out catalytic steps such as the hydroxylation of tyrosine to 3,4-dihydroxyphenylalanine (DOPA), the oxidation of DOPA to DOPA quinone, and the oxidation of 5,6-dihydroxyindole to 5,6-dihydroxuquinone [10, 11]. In addition to its catalytic features, tyrosinase is distinctive from other enzymes because it displays various inhibition patterns. Tyrosinase inhibition has been extensively studied for cosmetic, medicinal, and agricultural purposes [12].
The tyrosinase mechanism is complex, and this enzyme can catalyze multiple reactions. Despite several reported crystallographic structures of tyrosinase, the 3D structure and architecture of the active site are not well understood [22, 23]. Mechanistic studies must involve a variety of computational methods and kinetic analysis to derive the structure-function relationship between substrates and ligands. The inhibitory effect of compounds with sugar backbones on tyrosinase are of great interest [20, 24, 25]. D-(–)-arabinose, a potential tyrosinase inhibitor, is an aldopentose with one aldehyde and four hydroxyl groups, which was used to immobilize mushroom tyrosinase on a reusable glass bead preparation [26].
In the current study, we investigated the mechanism of tyrosinase inhibition by D-(–)-arabinose using computational simulation and kinetic analysis. We hypothesized that the aldehyde and hydroxyl groups of D-(–)-arabinose may block L-DOPA oxidation by binding to tyrosinase. Previous findings have shown the importance of aldehyde [27, 28] and hydroxyl [27, 29–32] groups in tyrosinase inhibition with regard to molecular position, number, and specific interactions of these groups with the enzyme. These findings further support our hypothesis that D-(–)-arabinose might have an inhibitory effect on tyrosinase, as D-(–)-arabinose has one aldehyde and four hydroxyl groups. D-(–)-arabinose exerted a mixed-type inhibition on tyrosinase. Kinetic parameters have consistently supported docking simulation results in which D-(–)-arabinose binds to residues at or near the active site, and measurements of intrinsic fluorescence have revealed great changes in tertiary protein structure. A combination of computational modeling and inhibition kinetics may facilitate the testing of potential tyrosinase inhibitors such as D-(–)-arabinose and the prediction of their inhibitory mechanisms.
2. Materials and Methods
2.1. Materials
Tyrosinase (M.W. 128 kDa) and L-DOPA were purchased from Sigma-Aldrich. D-(–)-arabinose was purchased from Tokyo Chemistry Industry. When L-DOPA was used as a substrate in our experiments, the purchased tyrosinase had a K m of 0.29 ± 0.11 mM (V max = 0.15 ± 0.03 mmol · min−1) using a Lineweaver-Burk plot. All kinetic reactions and measurements in this study were performed in 50 mM sodium phosphate buffer (pH 6.9) at the temperature of 25°C.
2.2. Computational Docking of Tyrosinase and D-(–)-arabinose and Molecular Dynamics Simulation
The tyrosinase structure was simulated according to the crystal structure of Agaricus bisporus tyrosinase (PDB ID: 2Y9X). The original structure of D-(–)-arabinose was derived from the PubChem database (ID: 66308), named (2S, 3R, 4R)-2, 3, 4, 5-tetrahydroxypentanal. At room temperature, D-(–)-arabinose exists in a ring structure because the terminal alcohol and aldehyde group interact with each other. Because the structure has an ambiguous chiral center in the ring, we generated two chemical forms (up and down). We used the Pck software to find the binding pocket [33] and found several neighboring residues from the binding pocket. Ten docking structures were generated from each neighboring residue. AutoDock Vina [34] was used for in silico protein-ligand docking. Using the final structure from the docking result, a 10 ns production of molecular dynamics simulation was performed by CHARMM [35]. Then, we measured the structure details of the protein-ligand interactions as a function of time to ensure that the interactions revealed by the docking study were conserved. The structures were saved every picosecond for trajectory analysis.
2.3. Tyrosinase Assay
A spectrophotometric tyrosinase assay was performed as previously described [16, 18, 36]. To begin the assay, a 10 μL sample of enzyme solution was added to 1 mL of reaction mix. Tyrosinase activity (υ) was recorded as the change in absorbance per min at 475 nm using a Helios Gamma spectrophotometer (Thermo Spectronic, UK). The final concentrations of L-DOPA and tyrosinase were 2 mM and 6.6 μg/mL, respectively. A range of D-(–)-arabinose from 0.05 M to 2 M was applied, according to the experimental conditions.
2.4. Kinetic Analysis for the Mixed-Type Inhibition
To describe the mixed-type inhibition mechanism, the Lineweaver-Burk equation in double-reciprocal can be written as follows:
| (1) |
Secondary plots can be constructed from
| (2) |
| (3) |
The V max, K m, α, and K i values can be derived from the above equations. The secondary plot of Slope or Y-intercept versus [I] is linearly fit, assuming a single inhibition site or a single class of inhibition site, as shown in Scheme 1.
Scheme 1.
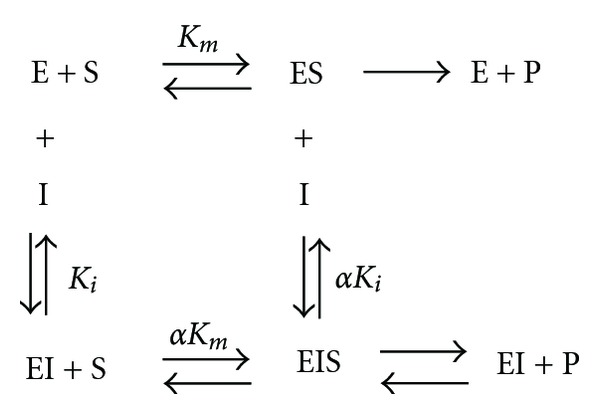
D-(–)-arabinose binds to the E and ES complex. E: enzyme tyrosinase; S: substrate L-DOPA; I: inhibitor D-(–)-arabinose; P: product dopachrome; K i: inhibitor dissociation constant; α: modifying factor.
2.5. Intrinsic and ANS-Binding Fluorescence Measurements
Fluorescence emission spectra were measured using a Hitachi F-2500 fluorescence spectrophotometer with a 1 cm path-length cuvette. An excitation wavelength of 280 nm was used for the tryptophan fluorescence measurements, and the emission wavelength was between 300 and 420 nm. Changes in the ANS-binding fluorescence intensity for tyrosinase were studied by labeling with 40 μM ANS for 30 min prior to measurement. An excitation wavelength of 390 nm was used for the ANS-binding fluorescence, and emission wavelength ranged from 420 to 600 nm. The final concentrations of tyrosinase was 6.6 μg/mL.
2.6. Determination of the Binding Constant and the Number of Binding Sites
According to a previous report [37], when small molecules are bound to equivalent sites on a macromolecule, the equilibrium between free and bound molecules is given by the following equation to evaluate the binding constant (K) and number of binding sites (n):
| (4) |
where F 0 and F are the relative steady-state fluorescence intensities in the absence and presence of quencher, respectively, and [Q] is the quencher (D-(−)-arabinose) concentration. The values for the binding constant (K) and number of binding sites (n) can be derived from the intercept and slope of a plot based on (4).
3. Results
3.1. Computational Docking and Molecular Dynamics Simulation
We searched for the D-(−)-arabinose binding pocket (Figure 1(a)) and common neighboring residues within tyrosinase and found several residues (Figure 1(b)), including HIS61, HIS85, HIS94 HIS244, GLU256, HIS259, ASN260, HIS263, PHE264, MET280, GLY281, SER282, VAL283, ALA286, PHE292, and HIS296. As the 1′-OH group of D-(−)-arabinose is flexible, there are two forms that exist: up and down (Figure 1(c)). The lowest docking energies of up-form and down-form D-(−)-arabinose were −4.5 kcal/mol and −4.4 kcal/mol, respectively. The negative energies indicated that the two chemicals were bound tightly to tyrosinase. Thermodynamically, the interactions between up-form D-(−)-arabinose and tyrosinase could be more quickly stabilized than those in down-form.
Figure 1.
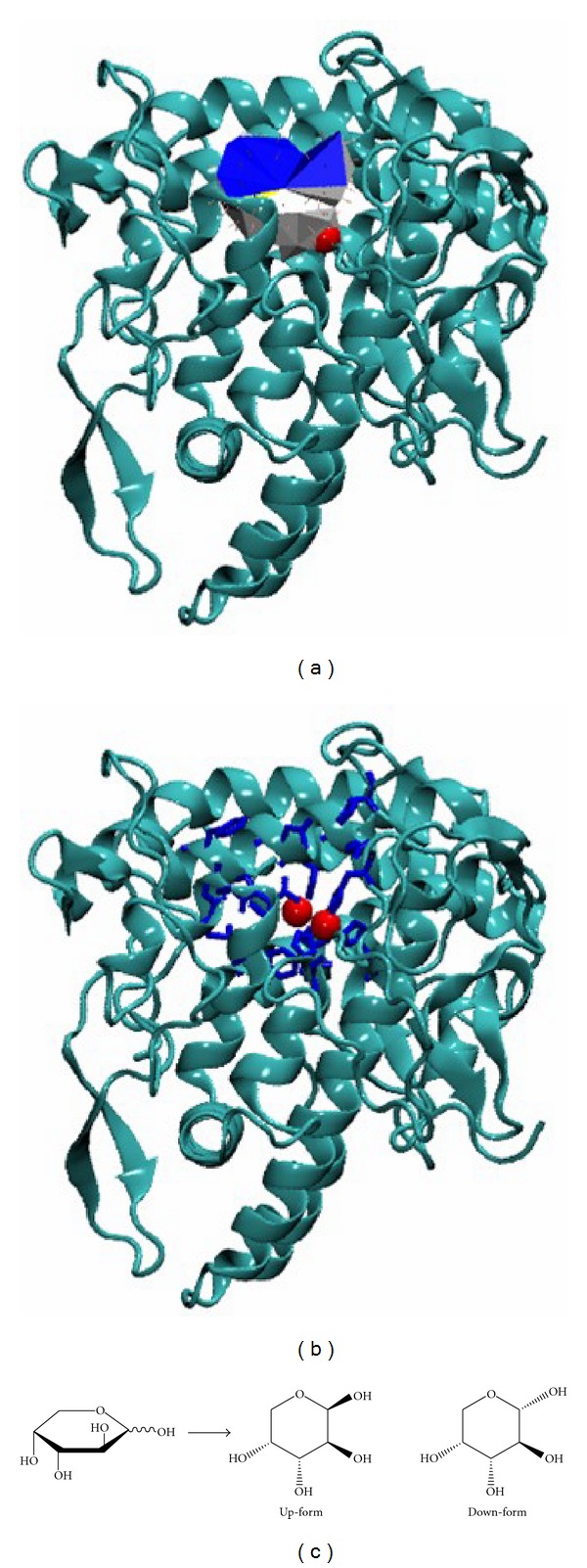
Computational simulations of tyrosinase and D-(–)-arabinose. (a) The binding pocket of tyrosinase was found using Pck software. Blue and white represent the pocket mouth, with the entrance colored in blue. The two red spheres indicate the two copper ions. (b) Plausible binding residues. Blue sticks indicate the pocket residues, and the two red spheres indicate the two copper ions. (c) The two forms of D-(–)-arabinose (up-form and down-form).
Next, the 10 ns molecular dynamics simulation was performed to examine structural changes during simulation. We verified the root mean square deviation (RMSD) of the alpha carbon, and the results showed that the structure was first rearranged and then stabilized (Figure 2(a)). Plausible candidate residues that may interact with D-(−)-arabinose were chosen, and the distances were measured via simulation. The interactions gradually stabilized after 1 ns as the simulation progressed (Figures 2(b) and 2(c)). The plausible residues determined by the molecular dynamics simulation were HIS85, HIS259, and HIS263 for both up-form (Figure 2(b)) and down-form (Figure 2(c)) D-(−)-arabinose. Both forms of D-(−)-arabinose were accessed to the core site of the pocket, and the binding of up-form D-(−)-arabinose with tyrosinase was slightly more stable than the down-form. The binding sites for both forms of D-(−)-arabinose with tyrosinase were predicted to be HIS85, HIS259, and HIS263 (Figure 3).
Figure 2.
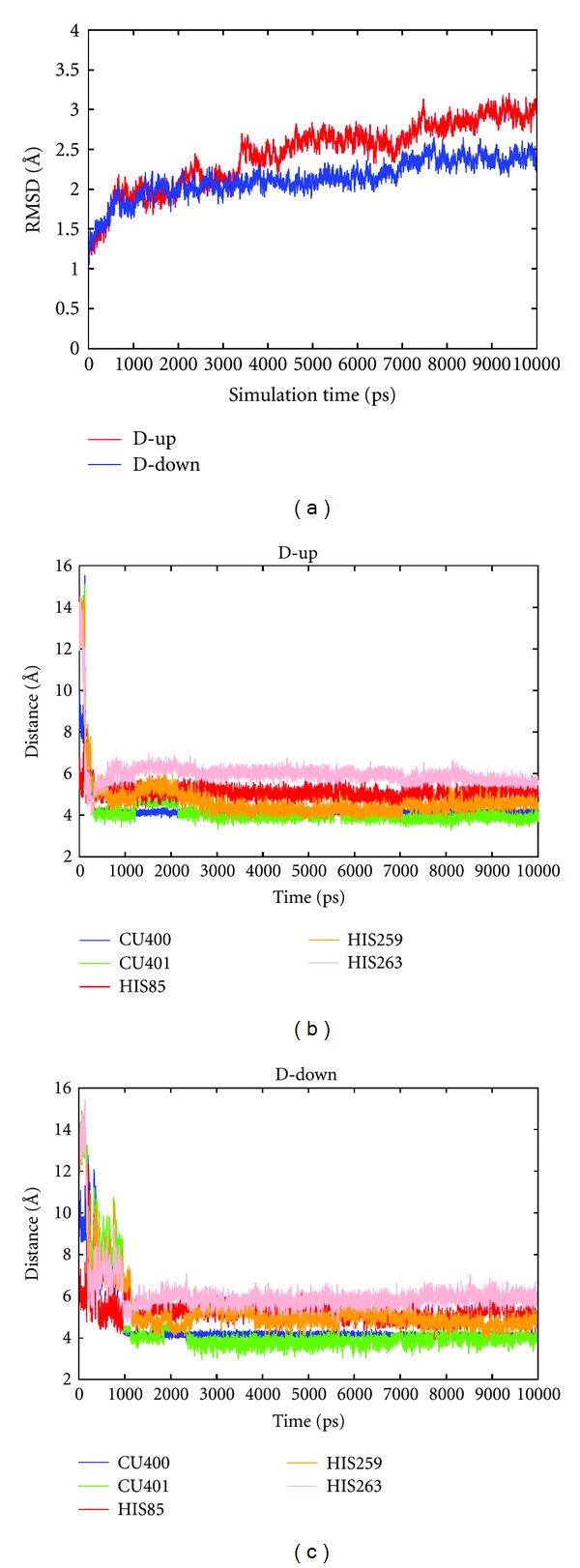
Molecular dynamics simulations between tyrosinase and D-(–)-arabinose. (a) RMSD time profiles of the alpha carbon. (b) Time profile of plausible interactions with up-form D-(–)-arabinose. (c) Time profile of plausible interactions with down-form D-(–)-arabinose. Cu400 and Cu401 represent the two copper ions.
Figure 3.
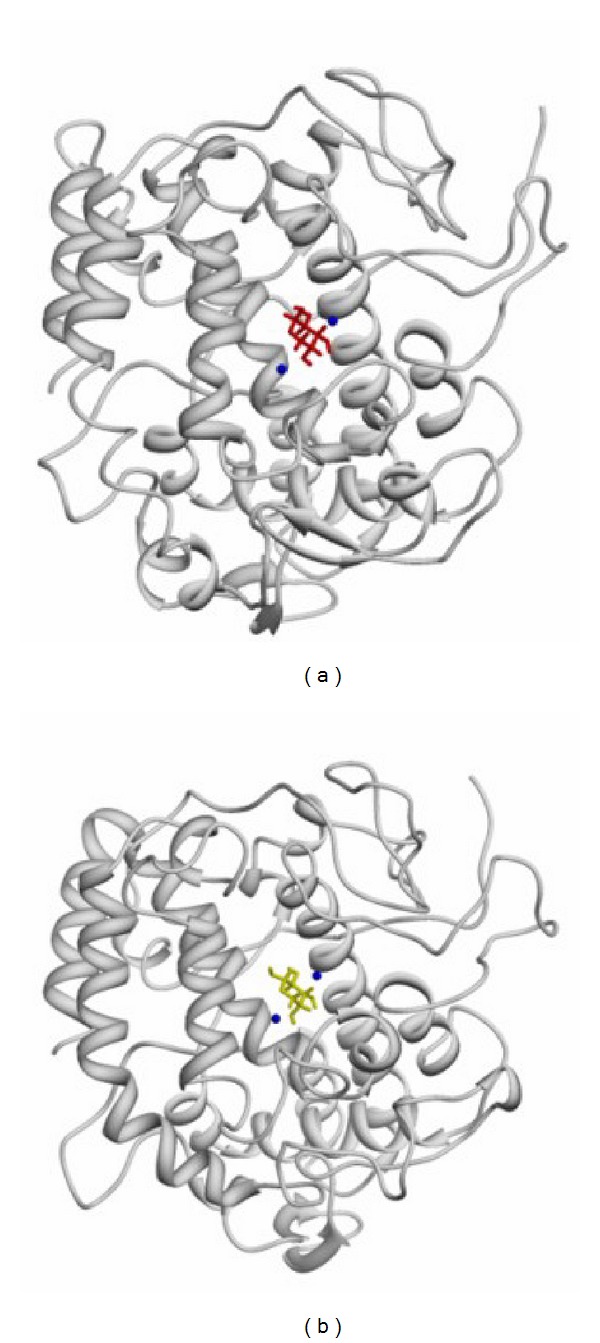
Binding regions predicted by molecular dynamics simulations. (a) Binding site for up-form D-(–)-arabinose. (b) Binding site for down-form D-(–)-arabinose.
In our studies, the identified plausible residues for docking and the molecular dynamics simulations were consistent.
3.2. The Effect of D-(−)-arabinose on Tyrosinase Activity
We assayed tyrosinase activity changes in the presence of D-(−)-arabinose. Tyrosinase activity was inactivated by D-(−)-arabinose in a dose-dependent manner (Figure 4). The inhibitor concentration leading to 50% tyrosinase activity loss (IC50) was estimated to be 0.61 ± 0.02 M (n = 3), and tyrosinase was almost completely inactive at 1.75 M D-(−)-arabinose (Figure 4(a)). When D-(−)-arabinose was absent from the assay system, the tyrosinase activity remained at 76%, even with 2 M D-(−)-arabinose in the preincubation step (Figure 4(b)). The difference is due to the dilution effect of inhibitor, indicating that D-(−)-arabinose reversibly inhibited tyrosinase. To confirm the reversibility of D-(−)-arabinose inhibition, the remaining enzyme activity at different inhibitor concentrations was tested. A plot of enzyme activity (v) versus enzyme concentration [E] resulted in a decrease in the slope of the line, indicating that the inhibition of D-(−)-arabinose on the enzyme was reversible (Figure 5).
Figure 4.
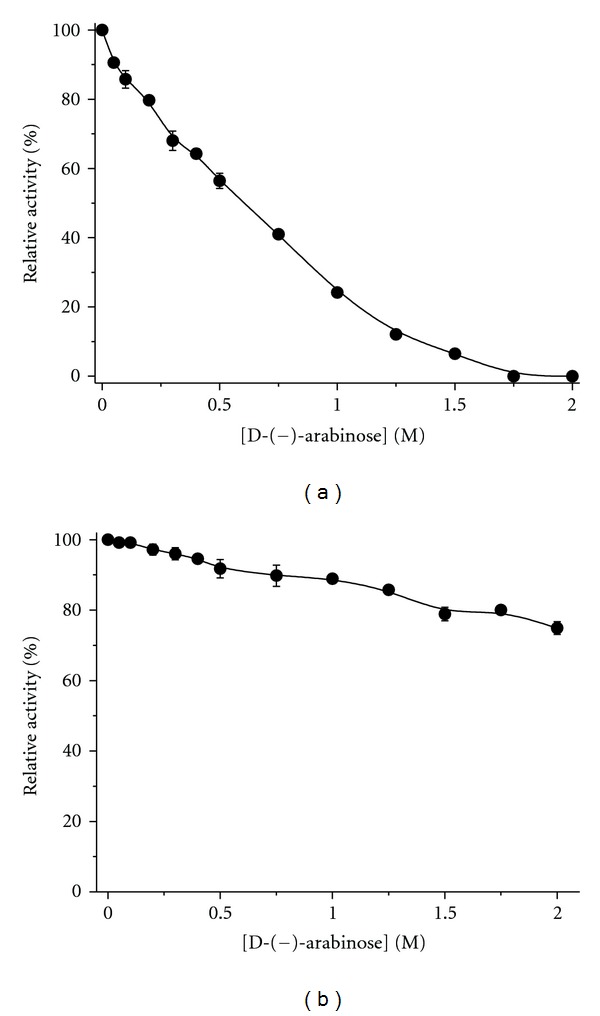
Inhibitory effect of D-(–)-arabinose on tyrosinase. Data are presented as the mean (n = 3). Tyrosinase was incubated with D-(–)-arabinose at various concentrations for 3 h at 25°C and then added to the assay system at the corresponding D-(–)-arabinose concentrations (a) or in the absence of D-(–)-arabinose (b). The final concentrations of L-DOPA and tyrosinase were 2 mM and 6.6 μg/mL, respectively.
Figure 5.
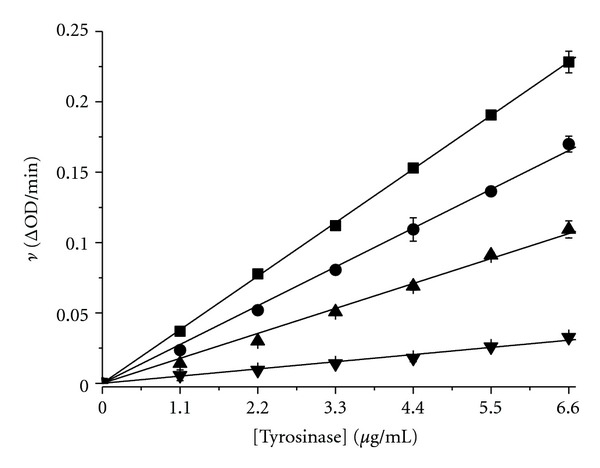
Plots of enzyme activity (v) versus enzyme concentration [E] at D-(–)-arabinose concentrations of 0 M (■), 0.2 M (●), 0.6 M (▲), and 0.8 M (▼). The ν value indicates the change in absorbance at 475 nm per minute. The final L-DOPA concentration was 2 mM.
Next, time courses for tyrosinase inhibition at different concentrations of D-(−)-arabinose were performed (Figure 6). The results showed that changes in the catalytic rate over time were not detectable at any of the tested D-(−)-arabinose concentrations (0.05 M to 1 M). Thus, D-(−)-arabinose appeared to bind tyrosinase very quickly and inhibit the oxidation of L-DOPA without an apparent change in the catalytic rate under these test conditions. These results indicated that the overall tertiary structural changes might not be synchronized with D-(−)-arabinose-induced inhibition due to rapid achievement of equilibrium states.
Figure 6.
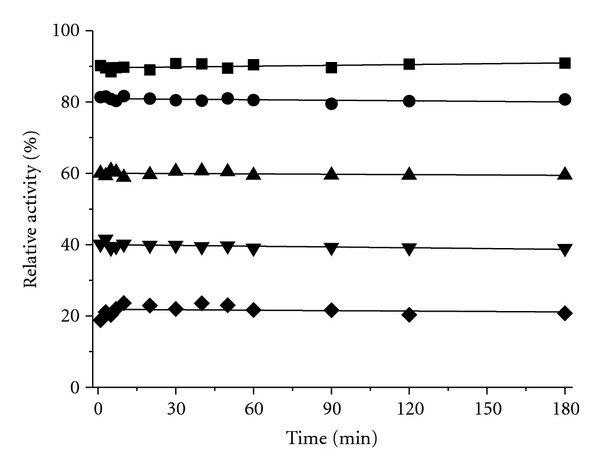
Time course of tyrosinase inhibition in the presence of D-(–)-arabinose. The enzyme solution was mixed with D-(–)-arabinose at 0.05 M (■), 0.2 M (●), 0.5 M (▲), 0.75 M (▼), and 1 M (◆) concentrations, and aliquots were collected for analysis at the indicated time intervals.
3.3. Determination of the D-(−)-arabinose Inhibition Type
The kinetics of tyrosinase in the presence of D-(−)-arabinose was studied using double-reciprocal Lineweaver-Burk plots. The results showed changes in the apparent V max and the K m, indicating that D-(−)-arabinose induced a mixed-type of inhibition (Figure 7). The secondary plots of Slope versus D-(−)-arabinose concentration and Y-intercept versus D-(−)-arabinose concentration were linearly fit (Figures 8(a) and 8(b), showing that D-(−)-arabinose has a single inhibition site or a single class of inhibition site on tyrosinase. Using (1)–(3), the α-value was found to be 6.11 ± 0.98 (n = 3), and the K i was 0.21 ± 0.19 M (n = 3).
Figure 7.
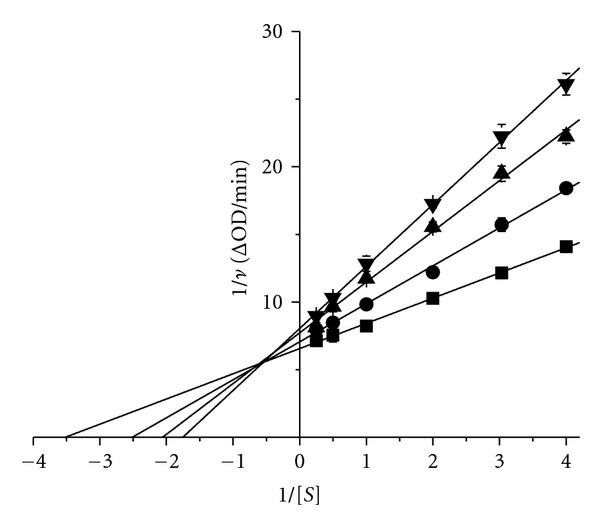
Lineweaver-Burk plot. The D-(−)-arabinose concentrations were 0 M (■), 0.1 M (●), 0.2 M (▲), and 0.3 M (▼). The final enzyme concentration was 6.6 μg/mL.
Figure 8.
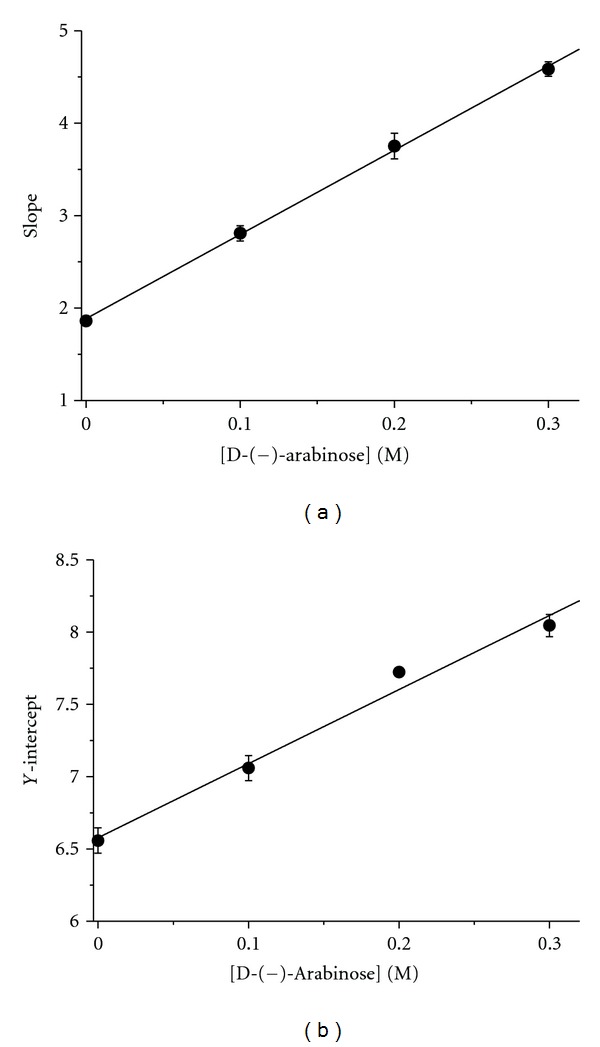
Secondary plots. (a) Plot of Slope versus D-(−)-arabinose concentration. All data were collected from Lineweaver-Burk plots. The plot was based on (2). (b) Plot of Y-intercept versus D-(−)-arabinose concentration. The plot was based on (3).
3.4. The Effect of D-(−)-arabinose on Tyrosinase Tertiary Structure
Tertiary structural changes of tyrosinase induced by D-(−)-arabinose were determined from measurements of the enzymatic intrinsic fluorescence changes. We found that the intrinsic fluorescence of tyrosinase was significantly changed. This was accompanied by a quenching effect that gradually decreased, with a significant shift of the maximum peak wavelength from 332 nm to 349 nm (Figures 9, 10(a), and 10(b)). At a 2 M concentration, D-(−)-arabinose almost completely quenched the fluorescence. To calculate the binding affinity, a double-reciprocal plot was evaluated according to (4) as shown in Figure 10(c). The plot revealed a linear relationship, and we calculated the binding constant (K = 1.58 ± 0.02 M−1) and the binding number (n = 1.49 ± 0.06) according to (4).
Figure 9.
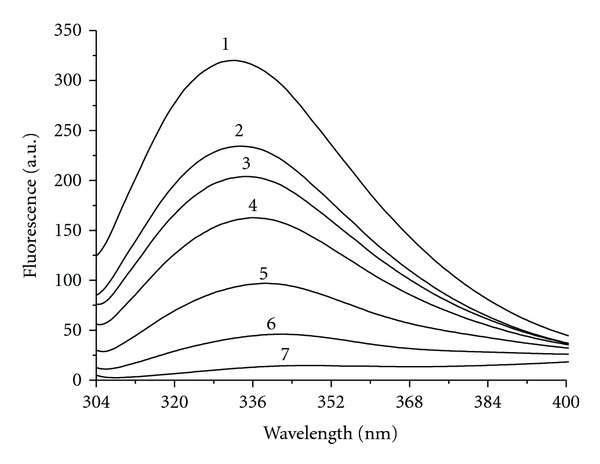
Intrinsic fluorescence changes by D-(−)-arabinose. Tyrosinase was incubated with D-(−)-arabinose for 3 h before the measurements were taken. Label 1 represents the native state. Labels 2 through 7 indicate D-(−)-arabinose concentrations of 0.2 M, 0.3 M, 0.5 M, 0.75 M, 1.25 M, and 2 M, respectively.
Figure 10.
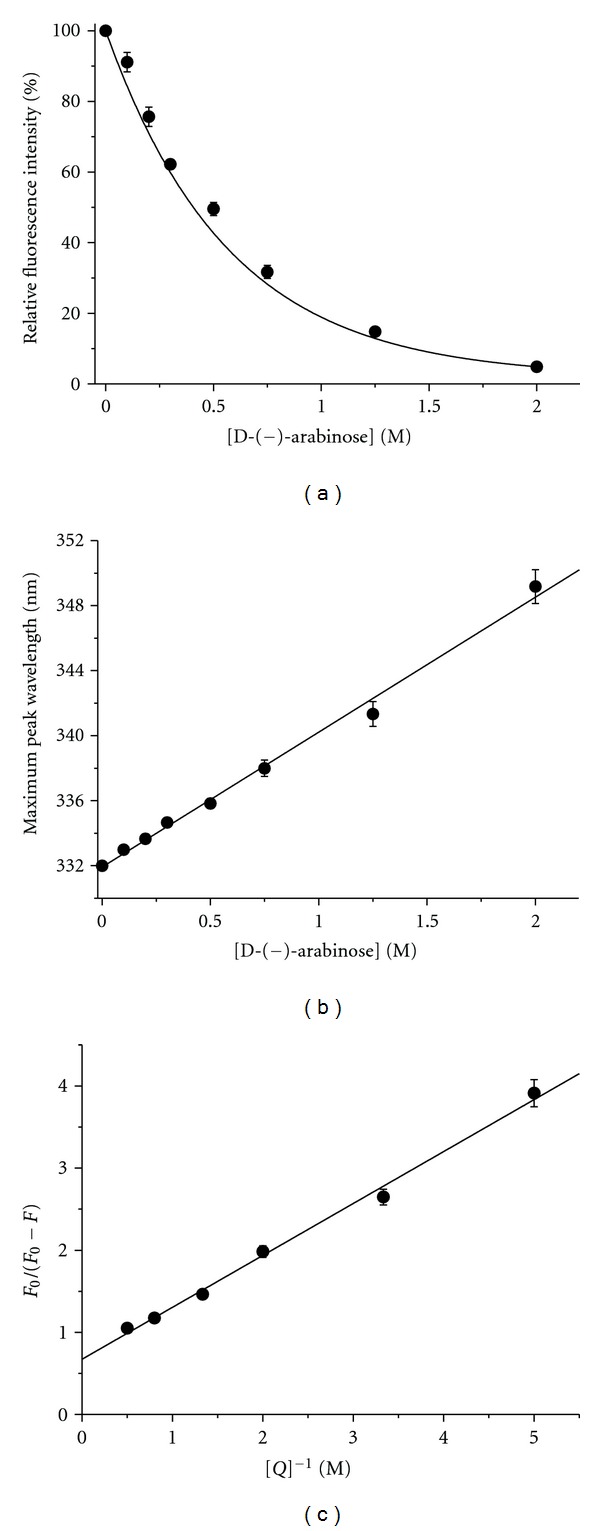
Tyrosinase tertiary structure changes in the presence of D-(−)-arabinose. Tyrosinase was incubated with various concentrations of D-(−)-arabinose (0 to 2 M). (a) Plot of maximum fluorescence intensity versus D-(−)-arabinose concentration. (b) Plot of maximum peak wavelength changes versus D-(−)-arabinose concentration. (c) Double reciprocal plot of F 0/(F 0 − F) versus [Q]−1. Data were treated according to (4). F 0: maximum native fluorescence intensity; F: maximum fluorescence intensity of sample; Q: quencher D-(−)-arabinose.
Next, we monitored changes in tyrosinase hydrophobicity in the presence of D-(−)-arabinose (Figure 11). Unfortunately, the ANS-binding fluorescence of D-(−)-arabinose alone was so strong that we could not tell whether it had influence on the ANS-binding fluorescence of tyrosinase.
Figure 11.
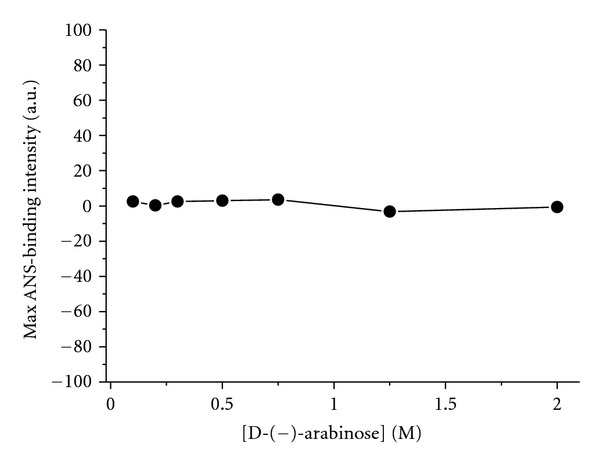
Changes in ANS-binding fluorescence of tyrosinase at different D-(−)-arabinose concentrations. ANS (40 μM) was incubated with tyrosinase for 30 min to label the hydrophobic enzyme surfaces prior to fluorescence measurements.
4. Discussion
Previous studies have identified compounds with aldehyde and hydroxyl groups to have a potent inhibitory effect on tyrosinase [38–40]. The hydroxyl groups in compounds carry out the nucleophilic attack on the coppers of tyrosinase in the active site and are directly involved in transferring protons during catalysis, which results in inactivation of tyrosinase [41, 42]. In this study, we hypothesized that D-(−)-arabinose could be a potent tyrosinase inhibitor due to its structure, with one aldehyde and four hydroxyl groups.
Using in silico docking, we predicted that D-(−)-arabinose could bind directly with amino acid residues inside the tyrosinase active site. Further molecular dynamics simulation showed a specific binding site consisting of HIS85, HIS259, and HIS263 for the up- and down-forms of D-(−)-arabinose. We found that the enzymatic activity of tyrosinase decreased as the concentrations of D-(−)-arabinose increased. D-(−)-arabinose inhibited tyrosinase in a reversible way and induced changes in the apparent V max and the K m, consistent with a mixed-type of inhibition through interactions with residues at or close to the active site. The second plots of Slope versus D-(−)-arabinose concentration and Y-intercept versus D-(−)-arabinose concentration were linearly fit. These results were consistent with previous computational simulation.
As conformational changes at or near the active site could influence access by the enzyme substrate, the conformational changes of tyrosinase induced by D-(−)-arabinose were detected by spectrofluorometry. Obvious changes were observed which indicated that the structure of the tyrosinase active site had been altered to reduce the access of the substrate L-DOPA.
Changes in catalytic rate were not detectable over time, which indicated that D-(−)-arabinose binding to the enzyme and decreased enzyme activity occurred very quickly. These results indicated that the overall tertiary structural changes might not be synchronized with D-(−)-arabinose-induced inhibition of tyrosinase due to rapid achievement of equilibrium states.
Our study had several principal findings: (1) D-(−)-arabinose binding to tyrosinase caused a mixed-type of inhibition; (2) D-(−)-arabinose inhibition of tyrosinase caused gross changes in tertiary structure, detected as unfolding; (3) putative D-(−)-arabinose-binding residues HIS85, HIS259, and HIS263 were predicted using computational simulation. Further molecular dynamics simulation predicted the specific binding site of HIS85, HIS259, and HIS263 for the up- and down-forms of D-(−)-arabinose.
Compared with other inhibitors, D-(−)-arabinose is a poor inhibitor of tyrosinase (Table 1). D-(−)-arabinose is less effective to tyrosinase than L-(−)-arabinose (K i = 0.22 ± 0.07 mM) [15]. Therefore, our study directly provides critical information that simple change of configuration is critical for the inhibition of tyrosinase and the results can provide basical information for the further designing or screening of tyrosinase inhibitors.
Table 1.
Comparison of inhibition constants between D-(−)-arabinose and other inhibitors of mixed-type inhibition for tyrosinase.
| Inhibitor | K i (mM) | Inhibitor | K i (mM) |
|---|---|---|---|
| N-Benzylbenzamides [13] | 1.3 × 10−3 | 5-Methoxysalicylic acid [14] | 5.45 |
| L-(−)-arabinose [15] | 0.22 | Terephthalic acid [16] | 11.01 |
| Isorhamnetin [17] | 0.235 | Isophthalic acid [18] | 17.8 |
| o-Toluic acid [19] | 1.73 | Phthalic acid [20] | 65.84 |
| Oxalic acid [21] | 3.16 | D-(−)-arabinose | 210 |
5. Conclusion
The inhibitory mechanism of D-(−)-arabinose resembles that of a copper chelator but differs in that D-(−)-arabinose does not directly bind to the copper atoms in the active site. Our study provides fresh insight into the role of active site residues in tyrosinase catalysis, and presents useful information regarding the 3D structure of tyrosinase. Minor change in the configuration of arabinose can lead to major difference of the inhibition effect on tyrosinase. These findings provide important information in the study of tyrosinase inhibitors in the development of hyperpigmentation treatment. A combination of computational modeling and inhibition kinetics may facilitate the testing of potential tyrosinase inhibitors and the prediction of their inhibitory mechanisms.
Authors' Contribution
H.-J. Liu and S. Ji contributed equally to this paper.
Acknowledgments
Dr. Y.-L. Wang was supported by funds from the Science and Technology Bureau of Jiaxing, Zhejiang (Grants 2009AZ1027 and 2011AZ1027). Dr. J. Lee was supported by a grant from Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program and the Korean Ministry of Education, Science and Technology (MEST) under Grant 2012R1A1A2002676. Dr. J.-M. Yang was supported by a Grant of the Korea Health 21 R&D Project (Ministry of Health, Welfare and Family Affairs, Republic of Korea, 01-PJ3-PG6-01GN12-0001) and a Grant from Samsung Biomedical Research Institute (GL1-B2-181-1). Dr. Hai-Meng Zhou was supported by a Grant from the 624 project supported by Zhejiang leading team of Science & Technology innovation (Team no. 2010R50019).
Abbreviations
- DOPA:
3,4-Dihydroxyphenylalanine
- ANS:
1-Anilinonaphthalene-8-sulfonate.
References
- 1.Jimbow K, Park JS, Kato F, et al. Assembly, target-signaling and intracellular transport of tyrosinase gene family proteins in the initial stage of melanosome biogenesis. Pigment Cell Research. 2000;13(4):222–229. doi: 10.1034/j.1600-0749.2000.130403.x. [DOI] [PubMed] [Google Scholar]
- 2.Olivares C, Solano F. New insights into the active site structure and catalytic mechanism of tyrosinase and its related proteins. Pigment Cell and Melanoma Research. 2009;22(6):750–760. doi: 10.1111/j.1755-148X.2009.00636.x. [DOI] [PubMed] [Google Scholar]
- 3.Kim YJ, Uyama H. Tyrosinase inhibitors from natural and synthetic sources: structure, inhibition mechanism and perspective for the future. Cellular and Molecular Life Sciences. 2005;62(15):1707–1723. doi: 10.1007/s00018-005-5054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rescigno A, Sollai F, Pisu B, Rinaldi A, Sanjust E. Tyrosinase inhibition: general and applied aspects. Journal of Enzyme Inhibition and Medicinal Chemistry. 2002;17(4):207–218. doi: 10.1080/14756360210000010923. [DOI] [PubMed] [Google Scholar]
- 5.Lai SC, Chen CC, Hou RF. Immunolocalization of prophenoloxidase in the process of wound healing in the mosquito Armigeres subalbatus (Diptera: Culicidae) Journal of Medical Entomology. 2002;39(2):266–274. doi: 10.1603/0022-2585-39.2.266. [DOI] [PubMed] [Google Scholar]
- 6.Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta . Immunological Reviews. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 7.Guerrero A, Rosell G. Biorational approaches for insect control by enzymatic inhibition. Current Medicinal Chemistry. 2005;12(4):461–469. doi: 10.2174/0929867053363126. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Wang Y, Jiang H, Deng J. Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(40):17002–17006. doi: 10.1073/pnas.0906095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon J, Fujii S, Solomon EI. Geometric and electronic structure differences between the type 3 copper sites of the multicopper oxidases and hemocyanin/tyrosinase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(16):6585–6590. doi: 10.1073/pnas.0902127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decker H, Tuczek F. Tyrosinase/catecholoxidase activity of hemocyanins: structural basis and molecular mechanism. Trends in Biochemical Sciences. 2000;25(8):392–397. doi: 10.1016/s0968-0004(00)01602-9. [DOI] [PubMed] [Google Scholar]
- 11.García-Molina F, Muñoz JL, Varón R, Rodríguez-López JN, García-Cánovas F, Tudela J. A review on spectrophotometric methods for measuring the monophenolase and diphenolase activities of tyrosinase. Journal of Agricultural and Food Chemistry. 2007;55(24):9739–9749. doi: 10.1021/jf0712301. [DOI] [PubMed] [Google Scholar]
- 12.Chang TS. An updated review of tyrosinase inhibitors. International Journal of Molecular Sciences. 2009;10(6):2440–2475. doi: 10.3390/ijms10062440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho SJ, Roh JS, Sun WS, Kim SH, Park KD. N-Benzylbenzamides: a new class of potent tyrosinase inhibitors. Bioorganic and Medicinal Chemistry Letters. 2006;16(10):2682–2684. doi: 10.1016/j.bmcl.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JP, Chen QX, Song KK, Xie JJ. Inhibitory effects of salicylic acid family compounds on the diphenolase activity of mushroom tyrosinase. Food Chemistry. 2006;95(4):579–584. [Google Scholar]
- 15.Hu WJ, Yan L, Park D, et al. Kinetic, structural and molecular docking studies on the inhibition of tyrosinase induced by arabinose. International Journal of Biological Macromolecules. 2012;50(3):694–700. doi: 10.1016/j.ijbiomac.2011.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Yin SJ, Si YX, Chen YF, et al. Mixed-type inhibition of tyrosinase from agaricus bisporus by terephthalic acid: computational simulations and kinetics. Protein Journal. 2011;30(4):273–280. doi: 10.1007/s10930-011-9329-x. [DOI] [PubMed] [Google Scholar]
- 17.Si YX, Wang ZJ, Park D, et al. Effects of isorhamnetin on tyrosinase: inhibition kinetics and computational simulation. Bioscience Biotechnology Biochemistry. 2012;76(6):1091–1097. doi: 10.1271/bbb.110910. [DOI] [PubMed] [Google Scholar]
- 18.Si YX, Yin SJ, Park D, et al. Tyrosinase inhibition by isophthalic acid: kinetics and computational simulation. International Journal of Biological Macromolecules. 2011;48(4):700–704. doi: 10.1016/j.ijbiomac.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Huang XH, Chen QX, Wang Q, et al. Inhibition of the activity of mushroom tyrosinase by alkylbenzoic acids. Food Chemistry. 2006;94(1):1–6. [Google Scholar]
- 20.Adrio JL, Demain AL. Genetic improvementof processes yielding microbial products. FEMS Microbiology Reviews. 2006;30(2):187–214. doi: 10.1111/j.1574-6976.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- 21.Yan L, Yin SJ, Park D, et al. Computational simulations integrating inhibition kinetics of tyrosinase by oxalic acid. International Research of Pharmacy and Pharmacology. 2011;1(6):126–135. [Google Scholar]
- 22.Gou L, Lü ZR, Park D, et al. The effect of histidine residue modification on tyrosinase activity and conformation: inhibition kinetics and computational prediction. Journal of Biomolecular Structure and Dynamics. 2008;26(3):395–401. doi: 10.1080/07391102.2008.10507254. [DOI] [PubMed] [Google Scholar]
- 23.Lü ZR, Shi L, Wang J, et al. The effect of trifluoroethanol on tyrosinase activity and conformation: inhibition kinetics and computational simulations. Applied Biochemistry and Biotechnology. 2010;160(7):1896–1908. doi: 10.1007/s12010-009-8730-9. [DOI] [PubMed] [Google Scholar]
- 24.Tran MN, Lee IS, Ha DT, Kim HJ, Min BS, Bae KH. Tyrosinase-inhibitory constituents from the twigs of Cinnamomum cassia . Journal of Natural Products. 2009;72(6):1205–1208. doi: 10.1021/np900031q. [DOI] [PubMed] [Google Scholar]
- 25.Kang J, Kim YM, Kim N, Kim DW, Nam SH, Kim D. Synthesis and characterization of hydroquinone fructoside using Leuconostoc mesenteroides levansucrase. Applied Microbiology and Biotechnology. 2009;83(6):1009–1016. doi: 10.1007/s00253-009-1936-5. [DOI] [PubMed] [Google Scholar]
- 26.Marín-Zamora ME, Rojas-Melgarejo F, García-Cánovas F, García-Ruiz PA. Effects of the immobilization supports on the catalytic properties of immobilized mushroom tyrosinase: a comparative study using several substrates. Journal of Biotechnology. 2007;131(4):388–396. doi: 10.1016/j.jbiotec.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Delogu G, Podda G, Corda M, Fadda MB, Fais A, Era B. Synthesis and biological evaluation of a novel series of bis-salicylaldehydes as mushroom tyrosinase inhibitors. Bioorganic and Medicinal Chemistry Letters. 2010;20(20):6138–6140. doi: 10.1016/j.bmcl.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Yi W, Cao R, Peng W, et al. Synthesis and biological evaluation of novel 4-hydroxybenzaldehyde derivatives as tyrosinase inhibitors. European Journal of Medicinal Chemistry. 2010;45(2):639–646. doi: 10.1016/j.ejmech.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Kanade SR, Suhas VL, Chandra N, Gowda LR. Functional interaction of diphenols with polyphenol oxidase: molecular determinants of substrate/inhibitor specificity. FEBS Journal. 2007;274(16):4177–4187. doi: 10.1111/j.1742-4658.2007.05944.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim D, Park J, Kim J, et al. Flavonoids as mushroom tyrosinase inhibitors: a fluorescence quenching study. Journal of Agricultural and Food Chemistry. 2006;54(3):935–941. doi: 10.1021/jf0521855. [DOI] [PubMed] [Google Scholar]
- 31.Shiino M, Watanabe Y, Umezawa K. Synthesis and tyrosinase inhibitory activity of novel N-hydroxybenzyl-N-nitrosohydroxylamines. Bioorganic Chemistry. 2003;31(2):129–135. doi: 10.1016/S0045-2068(03)00026-9. [DOI] [PubMed] [Google Scholar]
- 32.Yan Q, Cao R, Yi W, et al. Synthesis and evaluation of 5-benzylidene(thio)barbiturate-β-D-glycosides as mushroom tyrosinase inhibitors. Bioorganic and Medicinal Chemistry Letters. 2009;19(15):4055–4058. doi: 10.1016/j.bmcl.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 33.Le Guilloux V, Schmidtke P, Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10, article 168 doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trott O, Olson AJ. Software news and update AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brooks BR, Brooks C, III, Mackerell A, Jr., et al. CHARMM: the biomolecular simulation program. Journal of Computational Chemistry. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Si YX, Wang ZJ, Park D, et al. Effect of hesperetin on tyrosinase: inhibition kinetics integrated computational simulation study. International Journal of Biological Macromolecules. 2012;50(1):257–262. doi: 10.1016/j.ijbiomac.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Xie MX, Xu XY, Wang YD. Interaction between hesperetin and human serum albumin revealed by spectroscopic methods. Biochimica et Biophysica Acta. 2005;1724(1-2):215–224. doi: 10.1016/j.bbagen.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Gąsowska B, Kafarski P, Wojtasek H. Interaction of mushroom tyrosinase with aromatic amines, o-diamines and o-aminophenols. Biochimica et Biophysica Acta. 2004;1673(3):170–177. doi: 10.1016/j.bbagen.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Khatib S, Nerya O, Musa R, Shmuel M, Tamir S, Vaya J. Chalcones as potent tyrosinase inhibitors: the importance of a 2,4-substituted resorcinol moiety. Bioorganic and Medicinal Chemistry. 2005;13(2):433–441. doi: 10.1016/j.bmc.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Muñoz-Muñoz JL, Garcia-Molina F, Varon R, et al. Suicide inactivation of the diphenolase and monophenolase activities of tyrosinase. IUBMB Life. 2010;62(7):539–547. doi: 10.1002/iub.348. [DOI] [PubMed] [Google Scholar]
- 41.Espín JC, Varón R, Fenoll LG, et al. Kinetic characterization of the substrate specificity and mechanism of mushroom tyrosinase. European Journal of Biochemistry. 2000;267(5):1270–1279. doi: 10.1046/j.1432-1327.2000.01013.x. [DOI] [PubMed] [Google Scholar]
- 42.Muñoz-Muñoz JL, Garcia-Molina F, Garcia-Ruíz PA, et al. Phenolic substrates and suicide inactivation of tyrosinase: kinetics and mechanism. Biochemical Journal. 2008;416(3):431–440. doi: 10.1042/BJ20080892. [DOI] [PubMed] [Google Scholar]


