Abstract
Microcin B17 (MccB17) is a 3.1-kDa Escherichia coli antibiotic that contains thiazole and oxazole heterocycles in a peptide backbone. MccB17 inhibits its cellular target, DNA gyrase, by trapping the enzyme in a complex that is covalently bound to double-strand cleaved DNA, in a manner similar to the well-known quinolone drugs. The identification of gyrase as the target of MccB17 provides an opportunity to analyze the relationship between the structure of this unusual antibiotic and its activity. In this report, steady-state parameters are used to describe the induction of the cleavable complex by MccB17 analogs containing modified bisheterocyclic sites. The relative potency of these analogs corresponds to the capacity of the compounds to prevent growth of sensitive cells. In contrast to previously reported experiments, inhibition of DNA gyrase supercoiling activity by wild-type MccB17 also was observed. These results suggest that DNA gyrase is the main intracellular target of MccB17. This study probes the structure-function relationship of a new class of gyrase inhibitors and demonstrates that these techniques could be used to analyze compounds in the search for clinically useful antibiotics that block DNA gyrase.
Microcins are a class of small antibiotics (<10 kDa) produced and excreted by some strains of Enterobacteriaceae at the stationary phase of growth (1). Microcin B17 (MccB17) is a ribosomally synthesized compound that is produced in Escherichia coli harboring the plasmid-borne mcb operon, which contains the genes necessary for antibiotic synthesis, dedicated export, and immunity (2). The production of mature MccB17 (Fig. 1) involves posttranslational modification of a 69-residue polypeptide-precursor to generate two thiazoles, two oxazoles, and two 4,2-fused bisheterocycles (3, 4), followed by cleavage of the 26-residue leader sequence by an unidentified protease (5, 6). Exposure of sensitive bacteria to MccB17 results in inhibition of DNA synthesis, induction of the SOS response in cells with active replication, mutagenic effects, and DNA degradation (7). These observations suggest that MccB17 is either a DNA-damaging agent or targets a key DNA-processing pathway such as replication.
Figure 1.
Structure of MccB17 and the analogs. The two bisheterocyclic sites that were altered in the MccB17 analogs used in this study are labeled A and B and the changes made at those sites are drawn below each one. The serine residue that is heterocyclized in the MccB17Δ+1 analog is noted with an *, and the resulting third bisheterocyclic site is drawn at the top.
Genetic experiments designed to identify the target of MccB17 resulted in the isolation and characterization of a mutation in the B subunit of DNA gyrase (8). Strains carrying this mutation, a Trp-Arg substitution at position 751, exhibited increased resistance to MccB17 and a decrease in MccB17-induced DNA cleavage. Gyrase is a prokaryotic type II topoisomerase that introduces negative supercoils into DNA (9, 10). During the reaction, the enzyme passes through a transient intermediate in which two tyrosine hydroxyl groups form phosphodiester bonds with the exposed 5′ phosphates of double-strand cleaved DNA. This intermediate allows the enzyme to pull the ends of the DNA break apart to form a gate through which another DNA segment is passed. The quinolone family of drugs traps this covalent intermediate, known as the cleavable complex, and the accumulation of this complex is monitored in vitro by treatment with SDS and proteinase K, which releases linear DNA (11–13). The ternary complex blocks passage of the replication fork (14, 15), which may initiate the bactericidal activity of the quinolones, although the exact irreversible lethal event has not been firmly established (16). Recent in vitro analysis of the interaction between gyrase and MccB17 revealed that this antibiotic also causes accumulation of the cleavable complex in a manner similar to that of the quinolone drugs (17).
Gyrase is the target of several classes of clinically successful drugs, including the quinolones and the coumarins (11, 13); however, the emergence of resistant strains is an incentive to elucidate the mechanism of action of existing drugs and develop new candidates. The identification of gyrase as a target of MccB17 provides an opportunity to analyze the structure-function relationship of this unusual antibiotic. The experiments described here investigate the effects of modifying MccB17 at the sites of the tandem heterocycles, labeled A and B (Fig. 1). The interaction of MccB17 analogs with DNA gyrase was evaluated by measuring the rate of formation of the cleavable complex, providing information about the role of the bis-heterocyclic sites in the mechanism of gyrase inhibition. Furthermore, the ability of MccB17 to inhibit the negative supercoiling activity of gyrase was examined. These studies of MccB17 will not only lead to a better understanding of the mechanism of action of gyrase but may direct the design of a new class of antitopoisomerase agents.
Materials and Methods
Materials.
Acetylated BSA, spermidine, wheat germ tRNA, proteinase K, and ATP were purchased from Sigma. E. coli DH5α cells were purchased from GIBCO/BRL. The pH of Tris buffers was adjusted with HCl at room temperature, and all aqueous solutions were prepared with water that was deionized on a Milli-Q water system (Millipore). Relaxed plasmid pBR322 DNA was a gift from A. J. Howells (John Innes Centre). DNA gyrase A protein (GyrA) and DNA gyrase B protein (GyrB) were prepared as described (18). Ciprofloxacin was a generous gift from Bayer, and stocks were prepared in water.
Biosynthesis of MccB17 Analogs.
The pUC19-mccB17 plasmids that contain modifications in the mcbA gene and the methods of preparation of MccB17 and the analogs have been described (19). Purified compounds were lyophilized to yield white solids and analyzed by matrix-assisted laser desorption ionization/time of flight MS. The yields and masses from 6 liters were: 60 mg wild-type MccB17, Mobs = 3093.87, Mcalc = 3094.04; 5 mg MccB17Δ+1, Mobs = 3074.03, Mcalc = 3074.01; 25 mg A-GCC, Mobs = 3110.84, Mcalc = 3110.01; 18 mg B-GCC, Mobs = 3110.64, Mcalc = 3110.01; 18 mg B-GCG, Mobs = 3085.06, Mcalc = 3084.05; 5 mg B-GGG, Mobs = 3059.35, Mcalc = 3058.09. Because the solids contained significant amounts of water, the concentrations of MccB17 and analog stocks were determined in the following manner. MccB17 stocks were prepared in DMSO, and aliquots (1 μl) were dried in a Speedvac (Savant) and redissolved in 50% acetonitrile and water (500 μl). The absorbance at 254 nm was measured, and the concentrations of the stocks were calculated by using the published normalized absorbance unit (NAU) values. These values are based on the number of rings in each analog and were measured in conjunction with quantitative amino acid analysis (19). The concentration of the MccB17Δ+1 stock was calculated by using the NAU of wild-type MccB17 and adjusting it to the 9-rings of MccB17Δ+1.
Cleavage Assays.
GyrA and GyrB were added to solutions containing 35 mM Tris⋅HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 1.8 mM spermidine, 6.5% glycerol, 0.36 mg/ml BSA, 9 μg/ml tRNA, 5 mM DTT, 1.4 mM ATP, and 3.5 nM relaxed pBR322 DNA. Formal gyrase concentrations were 120 nM GyrA and 120 nM GyrB (or 60 nM A2B2 dimer). The amount of active enzyme for A and B, however, is not known. GyrB is the limiting component in this setup (data not shown). The reactions also contained MccB17 or an analog at varying concentrations, and the amount of DMSO was kept constant at 6.7%. The reactions were incubated at 5°C, and at each time point 15-μl aliquots were quenched with 0.2% SDS and 0.3 mg/ml proteinase K. Quenched aliquots were kept on ice until the experiment was completed, at which time they were incubated at 37°C for an hour. An equal volume of chloroform/isoamyl alcohol (24:1) and a half volume of loading buffer (40% sucrose/100 mM Tris⋅HCl, pH 7.5/1 mM EDTA/2 mg/ml bromophenol blue) were added, and the mixtures then were vortexed and centrifuged for 5 min at 13,000 rpm. The aqueous phase was loaded onto 1% agarose, TAE (40 mM Tris-acetate, 1 mM EDTA) gels that contained 1 μg/ml ethidium bromide and were run at 25 V overnight in TAE containing 1 μg/ml ethidium bromide. The gels were washed with multiple changes of TAE and then the data were quantitated on an AlphaImager 2200 (Alpha Innotech, San Leandro, CA). The amount of cleaved DNA was expressed as a percentage of total DNA in each lane. The data and errors listed in Table 1 were calculated by fitting the initial rates to the Michaelis–Menten equation with kaleidagraph (Synergy Software, Reading, PA). The KM(app) and kcat(app) are the Michaelis–Menten constants measured by using these experimental conditions. Larger errors cannot be excluded because the limited solubility of MccB17 analogs (and limited amounts available for B-GGG) precluded measurements well above KM(app), which would be necessary to determine steady-state parameters with more accuracy.
Table 1.
Michaelis–Menten parameters for the formation of a cleavable complex induced by MccB17 and analogs
| Analog | # Rings | Bioefficacy* | vmax(app), μM/min (×10−3) | kcat(app), min−1 | KM(app), μM | kcat/KM, μM−1⋅min−1 |
|---|---|---|---|---|---|---|
| MccB17 Δ+1 | 9 | 100 | 0.7 ± 0.2 | 0.012 ± 0.003 | 19 ± 8 | 0.0006 |
| MccB17 | 8 | 65 | 1.0 ± 0.1 | 0.017 ± 0.002 | 50 ± 7 | 0.0003 |
| A-GCC | 8 | 60 | 0.9 ± 0.3 | 0.015 ± 0.005 | 44 ± 31 | 0.0003 |
| B-GCC | 8 | 20 | 0.4 ± 0.1 | 0.007 ± 0.002 | 90 ± 36 | 0.00008 |
| B-GCG | 7 | 5 | — | — | >100 | |
| B-GGG | 6 | 0 | — | — | ≫30 |
The bioefficacy data correspond to the maximum growth inhibition obtained on a lawn of sensitive ZK4 cells and were normalized to the MccB17 Δ+1 analog (19).
Supercoiling Assays.
The supercoiling assays were performed at 5°C in the same manner as the cleavage assays except for the following changes. Samples were quenched with loading buffer containing 0.1 M EDTA, and the samples were incubated at 24°C for 30 min before the addition of chloroform/isoamyl alcohol (24:1). Alternatively, reactions were run at 37°C with the conditions described for the cleavage assay except that 2 mM ATP, 20 nM gyrase, and 14 nM pBR322 DNA was used. Reactions were stopped by the addition of an equal volume of chloroform/isoamyl alcohol (24:1) and a half volume of loading buffer containing 0.1 M EDTA. The reactions were analyzed on a 1% agarose TAE gel, which was stained for about 30 min in TAE containing 1 μg/ml ethidium bromide followed by destaining in multiple washes of TAE and quantitation as described above. The data were fitted to a single exponential equation.
Results
Steady-State Parameters for the Formation of the Cleavable Complex.
The treatment of gyrase-DNA reactions with MccB17 followed by SDS and proteinase K previously revealed gyrase-dependent accumulation of linear DNA, attributed to trapping of the cleavable complex by the antibiotic during supercoiling of closed-circular DNA (17). However, under the previous experimental conditions used, the initial rate of formation of this intermediate was too fast to measure with the standard in vitro cleavage assay. Therefore, reactions in this study were incubated at 5°C instead of 37°C, which slowed down the reaction ≈9-fold (data not shown) and permitted quantitation of the initial rate of linear DNA formation. All of the cleavage assays described below were performed under these conditions.
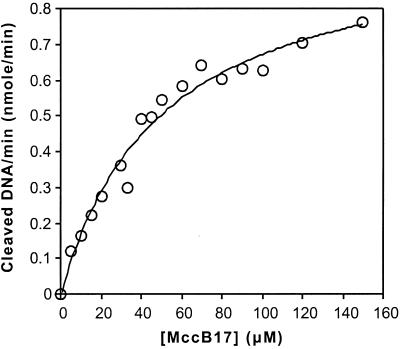
Experiments were performed with wild-type MccB17. Gyrase was incubated with relaxed DNA and the indicated concentrations of MccB17 (Fig. 2). Aliquots were removed, treated with Proteinase K, and analyzed for linear DNA. The formation of linear DNA over the first 3 min of each reaction was monotonic, with an R coefficient >0.99 for MccB17 concentrations up to 100 μM (data not shown). The plot of the initial rate of linear DNA formation versus MccB17 concentrations was fitted to a standard Michaelis–Menten model (Fig. 2), revealing a KM(app) of 50 μM and a kcat(app) of 0.017 min−1.
Figure 2.
DNA cleavage induced by MccB17. DNA gyrase was incubated with relaxed DNA and the indicated concentrations of MccB17 at 5°C as described in Materials and Methods. The amount of DMSO in each reaction was constant. For each concentration, aliquots were removed at 1, 2, and 3 min, quenched with SDS and Proteinase K, and extracted with chloroform. The DNA was resolved on 1% agarose gels containing 1 μg/ml ethidium bromide. The initial rate of cleavage at each concentration was determined and the data were fitted to a Michaelis–Menten equation with KM(app) = 50 ± 7 μM, Vmax(app) = 1.0 × 10−3 μM⋅min−1.
Inhibition of Gyrase by MccB17 Analogs.
To test whether modifications in the MccB17 structure affect its interaction with gyrase, cleavage assays were performed with several different analogs containing variations in the bisheterocycle sites A and B (Fig. 1). The A-GCC analog was produced by changing the serine residue in the A-site G39-S40-C41 sequence to a cysteine, resulting in a bis-thiazole. Similarly, B-GCC was produced by changing the serine residue in the B-site G54-C55-S56 to a cysteine. To produce the B-GCG and B-GGG analogs, one or both of the heterocycles in the B site was removed by replacing the appropriate residues with glycine. The final compound analyzed in these experiments, MccB17Δ+1, is a naturally occurring minor variant of MccB17 in which an additional serine residue, S52, is processed to produce a ninth heterocycle, resulting in a third 4,2-linked bisheterocycle (19).
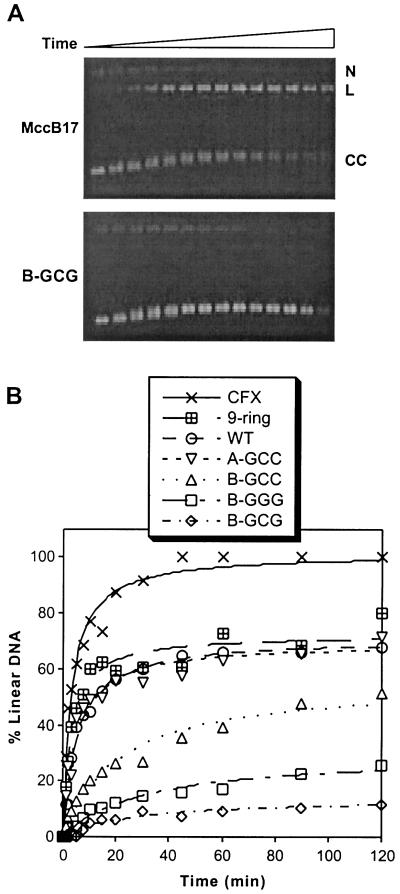
All of the MccB17 analogs cause accumulation of the covalent complex with increasing time, as indicated by the production of linear DNA. Representative gels for time-course experiments performed with MccB17 and the B-GCG analog are shown in Fig. 3A, and the quantitation of experiments for all of the MccB17 analogs is shown in Fig. 3B. Although the concentrations of the compounds used were kept constant, both the final amount of DNA cleavage observed as well as the time scale of its appearance varied with each analog. The most potent compounds were the MccB17Δ+1 analog, wild-type MccB17, and the A-GCC analog, whereas the least potent compounds were the ones with modifications to the B site. In comparison, experiments also were performed with ciprofloxacin, which was much more potent than any of the MccB17 analogs, with faster reaction times and a higher amount of cleavage at saturation. This result is consistent with our previously reported experiments (17).
Figure 3.
Time courses of DNA cleavage induced by MccB17 analogs. (A) DNA gyrase was incubated with relaxed DNA and 33 μM MccB17 (Upper) or B-GCG (Lower) at 5°C as described in Materials and Methods. At each time point, from 0 to 120 min, aliquots were treated as described in Fig. 2. The nicked-circular (N), linear (L), or closed circular (CC) DNA are indicated. (B) DNA gyrase was incubated with relaxed DNA and 33 μM MccB17, an analog, or ciprofloxacin (CFX) as described for A, and the amount of linear DNA was quantitated and plotted versus time. WT, wild type.
To quantify the effects of modifying the MccB17 structure on gyrase inhibition, we determined the steady-state parameters of MccB17-induced DNA cleavage (Fig. 4 and Table 1). The time points used to determine the initial rates of linear DNA formation were adjusted for each analog to ensure that the data were in the linear range of the reaction and the amount of cleaved DNA was large enough to quantify. The solubility of the analogs provided a constraint on the concentrations that could be used in the reactions and prohibited the determination of rate constants for the least potent compounds B-GCG and B-GGG. Furthermore, this assay is limited by the minimum amount of signal that could be detected above background, which we estimated to be about 5% of the DNA. Nevertheless, the trend is quite clear. The relative potencies characterized by KM(app) and kcat/KM reflect the bioefficacy of the compounds in inhibition of cell growth (Table 1). The KM(app) values vary between 19 μM and >100 μM and kcat(app) values between 0.017 and 0.007 min−1. Larger effects are revealed by the second-order rate constant kcat/KM, which shows a 7-fold difference between the MccB17Δ+1 and B-GCC analogs, compared with their 5-fold difference in bioefficacy (19).
Figure 4.
Michaelis–Menten curves of DNA cleavage induced by MccB17 analogs. DNA gyrase was incubated with relaxed DNA and the indicated concentrations of MccB17 or an analog at 5°C as described in Materials and Methods. The amount of DMSO in each reaction was constant. Aliquots were treated as described in Fig. 2. The rate of cleavage at each concentration was determined and the data were fitted to a Michaelis–Menten equation. The kinetic constants are summarized in Table 1. WT, wild type.
Inhibition of Gyrase Supercoiling Activity.
If MccB17 traps an intermediate in the gyrase-catalyzed supercoiling reaction then inhibition of this activity should be observable. We previously found that when MccB17 causes accumulation of cleaved DNA under normal reaction conditions, no inhibition of supercoiling was observed (17). One possible explanation for this result is that MccB17 acts inefficiently and a number of enzyme turnovers occur before the antibiotic stabilizes the cleaved complex, allowing the DNA to become supercoiled. In this work, two different reaction conditions were used to slow down the conversion of relaxed substrate to supercoiled product so that it more closely approximates the time required for MccB17-promoted formation of the cleavable complex.
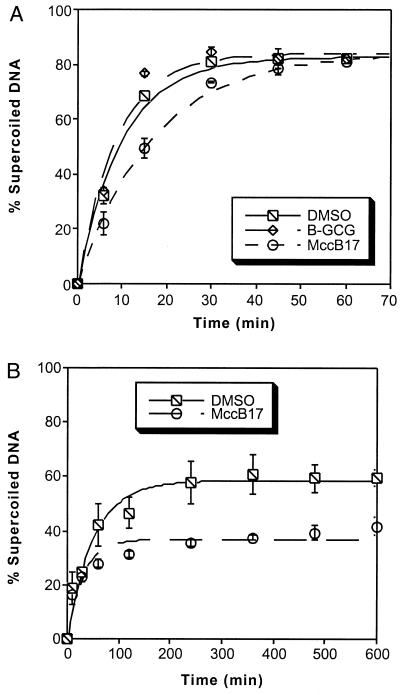
In the first set of reactions, gyrase was incubated at 5°C with relaxed DNA and 33 μM MccB17 or the corresponding amount of DMSO (Fig. 5A). Aliquots were removed at the indicated time points and analyzed on an agarose gel in the absence of ethidium bromide, and the amount of supercoiled DNA was quantitated. Under these conditions, MccB17 caused a 2-fold decrease in the rate of the supercoiling reaction. The same experiment also was performed with one of the least potent analogs as determined by the cleavage assays. Incubation of gyrase with 33 μM B-GCG did not produce any significant inhibition of supercoiling. As an alternative method of slowing down the gyrase reaction (Fig. 5B), the reactions were performed at 37°C but the amount of enzyme used was decreased and the amount of DNA was increased from the standard reaction conditions previously reported (17). Under these conditions, MccB17 causes premature cessation of the enzyme activity, producing a difference in the endpoint levels of supercoiled DNA.
Figure 5.
MccB17 inhibits DNA supercoiling. (A) DNA gyrase (60 nM) was incubated with 3.5 nM relaxed DNA and 33 μM MccB17, B-GCG, or the corresponding amount of DMSO at 5°C as described in Materials and Methods. Each point is the average of two experiments. The data were fitted to a single exponential equation. The half-lives of the supercoiling reactions were 6.9 ± 0.7 min for the DMSO control, 6.1 ± 0.9 min for B-GCG, and 11.6 ± 0.8 min for MccB17. (B) DNA gyrase (20 nM) was incubated with 14 nM relaxed DNA and 35 μM MccB17 or the corresponding amount of DMSO at 37°C as described in Materials and Methods. Each point is the mean of three experiments. Error bars represent ± 1 SD.
Discussion
Steady-State Analysis of Cleavable Complex Induced by MccB17 Analogs.
Based on the genetic evidence that the intracellular target of MccB17 is DNA gyrase (8), our previous study examined the interaction between MccB17 and the enzyme (17). These experiments demonstrated that MccB17 traps a covalently bound gyrase intermediate on double-strand cleaved DNA, and the IC50 values determined under equilibrium conditions at 37°C were reported (17). These measurements can provide some information about structure-activity relationships of gyrase poisons but they do not reflect a kinetic process and thus do not provide details about the mechanism of inhibition. In an effort to understand the mechanism of action of MccB17, the present investigation uses initial rate measurements to examine the relation between the structural motifs of MccB17 and the antigyrase activity. Slowing the reaction down by decreasing the temperature to 5°C revealed kinetic differences between the MccB17 analogs that may be masked in the comparison of the IC50 values. At this point, it is not clear to which gyrase-DNA intermediate MccB17 binds or how it traps the cleavable complex. Thus the apparent kinetic constants reported here may be a compilation of rates from multiple steps.
Several MccB17 analogs that span the spectrum of bioefficacy, defined as quantitative growth inhibition of sensitive cells (19), were examined. The relative potency of these analogs as gyrase poisons correlates with the bioefficacy; a 7-fold decrease in kcat/KM was observed when comparing MccB17 analogs that span a 5-fold decrease in growth inhibition activity. A large part of this effect is due to an increase in the KM(app) for formation of cleaved DNA. One explanation for this observation is that changes in the heterocycle pattern of MccB17 affects binding to the gyrase-DNA complex. This hypothesis accounts for the previously reported effects of the MccB17 analogs on DNA synthesis in permeabilized cells (19). In these experiments, wild-type MccB17 could completely inhibit DNA replication whereas the less toxic B-GCC analog did not cause more than 20% inhibition. Trapping of gyrase as a covalent complex on DNA can block progression of the replication fork (14, 15), and this effect of MccB17 may be the cause of replication inhibition observed in vivo. However, in the case of the B-GCC analog, if the compound has very weak binding and a limited effect on gyrase, then only a small amount of replication inhibition would be observed.
Structure-Reactivity Relationship of MccB17.
The structure of MccB17, aside from the oxazoles and thiazoles, is essentially a glycine linker (3, 4), suggesting that the heterocycles are the important structural determinants of the antibiotic activity. Oxazoles and thiazoles are found in a wide variety of natural products that target a diverse array of cellular molecules (20). For example, the anticancer drug bleomycin has a 4,2-bisthiazole tail that can intercalate into DNA (21) and also may be involved in binding the minor groove (22). By analogy with bleomycin, the bisheterocycles of MccB17 could play a similar role and direct the antibiotic to bind DNA and block gyrase. It is not known when in the gyrase pathway MccB17 binds, or whether it binds to the protein, DNA, or complex. Initial studies with quinolone-resistant mutants suggested that MccB17 interacts with a region of gyrase that overlaps with the quinolone-binding site (17). Quinolones can bind nonspecifically to DNA alone (23), but binding to the DNA-gyrase complex is specific and correlates with inhibition of gyrase (23–25).
Modification to the A site produced a modest change, if any, in the effects of MccB17 in either the in vitro gyrase experiments or the growth inhibition studies (19). No detectable material, even partially modified MccB17 precursors, were produced in vivo during attempts to express analogs with only a single heterocycle at site A (19), suggesting that this site is important for the initial stages of the mature antibiotic production. In contrast, all of the mutants at site B could be expressed and purified, but modifications at this site caused a significant change in potency. These results suggest that the B site plays a larger role in the activity of MccB17. The very low potencies of the B-GCG and B-GGG analogs reveal that the intact bisheterocycle is a major determinant for activity. Furthermore, the decrease in activity produced by changing the B-GCS site of wild-type MccB17 to B-GCC suggests that the composition of this site modulates the interaction with the molecular target. The change from thiazole to oxazole leads to an increase in KM(app) by a factor of 2, corresponding to ≈0.4 kcal/mol for the specific interaction of the oxygen heterocycle if the KM(app) values correlate with the KDs. Based on the observed KM(app)s for MccB17Δ+1 and wild-type MccB17, the additional binding interactions of an extra oxazole is worth a similar amount. The extra oxazole of this minor variant of MccB17 is in a third bisheterocyclic site, and this analog is more potent than any of the other compounds tested, including wild-type MccB17. This result is further evidence that the bisheterocycles are important for the activity of the antibiotic.
Supercoiling Inhibition.
Under the standard reaction conditions, inhibition of the supercoiling activity of gyrase by MccB17 was not detectable (17), suggesting that MccB17 inhibits the enzyme very slowly or inefficiently. Consistent with this hypothesis, inhibition of supercoiling was observed when the reaction was slowed down by decreasing the reaction temperature. MccB17 caused a 2-fold decrease in the rate of the reaction. This result demonstrates that not only does MccB17 trap an intermediate in the reaction pathway but it also can inhibit the overall reaction pathway of this enzyme. In contrast, when the supercoiling reactions were performed by decreasing the enzyme and increasing the amount of DNA substrate, MccB17 inhibition also was observed but it took the form of blocking the enzymatic reaction, reducing the final amount of supercoiled DNA. A similar profile was observed if the reactions performed at 5°C (Fig. 5A) were treated with SDS and Proteinase K (data not shown). These results may suggest that MccB17 can interact with gyrase at two different stages in the reaction pathway. Quinolones bind to the gyrase-DNA complex in the absence of DNA cleavage (26), and the effects of ciprofloxacin can be separated into two components, suggesting that there are two sequential steps in the mechanism of action of quinolones, binding followed by trapping of the covalent complex (27). Further experiments will be required to characterize the mechanism of action of MccB17 and the nature of the complexes formed.
Comparison of in Vitro and in Vivo Data.
The fact that in vitro trapping of the cleavable gyrase complex by MccB17 analogs correlates with the in vivo bioefficacy data suggests that gyrase is the main target of this antibiotic. Quinolones target both gyrase and the related topoiosomerase IV, the other prokaryotic topoisomerase (12). However, preliminary studies suggest that MccB17 does not affect topoisomerase IV (data not shown). Trapping of the cleavable complex was observed with the in vitro gyrase assay with the B-GGG analog but the low level of activity was not enough to produce detectable cell growth inhibition (19). By focusing on the putative target of MccB17, the in vitro gyrase assay provides a more sensitive means of comparing small structural differences between the analogs.
Thus, the experiments reported here start to map out the functional groups involved in the mechanism of action of MccB17 and they suggest that the bisheterocyclic groups are key to blocking gyrase. Additional studies of MccB17 as well as analogs that are modified at the monoheterocyclic sites will provide more information about the mechanism of action of this antibiotic. In particular, this assay could be useful to determine the minimal bisheterocyclic composition and sequence of MccB17 that induces cleaved DNA in the gyrase catalytic cycle. Although in vivo assays will remain the method of choice for drug screening, this targeted in vitro assay also may prove useful for drug development by uncovering more subtle structural variations that are masked under equilibrium conditions or in vivo.
Acknowledgments
We thank Stephen J. Blance for some preliminary experiments. This research was supported by grants from the Wellcome Trust and Biotechnology and Biological Sciences Research Council (to A.M.) and by National Institutes of Health Grant GM 20011 (to C.T.W.). D.B.Z. is a National Institutes of Health Postdoctoral Fellow, F.H. is a Wellcome Trust International Prize Fellow, and J.G.H. was supported by the Biotechnology and Biological Sciences Research Council and the Wellcome Trust.
Abbreviations
- GyrA
DNA gyrase A protein
- GyrB
DNA gyrase B protein
- MccB17
microcin B17
- TAE
Tris-acetate EDTA
References
- 1.Kolter R, Moreno F. Annu Rev Microbiol. 1992;46:141–163. doi: 10.1146/annurev.mi.46.100192.001041. [DOI] [PubMed] [Google Scholar]
- 2.Moreno F, San Millán J L, Hernández-Chico C, Kolter R. In: Genetics and Biochemistry of Antibiotic Production. Vining L C, Stuttard C, editors. Boston: Butterworth–Heinemann; 1995. pp. 307–321. [Google Scholar]
- 3.Yorgey P, Lee J, Kördel J, Vivas E, Warner P, Jebaratnam D, Kolter R. Proc Natl Acad Sci USA. 1994;91:4519–4523. doi: 10.1073/pnas.91.10.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer A, Freund S, Jung G. Eur J Biochem. 1995;234:414–426. doi: 10.1111/j.1432-1033.1995.414_b.x. [DOI] [PubMed] [Google Scholar]
- 5.Yorgey P, Davignino J, Kolter R. Mol Microbiol. 1993;9:897–905. doi: 10.1111/j.1365-2958.1993.tb01747.x. [DOI] [PubMed] [Google Scholar]
- 6.Madison L L, Vivas E I, Li Y-M, Walsh C T, Kolter R. Mol Microbiol. 1997;23:161–168. doi: 10.1046/j.1365-2958.1997.2041565.x. [DOI] [PubMed] [Google Scholar]
- 7.Herrero M, Moreno F. J Gen Microbiol. 1986;132:393–402. doi: 10.1099/00221287-132-2-393. [DOI] [PubMed] [Google Scholar]
- 8.Vizán J L, Hernández-Chico C, del Castillo I, Moreno F. EMBO J. 1991;10:467–476. doi: 10.1002/j.1460-2075.1991.tb07969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reece R J, Maxwell A. CRC Crit Rev Biochem Mol Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- 10.Roca J. Trends Biochem Sci. 1995;20:156–160. doi: 10.1016/s0968-0004(00)88993-8. [DOI] [PubMed] [Google Scholar]
- 11.Maxwell A. Trends Microbiol. 1997;5:102–109. doi: 10.1016/S0966-842X(96)10085-8. [DOI] [PubMed] [Google Scholar]
- 12.Drlica K, Zhao X. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper D C. Clin Infect Dis. 1998;27:S54–S63. doi: 10.1086/514923. [DOI] [PubMed] [Google Scholar]
- 14.Hiasa H, Yousef D O, Marians K J. J Biol Chem. 1996;271:26424–26429. doi: 10.1074/jbc.271.42.26424. [DOI] [PubMed] [Google Scholar]
- 15.Wentzell L M, Maxwell A. J Mol Biol. 2000;304:779–791. doi: 10.1006/jmbi.2000.4266. [DOI] [PubMed] [Google Scholar]
- 16.Drlica K. Curr Opin Microbiol. 1999;2:504–508. doi: 10.1016/s1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 17.Heddle J G, Blance S J, Zamble D B, Hollfelder F, Miller D A, Wentzell L M, Walsh C T, Maxwell A. J Mol Biol. 2001;307:1223–1234. doi: 10.1006/jmbi.2001.4562. [DOI] [PubMed] [Google Scholar]
- 18.Maxwell A, Howells A J. In: Protocols of DNA Topoisomerases I: DNA Topology and Enzyme Purification. Bjornsti M-A, Osheroff N, editors. Towata, NJ: Humana; 1999. pp. 135–144. [Google Scholar]
- 19.Sinha Roy R, Kelleher N L, Milne J C, Walsh C T. Chem Biol. 1999;6:305–318. doi: 10.1016/s1074-5521(99)80076-3. [DOI] [PubMed] [Google Scholar]
- 20.Sinha Roy R, Gehring A M, Milne J C, Belshaw P J, Walsh C T. Nat Prod Rep. 1999;16:249–263. doi: 10.1039/a806930a. [DOI] [PubMed] [Google Scholar]
- 21.Stubbe J, Kozarich J W, Wu W, Vanderwall D E. Acc Chem Res. 1996;29:322–330. [Google Scholar]
- 22.Zuber G, Quada J C J, Hecht S M. J Am Chem Soc. 1998;120:9368–9369. [Google Scholar]
- 23.Shen L L, Baranowski J, Pernet A G. Biochemistry. 1989;28:3879–3885. doi: 10.1021/bi00435a038. [DOI] [PubMed] [Google Scholar]
- 24.Shen L L, Kohlbrenner W E, Weigl D, Baranowski J. J Biol Chem. 1989;264:2973–2978. [PubMed] [Google Scholar]
- 25.Yoshida H, Nakamura M, Bogaki M, Ito H, Kojima T, Hattori H, Nakamura S. Antimicrob Agents Chemother. 1993;37:839–845. doi: 10.1128/aac.37.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Critchlow S E, Maxwell A. Biochemistry. 1996;35:7387–7393. doi: 10.1021/bi9603175. [DOI] [PubMed] [Google Scholar]
- 27.Kampranis S C, Maxwell A. J Biol Chem. 1998;273:22615–22626. doi: 10.1074/jbc.273.35.22615. [DOI] [PubMed] [Google Scholar]