Abstract
B cell development is controlled by a series of checkpoints that ensure that the immunoglobulin (Ig)-encoding genes are assembled in frame to produce a functional B cell receptor (BCR) and antibodies. The BCR consists of Ig proteins in complex with the immunoreceptor tyrosine-based activation motif (ITAM)-containing Igα and Igβ chains. Whereas the activation of Src and Syk tyrosine kinases is essential for BCR signaling, the pathways that act downstream of these kinases are incompletely defined. Previous work has revealed a key role for the p110δ isoform of phosphoinositide 3-kinase (PI3K) in agonist-induced BCR signaling; however, early B cell development and mature B cell survival, which depend on tonic BCR signaling, are not substantially affected by a deficiency in p110δ. Here, we show that in the absence of p110δ, p110α, but not p110β, can compensate to promote early B cell development in the bone marrow and B cell survival in the spleen. In the absence of both p110α and p110δ activities, pre-BCR signaling fails to suppress the production of recombination-activating gene (Rag) protein and to promote developmental progression of B cell progenitors. By contrast, p110α does not contribute to agonist-induced BCR signaling. These studies indicate that either p110α or p110δ can mediate tonic signaling from the BCR, but that only p110δ can contribute to antigen-dependent activation of B cells.
Introduction
B cell development occurs in the bone marrow, where the gradual acquisition of B cell characteristics correlates with the loss of potential for differentiation into other blood cell lineages (1). B cells are defined by the surface expression of the B cell receptor (BCR), which is encoded by rearranged immunoglobulin (Ig) heavy chain (Igh) and Ig light chain (Igκ or Igλ) genes. The Igh locus comprises multiple Variable (V), Diversity (D) and Joining (J) gene segments. First a D segment is joined to a J segment and then a V segment is joined to a DJ segment to form a VDJH recombined Igh gene. Before this can occur, the interleukin-7 receptor (IL-7R) stimulates chromatin changes in the Igh locus rendering it accessible for recombination activating gene (Rag1 and Rag2) proteins that catalyze VDJH recombination (2). If the Igh gene segments are rearranged in-frame, then the Igμ heavy chain forms a pre-BCR in association with the surrogate light chains λ5 and VpreB on the cell surface. After several rounds of division, during which the Rag genes are temporarily turned off, the Igκ or Igλ locus, each of which comprises multiple V and J gene segments, is rearranged to form Igκ or Igλ genes. Igκ or Igκ light chain proteins replace the surrogate light chains to form the mature BCR with the Igμ heavy chain. B cell precursors that lack Rag1, Rag2 or the transmembrane domain of Igμ (μMT) are blocked in their development at the pro-B cell stage (3-5). These observations demonstrate the existence of a developmental checkpoint that only permits pre B cells with in-frame rearranged Igμ heavy chains to develop further. There is increasing evidence that the pre-BCR transmits signals without being clustered by specific agonists (6).
Pre-BCR signaling is initiated by the activation of Src family tyrosine kinases that phosphorylate immunoreceptor tyrosine-based activation motifs (ITAMs) within the invariant Igα and Igβ transmembrane proteins that form a complex both with the pre-BCR and later with the BCR (6). The tyrosine kinase Syk is recruited to phosphorylated Igα and Igβ and it plays an important role in the development of immature B cells in the spleen (7). Together with the related tyrosine kinase ζ chain–associated protein kinase of 70 kD (ZAP-70), Syk is essential for pre-BCR signaling (8). Src homology 2 (SH2) domain–containing leukocyte adaptor protein of 65 kD (SLP-65, also known as BLNK) is an adaptor protein that links Syk to the activation of phospholipase c γ (PLC-γ). SLP-65-deficient pre-B cells are partially blocked at pre-B cell stage of development; however, the pre-B cells continue to proliferate and eventually develop into pre-B tumor cells (9-11). These results implicate additional signals downstream of Syk that are also important for pre-BCR signaling.
Phosphoinositide 3-kinases (PI3Ks) are a family of enzymes that phosphorylate the 3-position of the phosphatidylinositol (PtdIns) ring. Class I PI3Ks use the substrate PtdIns-4,5-bisphosphate (PIP2) to generate PtdIns-3,4,5-trisphosphate (PIP3) (12, 13). PIP3 acts as a membrane tether for proteins such as Akt and Btk in B cells. Akt can stimulate the serine and threonine kinase mammalian target of rapamycin (mTOR ) and suppress Foxo transcription factors, whereas Btk contributes to the activation of PLC-γ. Class I PI3Ks integrate a number of signaling events that are controlled by Syk, because key proteins that are phosphorylated by Syk, including CD19, B-cell adapter for phosphoinositide 3-kinase (BCAP), and the guanine nucleotide exchange factor Vav, contribute to the activation of PI3K as initiated by the pre-BCR or the BCR (14, 15). Syk may also directly regulate the activity of PI3K (16); however, the precise role of PI3K signaling, especially downstream of the pre-BCR is incompletely understood. Tyrosine kinases are linked to the activation of subset of PI3Ks (class IA), which are associated with p85 regulatory subunits that can bind to proteins that contain phosphorylated tyrosine residues. Mammals have three genes, Pik3r1, Pik3r2, and Pik3r3, which encode the class IA PI3K regulatory subunits p85α, p85β. and p55γ respectively. The subunits p55α and p50α are generated by alternative start codon usage from Pik3r1. Pik3ca, Pik3cb, and Pik3cd encode the class IA PI3K catalytic subunits p110α, p110β, and p110δ, respectively, each of which can bind to any of the regulatory subunits.
Studies to date suggest a non-essential role for PI3K in early B cell development. Thus both p85α and p110δ-deficient mice showed near normal B cell development in the bone marrow (17-24). After development in the bone marrow B cells circulate through the blood and lymph and populate the spleen, lymph nodes and plural cavities where they continue their development. There are three main subtypes of mature B cell populations which are defined by their preferred anatomical location and cell surface phenotypes: follicular (FO) B cells, also known as B2 cells, are found in the follicles in the spleen and lymph nodes, marginal zone (MZ) B cells are found in the MZ at the perimeter of the follicles of the spleen, whereas B1 cells are found primarily in the pleural cavities such as the peritoneum. In both p85α-deficient and p110δ-deficient mice there were reduced numbers of follicular B cells, and peritoneal B1 and splenic MZ B cells were almost absent (17-24). Additional studies have shown that “tonic” PI3K signaling is required to suppress the expression of Rag and to promote the survival of immature and mature B cells (25-28). PI3K suppresses the expression of Rag by virtue of its capacity to terminate transcription of Foxo which binds to the promoters of Rag1 and Rag2 (29, 30). Thus, a picture emerged in which p85α recruits p110δ to a BCR-associated signaling complex, and this interaction is essential for the development of B1 and MZ B cells, but is not essential for the development of B cell precursors in the bone marrow and follicular (FO) B cells in the spleen.
Here, we report that similar to what occurs in p110δ-deficient mice, the development of B cells in the bone marrow occurred normally in mice with lymphocytes that lacked either p110α or p110β; however, the combined loss of p110α and p110δ resulted in a near complete block in B cell development at the pre-B cell stage. Single-cell analysis revealed that B cells blocked at this stage contained Rag and had rearranged their heavy chain genes. Moreover, p110α and p110δ doubly-deficient pre-B cells failed to inhibit the expression of Rag in Igμ+ cells. Consequently, we observed increased proportions of pre B cells with both loci rearranged and which contained Igμ and Rag. Thus, p110α, but not p110β, compensated for the lack of p110δ in pre-B cells to provide PI3K activity that was essential for developmental progression; however, antigen-dependent activation of mature B cells was strictly dependent on p110δ.
Results
CD2Cre-p110αfl mice exhibit a normal B cell phenotype
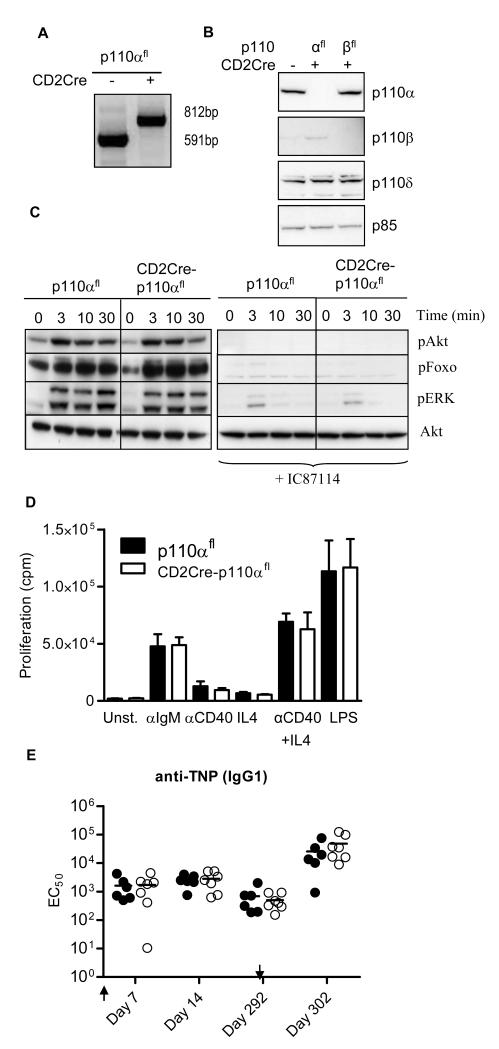
Because germline deletion of p110α causes embryonic lethality, we crossed p110αfl mice, in which exons 18 and 19 of Pik3ca are flanked by loxP sites (31), with CD2-Cre transgenic mice to examine the role of this PI3K isoform in lymphocytes. The CD2 transgene is expressed from the earliest observable stages during B cell and T cell development (32). Polymerase chain reaction (PCR) analysis of genomic DNA obtained from purified B cells revealed that Cre-mediated excision of Pik3ca was complete (Fig. 1A). Similarly, Western blotting analysis revealed a complete loss of p110α protein, suggesting that these were effectively null alleles (Fig. 1B). In addition, CD2-Cre efficiently deleted p110β from p110βfl B cells in which exons 21 and 22 of Pik3cb were flanked by loxP sites (33) (Fig. 1B).
Fig. 1.
Intact PI3K signaling in B cells from CD2Cre-p110αfl mice. (A) PCR analysis of genomic DNA from purified B cells. The 812 kb band is from a recombined allele, whereas the 591 bp band is from an allele with both loxP sites intact. (B) Western blotting analysis of p110α, p110β, p110δ, and p85 from B cells purified from wild-type, CD2Cre-p110αfl, and CD2Cre-p110βfl mice. (C) Western blotting analysis of B cells stimulated with antibody against IgM F(ab’)2 for 3, 10, or 30 min. Blots were incubated with antibodies against pAkt (Ser407), pFoxo1 (Thr24)/Foxo3(Thr32), pERK (Ser202Tyr204), and total Akt. The cells in the panel on the right had been treated with IC87114 (5 μM). These data are representative of three experiments. (D) Proliferation of CD2Cre-p110αfl B cells in response to antibody against IgM F(ab’)2 (αIgM, 10 μg/ml), antibody against CD40 (αCD40,10 μg/ml), IL-4 (20 ng/ml), antibody against CD40 with IL-4, or LPS (10 μg/ml). The cpm represents the amount of 3H thymidine incorporated during the last 6 hours of the 48 hours of culture. The mean values represent averages from 6 mice analyzed in 3 independent experiments. The error bars represent the standard error of the mean (SEM). (E) Specific IgG1 titers after primary and secondary challenge. p110αfl (filled circles) and CD2Cre-p110αfl (open circles) mice were immunized with TNP-KLH on days 0 and day 292 and bled on days 7, 14, 292, and 302. EC50 values were calculated from serially diluted serum samples on TNP-BSA–coated plates. Each dot represents EC50 IgG1 values obtained from one of six mice per group.
Loss of p110α in CD2Cre-p110αfl B cells had no impact on the phosphorylation of Akt induced by antibody against IgM, which instead was completely blocked by the p110δ-specific inhibitor IC87114 (Fig. 1C). Similarly, phosphorylation of Foxo and extracellular signal–regulated kinase (ERK) proteins was selectively ablated by the inhibition of p110δ, whereas the loss of p110α had no obvious effect (Fig. 1C). In contrast to the reduced proliferation of p110δ-deficient B cells (20-22), B cells from CD2Cre-p110αfl mice proliferated normally in response to antibody against IgM, IL-4, CD40, or lipopolysaccharide (LPS) (Fig. 1D). CD2Cre-p110αfl mice produced normal primary and secondary specific antibody titers in response to immunization with a haptenated protein (Fig. 1E). Thus, p110α was dispensable for the development and activation of mature B cells in which p110δ appeared to be both necessary and sufficient to provide optimal PI3K signaling.
Loss of both p110α and p110δ blocks B cell development
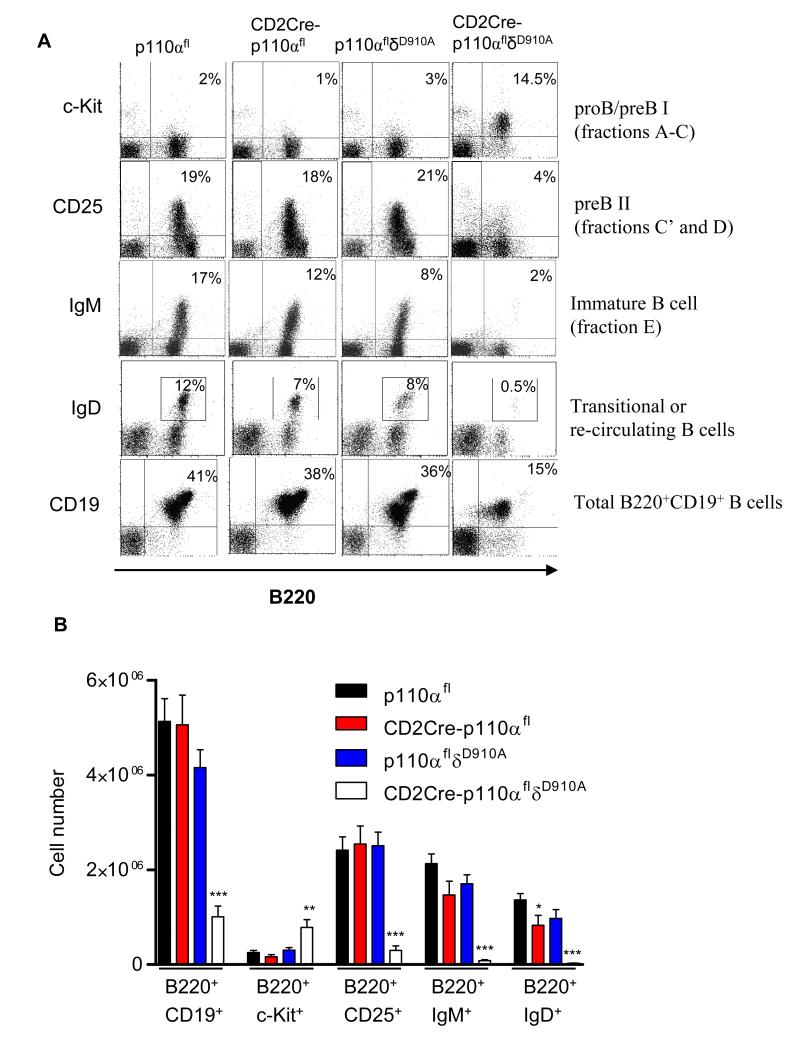
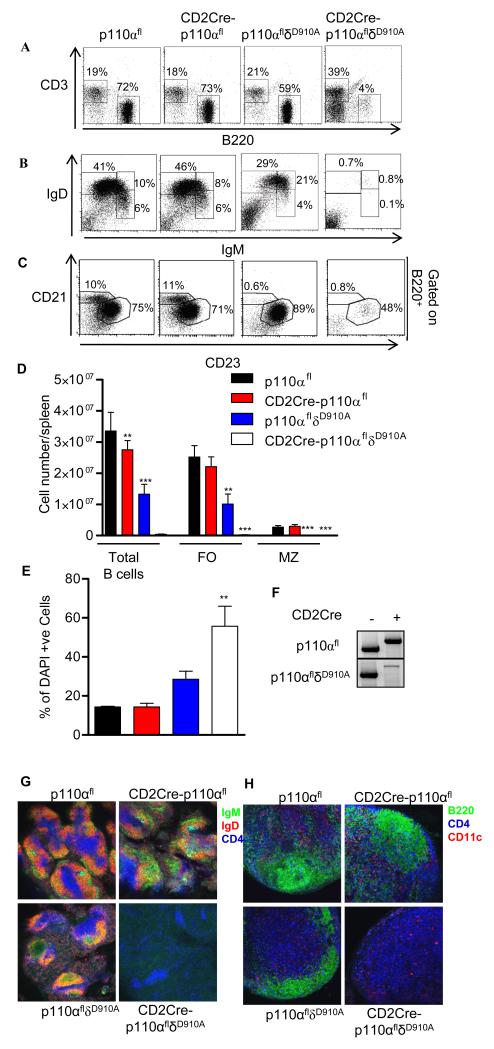
Although p110δ contributes to most of the PI3K activity in mature cells, we considered whether there was a greater extent of redundancy between the p110 isoforms during B cell development in the bone marrow, in which p110δ deficiency alone has minimal effect (20). B cell development in the bone marrow proceeds through several well-defined stages referred to as pro B cells, pre B cells, and immature B cell stages or fractions A to F (1). We crossed p110δD910A mice, in which the p110δ catalytic subunit has been rendered catalytically inactive by point mutation (20), with CD2Cre-p110αfl or CD2Cre-p110βfl mice. Because the loxP sites themselves did not affect the function of p110α or p110β, the p110αfl or p110βfl mice (not expressing Cre) were used as “wild-type controls” and p110αflδD910A or p110βflδD910A mice (that do not express Cre) are referred to in the text simply as p110δD910A. CD2Cre-p110αfl/fl;p110δD910A/D910A mice (hereafter referred to as CD2Cre-p110αflδD910A or p110αβ-deficient mice) showed a profound block at the B220+CD19+cKit+CD25− stage of B cell development, with a substantial loss of CD25+ B cells (Fig. 2, A and B). A high percentage of the CD2Cre-p110αflδD910A B220+ cells were also BP1+ (Fig. 3A), which indicated that development was blocked at the pre-B cell fraction C stage (1). Consequently, the numbers of IgM+B220highCD19high immature B cells and mature B cells were reduced by more than 95% compared to those in p110αfl mice (Fig. 2B). In contrast to these results, CD2Cre-p110βflδD910A mice showed no evidence of impaired B cell development (fig. S1). Therefore, a PI3K-signaling complex containing either p110α or p110δ was essential for early B cell development.
Fig. 2.
B cell development requires the activity of p110α or p110δ. (A) Flow cytometric analysis of lymphocytes from the bone marrow of the indicated mice. The percentages in each quadrant or gate are averages; p110αfl (n = 10 mice), CD2Cre-p110αfl (n = 6 mice), p110δD910A (n = 8 mice), CD2Cre-p110αflδD910A (n = 13 mice). (B) Total numbers (± SEM) of each B cell subset from two femurs of the indicated mice.
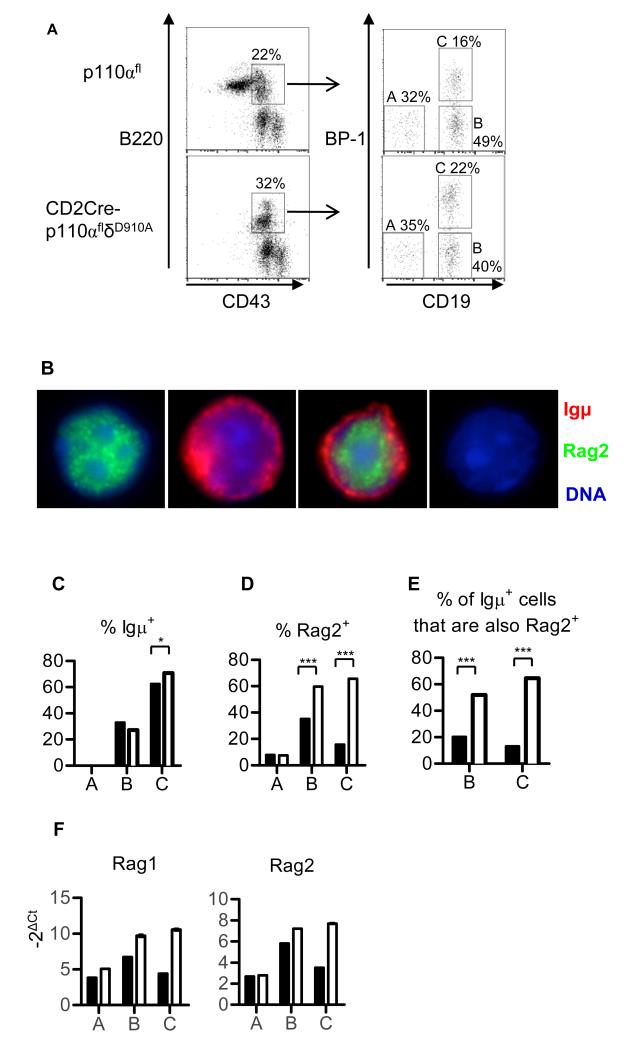
Fig. 3.
PI3K is required for pre-BCR-dependent inhibition of Rag expression. Immunofluorescent analysis of fraction A, B, or C cells purified from WT (p110αfl) or CD2Cre-p110αflδD910A mice. (A) Flow cytometric analysis showing the gating strategy used to sort B220+CD43+ bone marrow cells (depleted of Ter119+, Mac1+, and Gr1+ cells) into fractions A (CD19−BP1−), B (CD19+BP1−), and C (CD19+BP1+). (B) Representative images of cells incubated with antibodies against IgM (red) and Rag2 (green). The first cell imaged contained only Rag2, the second cell contained only Igμ, the third cell contained both proteins, whereas the fourth cell was negative for both proteins. (C to E) Percentages of p110αfl or CD2Cre-p110αflδD910A cells that stained positively for Igμ (C), Rag2 (D), or both Igμ and Rag2 (E); 200 to 500 cells from one experiment were scored per condition. (F) Gene-specific quantitative, real-time, reverse transcription PCR (QRT-PCR) analysis of cDNA prepared from fraction A, B, and C cells from p110αfl and CD2Cre-p110αflδD910A mice. ΔCt=Ct(Rag)-Ct(Hprt). Data is representative of two experiments.
p110α and p110δ regulate Rag expression and VDJ recombination
We next sorted B220+CD43+ pre-B cells into fractions A (CD19−BP1−), B (CD19+BP1−), and C (CD19+BP1+) (Fig. 3A) and incubated the cells with antibodies against Rag2 and Igμ (Fig. 3B). Igμ was detected in wild-type and CD2Cre-p110αflδD910A fraction B (~30%) and fraction C (~70%) cells (Fig. 3C). The appearance of Rag2 protein preceded that of Igμ and was apparent in a small proportion of fraction A cells (<10%, both genotypes) (Fig. 3D). Approximately 40% of the wild-type fraction B cells had Rag2, but fewer than 20% of the wild-type fraction C cells had Rag2, consistent with pre-BCR–dependent suppression of the expression of Rag. By contrast, more than 60% of p110αδ-deficient cells from fractions B and C contained Rag2. Moreover, although fewer than 20% of wild-type fraction B or C cells contained both Igμ and Rag2, such co-expression was observed in more than 50% and 60% percent of CD2Cre-p110αflδD910A fraction B and fraction C cells, respectively (Fig. 3E). Quantitative PCR analysis of complementary DNA (cDNA) prepared from cells sorted as described earlier (Fig 3A) showed that the expression of both Rag1 and Rag2 failed to be suppressed in CD2Cre-p110αflδD910A fraction C cells relative to that observed in wild-type fraction C cells (Fig. 3F).
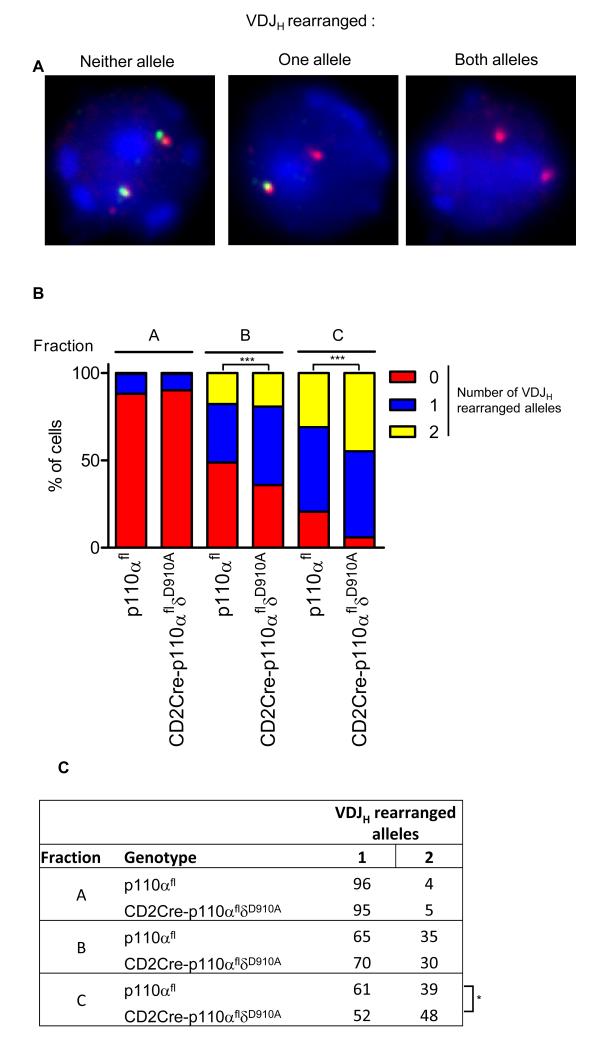
The failure to inhibit the expression of Rag could lead to excessive VDJH recombination. This possibility was explored by monitoring heavy chain gene rearrangements in single sorted B cell progenitors by fluorescent in situ hybridization (FISH). This assay measures the loss of a DNA fragment flanked by the VH and DH segments such that VDJH-rearranged alleles can be distinguished from DJH-rearranged or germline alleles (34) (Fig. 4A). We found that the percentages of cells that had undergone VDJH rearrangement were higher in fraction B and C cells from CD2Cre-p110αfloxδD910A mice than in fraction B cells from wild-type, consistent with the enhanced expression of Rag (Fig. 4B). Consequently, the ratio of cells that had undergone VH to DJH rearrangement on one allele to those having rearrangements on both alleles was higher for wild-type fraction C cells (61:39) than for p110α and p110δ doubly-deficient fraction C cells (Fig. 4C) (52:48).
Fig. 4.
Enhanced VDJH recombination in the absence of PI3K signaling. FISH analysis of intergenic sequences in the Igμ locus. (A) Representative images of cells incubated with a probe that hybridizes between the VH and DH loci (green) or within the CH locus (red). The image on the left shows a cell with both loci intact. The image in the middle shows a cell that has lost the inter VH-JH segment (that is, it has undergone VH to DJH recombination) on one allele, whereas the image on the right shows a cell that has undergone VH to DJH recombination on both alleles. (B) Percentages of p110αfl or p110αflδD910A cells with zero (red), one (blue), or two (yellow) VDJH recombined alleles. More than 200 cells from two independent experiments were scored. (C) The ratio of cells that had undergone VH to DJH rearrangement on one versus two alleles is shown.
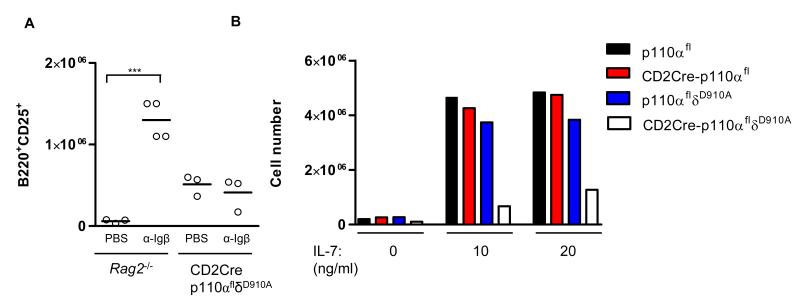
The enhanced VDJH recombination and delayed decrease in the abundances of Rag proteins suggested a defect in pre-BCR signaling. To directly test pre-BCR signaling, we injected Rag2−/− or CD2Cre-p110αflδD910A mice with an antibody against Igβ, which provides signals that promote the development of Rag2−/− fraction B cells to become CD25+ fraction C’ cells (35). Consistent with previous results, we found increased proportions of Rag2−/− cells that had surface expression of CD25 after injection with antibody against Igβ (Fig. 5A). Although a small proportion of CD2Cre-p110αflδD910A pre-B cells expressed CD25 on the cell surface, this proportion did not increase further upon administration of antibody against Igβ (Fig. 5A). Together, these results suggest that CD2Cre-p110αflδD910A pre-B cells can assemble Igμ to form a pre-BCR, but that this receptor fails to inhibit the expression of Rag and provide the necessary signals for the development of large preB-II/fraction C’ cells.
Fig. 5.
Pre-BCR-dependent development and IL-7-dependent proliferation require PI3K activity. (A) Rag2−/− or CD2Cre-p110αflδD910A mice were injected with 1 mg of antibody against Igβ (HM-79) or with PBS as a negative control, and bone marrow cells were harvested 8 days later. The plots represent the number of B220+CD25+ cells in the lymphocyte gate. Three or four mice were analyzed per group in one experiment. (B) 5 × 105 FACS-sorted B220+cKit+ B cells were incubated in duplicate on OP9 stromal cell layers in the presence of 10 or 20 ng of IL-7. After 5 days, the cell numbers in each well were determined. Data in B is representative of two experiments.
In addition to the pre-BCR, the IL-7 receptor (IL-7R) activates PI3K, and IL-7 contributes synergistically with the pre-BCR to the proliferation of pre-B1 cells (36, 37). We therefore measured the ability of CD2Cre-p110αflδD910A cKit+ pro-B cells to proliferate in vitro by culturing the cells with OP9 stromal cells and IL-7. Although the CD2Cre-p110αfl and lp110δD910A cells proliferated normally, CD2Cre-p110αflδD910A cells showed impaired proliferation (Fig. 5B). Thus, although IL-7 appears capable of supporting the development of pro B cells, as evidenced by enhanced VH-DJH recombination, IL-7-dependent proliferative signals were compromised in the absence of p110α and p110δ activities.
p110α can only partially compensate for the lack of p110δ in mature B cells
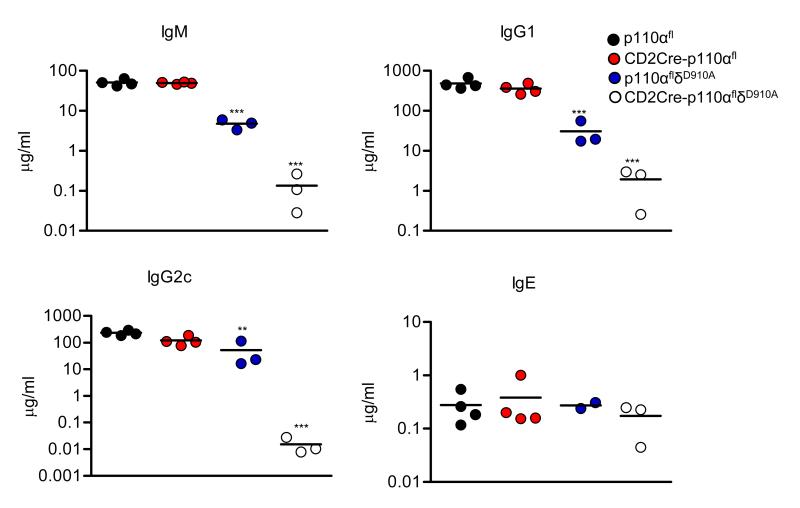
Flow cytometric analysis confirmed the reduced numbers of mature B cells and a near complete absence of CD21highCD23low MZ B cells in the spleens of p110δD910A mice (Fig. 6, A to D); deficiency in p110α alone had no impact on either of these populations. However, in the CD2Cre-p110αflδD910A mice both the FO and MZ populations were virtually absent (Fig. 6, B and D). Consistent with a key role for PI3K in transmitting essential survival signals from the BCR (28), we found that a large percentage of CD2Cre-p110αflδD910A B cells were apoptotic (Fig. 6E). The few B cells that we were able to recover from the spleens of CD2Cre-p110αflδD910A mice had undergone Cre-mediated deletion in the p110α gene as detected by PCR analysis of their genomic DNA (Fig. 6F). Similar results were obtained by immunohistochemical analysis of spleen slices, which revealed a virtual absence of IgM+ or IgD+ B cells in the spleens of CD2Cre-p110αflδD910A mice (Fig. 6G). This assay also revealed the selective loss of IgMhighIgDlow MZ B cells in the spleens of p110δD910A mice, whereas MZ B cells were clearly detected in the spleens of CD2Cre-p110αfl mice. Moreover, the lymph nodes of CD2Cre-p110αflδD910A mice lacked B cell follicles and were instead uniformly populated by T cells (Fig. 6H). In contrast, deficiency in p110β did not lead to a further reduction in B cell numbers, even in absence of p110δ activity (fig. S2). Serum Ig, which was already reduced in p110δD910A mice compared to that in wild-type mice, was nearly undetectable in CD2Cre-p110αflδD910A mice, with the exception of IgE which was similarly abundant in both mice (Fig. 7). Thus, in the absence of p110α and p110δ activities, the very few B cells that developed and survived appeared to produce disproportionately large amounts of IgE.
Fig. 6.
Defective development of mature B cells in CD2Cre-p110αflδD910A mice. Splenocytes from p110αfl (n = 11 mice), CD2Cre-p110αfl (n = 6 mice), p110αflδD910A (n = 11 mice), and CD2Cre-p110αflδD910A (n = 11 mice) mice were analyzed by flow cytometry for the indicated markers. (A) B220 (B cells) versus CD3 (T cells). (B) Splenocytes incubated with antibodies against IgM and IgD to detect immature transitional 1 (IgMhighIgDlow), transitional 2 (IgMhighIgDhigh), and mature (IgMlowIgDhigh) B cells. (C) B220+ cells were incubated with antibodies against CD21 and CD23 to distinguish FO cells (CD21lowCD23high) from marginal zone B cells (CD21highCD23low). (D) The mean total numbers of CD19+ B cells, FO B cells (CD21lowCD23high), and MZ B cells (CD21highCD23low) per spleen as calculated from the data in panels A to C (± SEM). (E) Percentages of viable B cells analyzed by DAPI staining. (F) PCR analysis of genomic DNA from purified B cells. The 812 kb band is from a recombined allele, whereas the 591 bp band is from an allele with both loxP sites intact. The lower band in each panel is from the floxed allele, whereas the upper band is from the recombined allele. (G) Detection of IgM+ (green) and IgD+ (red) B cells and CD4+ T cells (blue) in spleen slices from the indicated mice. (H) Detection of B cells (green), CD4+ T cells (blue), and dendritic cells (red) in slices of lymph nodes from the indicated mice. Data in G and H are representative images from two experiments with 2 mice per group.
Fig. 7.
Reduced concentrations of serum Ig in CD2Cre-p110αflδD910A mice. Concentrations of IgM, IgG1, IgG2C and IgE in the sera from 3-4 mice per group was measured by ELISA and the concentration of each determined from a concentration curve using a know standard.
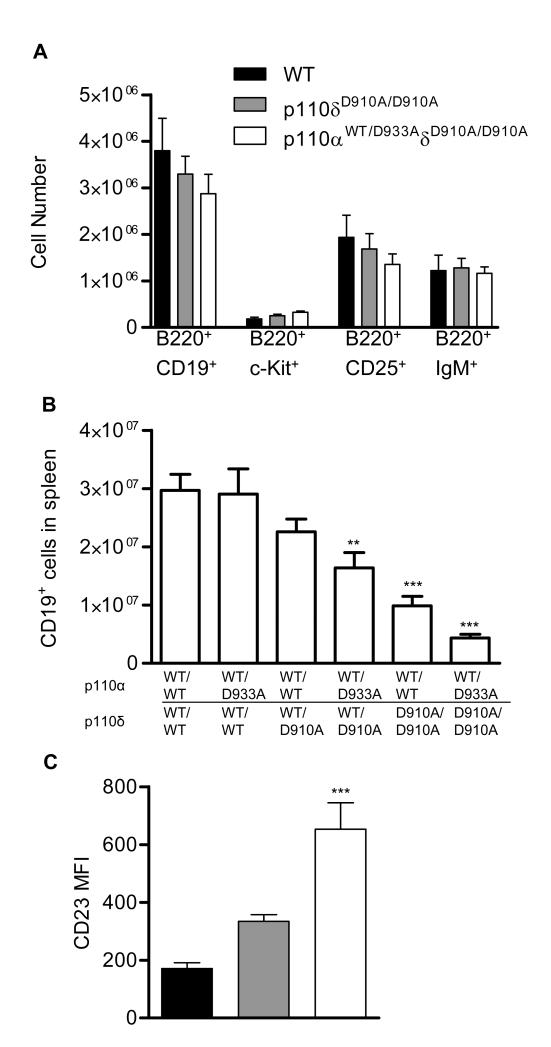
In the preceding experiments, we compared the effects of a kinase-defective knockin mutation in p110δ to a p110α null allele. The presence of a kinase-defective protein can, in some circumstances, reveal phenotypes that are not apparent when analyzing null alleles, especially in heterozygous animals (38). Therefore, to examine whether a kinase-defective allele of p110α might have a stronger impact on B cell development than a null allele, we crossed p110δD910A mice with p110αD933A mice to generate an allelic series of mice expressing one or more kinase-defective p110 alleles. B cell development in the bone marrow was largely intact in p110αWT/D933AδD910A/D910A mice, demonstrating that just one of the four p110α or p110δ alleles was sufficient to promote B cell development in the bone marrow (fig. 8A). In the spleen, p110δWT/D910A heterozygosity led to a reduction in B cell numbers, whereas p110αWT/D933A heterozygosity did not; however, p110αWT/D933A on a p110δWT/D910A or p110δD910A background revealed synergy between these isoforms (fig. 8B). The surface expression of CD23 is thought to be directly regulated by Foxo transcription factors (28). Accordingly, as was also apparent in CD2Cre-p110αflδD910A B cells (Fig. 6C), the abundance of CD23 was higher in p110αWT/D933AδD910A B cells than in p110δD910A B cells, which had a higher abundance of CD23 than did wild-type cells (fig. 8C). We conclude that p110 protein made by a single Pik3ca or Pik3cd locus is sufficient to promote pre-B cell development, but that optimal mature B cell development, survival, or both requires full expression of p110δ whose absence can only partially be compensated for by p110α.
Fig. 8.
Intact B cell development in the bone marrow, but reduced spleen B cell numbers in p110αWT/D933AδD910A/D910A mice (A) Bone marrow cells from WT, p110δD910A/D910A and p110αWT/D933AδD910A/D910A mice were analyzed as in figure 2. (B) Total spleen cell numbers from each of the six possible genotypes resulting from p110αD933A and p110δD910A mouse crosses. (C) Mean fluorescent intensity of CD23 on WT, p110δD910A/D910A and p110αWT/D933AδD910A/D910A mice.
Discussion
Here, we describe an essential role for the p110α and p110δ proteins at the pre-BCR–dependent stage of B cell development in the bone marrow. Previous studies had shown that the pre-BCR needs to activate Syk and ZAP-70 at this stage (8). In addition, the Syk substrate SLP-65 is required for further differentiation, but not for proliferation of pre-B cells (6). Here, we help to complete our understanding of pre-BCR signaling by demonstrating that PI3K activity is required for pre-BCR–dependent suppression of Rag expression and, possibly in concert with SLP-65, for differentiation.
A question that arises from these studies is whether the failure to inhibit Rag expression is sufficient to cause a complete block in B cell development. There are at least two reasons why it might be important to inhibit the expression of Rag during the time between heavy chain and light chain gene rearrangements. First, maintaining the expression of Rag during cell division that occurs between the stages of heavy chain and light chain gene rearrangements might increase the risk of accumulating DNA breaks (39). Second, it may be important to actively suppress Rag expression to prevent recombination of the second heavy chain allele if the first heavy chain allele were successfully recombined in frame (40). Typically, the ratio of cells having one rather than two VHDJH rearranged loci is 60:40 (41). Consistent with this, we observed a ratio of 61:39 of singly- to doubly-recombined loci in wild-type fraction C cells. However, in CD2Cre-p110αflδD910A fraction C cells, the ratio was reduced to 52:48 (from 70:30 in the fraction B cells). Our results therefore raise the possibility that allelic exclusion is compromised in CD2Cre-p110αflδD910A pre-B cells, which fail to suppress the expression of Rag; however, this remains to be tested definitively by examining the frequency of productively rearranged Igh loci at the single-cell level.
The pre-BCR is thought to provide a feedback signal that informs the cell that Igμ is functionally expressed and that further development may proceed (6). The failure of anti-Igβ to induce the formation of CD25+ pre-B cells in CD2Cre-p110αflδD910A mice suggests that the p110α and p110δ subunits are essential for pre-BCR signaling. However, IL-7-dependent proliferation was also attenuated in p110αδ-deficient pre-B cells. There are two possible mechanisms that might explain this result. The most direct is based on the observation that a tyrosine within the cytoplasmic domain of the IL-7Rα chain can bind to the p85 subunits of PI3K. Indeed, the introduction of a tyrosine-to-phenylalanine mutation in IL-7Rα, which uncouples the IL-7Rα from PI3K activity, interferes with IL-7-dependent proliferation but has no impact on IL-7-dependent differentiation (36). Presumably the signals downstream of the IL-7R that regulate chromosome accessibility for Rag recombination (2) are also intact in the p110α and p110δ doubly-deficient pre-B cells, which show an enhanced proportion of cells undergoing VDJH recombination. However, the pre-BCR lowers the threshold of activation by IL-7 in pre-B cells (37). Thus, another possibility is that defective pre-BCR signaling in CD2Cre-p110αflδD910A cells also contributes to a reduced responsiveness to IL-7. Either way, compromised pre-BCR signaling may be confounded by reduced IL-7 responsiveness in CD2Cre-p110αflδD910A pre-B cells to block development at the pre-BCR checkpoint.
An analysis of mature B cells and T cells indicated a nonredundant role for p110δ in antigen receptor signaling (24). The reason that p110β does not contribute substantially to lymphocyte antigen receptor signaling may be explained by two independent observations. First, p110β is found at low abundance in B cells (Fig. 1) (42). Second, p110β responds preferentially to G protein–coupled receptors (GPCRs) and poorly, if at all, to tyrosine kinase–induced signaling (33). In this context, it is worth noting that although double deficiency in p110δ and p110γ has a major impact on early T cell development, B cell development is normal in p110δ and p110γ double knockout mice (43). Although a subsequent report argued that deficiency in p110γ potentiates the effect of p110δ-deficiency on B cell development, this is likely due to indirect effects of p110γ deficiency in non-B cells (44). Instead, activation of PI3K provided by the chemokine receptor CXCR4 (a GPCR) appears to be essential for T cell development, but not for B cell development (45).
The lack of effect of p110α deficiency on antigen receptor signaling in mature B cells is more difficult to explain. In other cell types, such as muscle cells, liver cells, fat cells, fibroblasts, and endothelial cells, p110α produces PIP3, which leads to the robust phosphorylation of Akt in response to diverse tyrosine kinase–activating stimuli (31, 38, 46). In vitro, up to 60% of class IA PI3K activity is lost in p110δD910A B cells (47), which suggests that the remaining 40% activity, based on the results in this report, is mostly provided by p110α. Consistent with this notion is the observation that p110δ-deficiency does not entirely restore the gain-of-function phenotype of Pten-deficient B cells (42). However, in agonist-stimulated cells, taking the detection of pAkt as a readout for PI3K activity, p110δ-deficiency results in an estimated 90% or greater loss of PI3K activity (24). Thus, there seems to be a discrepancy between the amount of PI3K activity that can be detected in immune complexes in vitro and that by signaling analysis in cells.
One mechanism to explain these paradoxical results involves the selective recruitment of p110δ or the selective activation of recruited p110δ in antigen receptor complexes. This may be the case if p110α and p110δ bind to different Ras isoforms, for example; however, the present demonstration that the pre-BCR shows no obvious preference for p110α versus p110δ even though the pre-BCR is presumed to use a similar signaling machinery to that of the mature BCR, suggests that p110α and p110δ can be effectively recruited and activated by BCR-associated signaling proteins. The BCR can engage the PI3K pathway in the absence of obvious agonist-stimulation (that is, antigen). Such so-called tonic signaling to PI3K signaling is essential for BCR-dependent survival of B cells and can be provided by a constitutively active p110α transgene (28). However, transgenic expression of the p110α mutant fails to induce the phosphorylation of Akt to an extent comparable to that in wild-type B cells stimulated by BCR crosslinking (28). Together with the observation that a deficiency in p110α had no impact on the phosphorylation of Akt induced by antibody against IgM, these results indicate that cross-linking of the BCR fails to increase p110α activity beyond that which is achieved by tonic BCR signaling. This might help to explain the selective role for p110α in pre-BCR signaling, which is thought to involve tonic rather than specific agonist-induced signaling (6). In this context, it is noteworthy that whereas tonic BCR signaling is likely to be sufficient for the development and survival of FO B cells, the development of B1 cells and MZ cells is thought to require crosslinking of the BCR by autoantigens (48). Thus, it seems that where agonist-induced aggregation of the BCR is required, p110α cannot substitute for the loss of p110δ activity. Currently, inhibitors against p110α, p110δ, or both are being developed for therapeutic purposes. Based on our findings, we suggest that whereas p110δ-specific inhibitors should be able to target immune cells with high selectivity to alleviate autoimmunity, p110α-specific inhibitors should be able to target tumor cells with minimal impact on protective immune responses against pathogens.
Materials and Methods
Mice
p110αfl (31), p110αD933A (38), p110βfl (33), p110δD910A (20) and CD2Cre mice (32) were maintained on a mixed B6:129 background (backcrossed to B6 for 2 to 4 generations). The p110αfl and p110βfl alleles were bred to homozygosity, whereas the CD2Cre transgene was maintained heterozygously. The p110δD910A allele was also bred to homozygosity in all experiments except those shown in Fig. 8, in which heterozygous mice were also analyzed as indicated. The p110αD933A allele was only maintained heterozygously because homozygous animals die as embryos (31, 38). Rag2−/− mice (3) were maintained on the B6 background. All mice were bred under specific pathogen-free conditions, and all experiments were approved by a local ethical review committee and the Home Office. Deletion of exons 18 and 19 form the Pik3ca allele was confirmed by PCR analysis of genomic DNA with the following primers: Ma9, 5′-ACACACTGCATCAATGGC-3′; Ma50, 5′-CTAAGCCCTTAAAGCCTTAC-3′; Ma51, 5′-CAGCTCCCATCTCAGTTCA-3′. The Ma50 and Ma51 primers amplify a 591 bp fragment from floxed alleles, whereas the Ma9 and Ma51 primers amplify an 812 bp fragment after Cre-mediated recombination.
Western blotting analysis
B cells were purified by negative selection with antibody against CD43 and Miltenyi beads. B cells were stimulated with antibody against IgM F(ab’)2 (10 μg/ml, Jackson Immunoresearch) for the indicated times in the presence or absence of the p110δ-selective inhibitor IC87114 (49). The cells were lysed in ice cold buffer [50 mM Hepes (pH x.x), 150 mM NaCl, 10 mM NaF, 10 mM iodoacetamide, 1% NP-40, 1 mM phenylmethyl sulfonyl fluoride (PMSF) and protease inhibitors (Roche)]. Lysates were resolved on NuPage 4 to 12% BisTris gels (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked in 5% milk and incubated with the following antibodies: antibody against p110α (Cell Signaling Technology, CST 4249), antibody against p110β (CST 3011), antibody against p110δ (Abcam, ab1678), antibody against Akt (CST 9272), antibody against Akt pSer473 (CST 4058), antibody against pERK1/2 (CST 4377), and antibody against pFoxo3a (CST 9464).
Proliferation assays
Purified B cells were stimulated with anti-IgM F(ab’)2, antibody against CD40 (10 μg/ml, Clone 3/23, Becton Dickinson), IL-4 (20 ng/ml, R&D systems), or LPS (10 μg/ml, Sigma) in round-bottomed 96-well plates in RPMI-1640 (Invitrogen) supplemented with 10% fetal calf serum (FCS), 1% penicillin and streptomycin (Invitrogen) and 50 μM β-mercaptoethanol (Sigma). 1 μCi of 3H-thymidine was added during the last 6 hours of a 48-hour culture after which the DNA was harvested and the incorporated radioactivity was measured. FACS-sorted B220+c-Kit+ bone marrow B cells were added to a monolayer of OP9 cells (50) in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented as above with the addition of 10 or 20 ng/ml of IL-7 (Peprotech). The number of B cells was counted 5 days later.
Immunizations and detection of serum antibody
Wild-type (p110αfl) or CD2Cre-p110αfl mice were immunized intraperitoneally with 100 μg of (2,4,6) trinitrophenyl TNP coupled to keyhole limpet hemocyanin (KLH) (Biosearch) adhered to alum. TNP-specific antibodies were detected on plates coated with TNP-BSA (Biosearch) with an enzyme-linked immunsorbent assay (ELISA) kit (Southern Biotech). Antibodies from nonimmunized mice were detected on plates coated with antibody against Ig and an ELISA kit and Ig standards (Southern Biotech), except for IgE, which was measured with a kit from Biolegend.
Flow cytometry
Single-cell suspensions from bone (femur and tibia) or spleen were incubated with the indicated antibodies and analyzed on a Becton Dickinson (BD) FACSCalibur or LSRII, or were sorted with a BD FACSaria instrument. Data were analyzed with FlowJo software. All antibodies were from eBioscience, except where otherwise stated; clone numbers are shown in parentheses: B220 (RA3-6B2), c-Kit (ack45), CD25 (PC61), IgM (AF6-78), IgD (11-26c2a), CD19 (1D3), CD43 (S7) (BD), Ly-51 (BP-1) (BD), Igβ (HM-79) (Abcam or produced in our laboratory).
Gene expression analysis
RNA was prepared with Trizol and first strand complementary DNA (cDNA) was synthesized with random hexamers and reverse transcriptase. The PCR primers used were as described by Hsu et al. (51).
DNA FISH and immunohistochemistry
DNA FISH was performed as previously reported (52). Briefly, FACS-sorted cells were allowed to settle on poly-L-lysine-coated slides (Sigma) before being fixed in 4% formaldehyde. Slides were quenched in 0.1 M Tris-HCl (pH 7.2) for 10 min at room temperature, extracted with 0.1% Triton X-100 containing 0.1% saponin in phosphate-buffered saline (PBS) for 10 min, rinsed in PBS, and then incubated in 20% glycerol in PBS for 20 min. Slides were then immersed in liquid nitrogen, allowed to thaw, and were then placed back in PBS containing 20% glycerol. Two additional freeze-thaw cycles were performed. The slides were then rinsed in PBS and incubated in a 0.1N HCl solution for 30 min at room temperature. After further rinsing in PBS, the slides were treated with RNase A in 2×SSC at 37°C for 1 hour. Slides were then rinsed in PBS before further extraction in 0.5% Triton X-100 containing 0.5% saponin in PBS. Following rinsing in PBS, slides were equilibrated in 50% formamide in 2× SSC for 20 min. A coverslip containing 100 to 200 ng of each probe, 6 μg of mouse CotI DNA, and 10 μg of sheared salmon sperm DNA in 50% formamide, 2× SSC and 10% dextran sulphate was then inverted on to the cell spot. The genomic DNA and probe were co-denatured by placing the sealed slide on a hot plate set at 78°C for 2 min. Hybridization was performed for 16 hours at 37°C. Coverslips were carefully removed and slides were washed in 50% formamide in 2× SSC at 45°C for 15 min, 0.2× SSC at 63°C for 15 min, then 2× SSC at 45°C for 5 min before equilibrating at room temperature in 2× SSC for 5 min. Slides were blocked in 3% BSA in 2× SSC for 30 min at room temperature before detection with the antibodies diluted in the same solution. Washes between antibody steps were with 0.1% Triton X-100 in 2× SSC. After immunodetection, slides were counterstained with DAPI and mounted with Vectashield. The mouse Igh constant region BAC RP24-258E20 BAC was labeled with DNP-11-dUTP (Enzo) by standard nick translation and was detected with rat antibody against DNP (Serotec) and then with donkey antibody against rat Ig conjugated with rhodamine red-X (Jackson). For the V-D region probes, 7 repeat-free regions in the V-D interval of 1 to 3 kb were amplified by PCR and cloned into pGEM T-easy (Promega). These were then labeled with DIG-11-dUTP (Roche) by standard nick translation. DIG probes were detected with sheep antibody against DIG (Roche), fluorescein isothiocyanate (FITC)-conjugated rabbit antibody against sheep Ig (Calbiochem), and then with Alexa Fluor 488-conjugated goat antibody against rabbit Ig (Invitrogen). Slides were counted on an Olympus BX61 epifluorescence miscroscope. For immunohistochemistry, cells were fixed to poly-L-lysine slides as for DNA FISH. Slides were washed three times in PBS and then extracted with 0.5% Triton-X in PBS for 10 minutes. Surface μ was detected with a biotinylated goat antibody against mouse IgM (Sigma) and Alexa 647-conjugated streptavidin (Invitrogen). Mouse Rag2 was detected with rabbit polyclonal ab432 (a gift from S. Desiderio) (53) and Alexa 488-conjugated goat antibody against rabbit Ig (Invitrogen). Nuclei where counterstained with DAPI and mounted with Vectashield before visualization on an Olympus BX61 epifluorescence miscroscope. Lymph node and spleen slices were prepare as previously described (54). Briefly, spleens and lymph nodes were embedded in 4% agarose prepared in PBS. 320 μm slices were cut from each lymph node with a vibratome. Slices were fixed with PBS containing 4% PFA for 30 min. Lymph node slices were incubated for 1 hour with B220-FITC-conjugated antibody against B220, Alexa fluor 647-conjugated antibody against CD4, and phycoerythrin (PE) conjugated antibody against CD11c. Spleen slices were incubated overnight with FITC-conjugated antibody against IgM, PE-conjugated antibody against IgD, and Alexa fluor 647-conjugated antibody against CD4.
Pre-BCR stimulation in vivo
Rag2−/− and CD2Cre-p110αfl-p110δD910A mice were injected intraperitoneally with 1 mg of purified anti-Igβ (HM-79) as previously described (35), and bone marrow was analyzed 8 days later.
Statistical analysis
Statistical significance was calculated with the Graphpad Prism software. One-way ANOVA with Tukey post test was used when four groups were compared. When two groups were compared, we used the Student’s t test, except for Figs. 3 and 4, where we used chi squared analysis. The following symbols are used to indicate if the mean value for a mutant is statistically significant different from that of the wild-type: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Supplementary Material
Acknowledgments
We thank M. Janas and Martin Turner for critical review of the manuscript. We also thank D. Patton for help with RNA purification and cDNA synthesis. We are grateful to the SABU staff for expert animal husbandry, to G. Morgan for help with FACS sorting, and to D. Kioussis for providing the CD2Cre mice. Funding: Work described in this study was funded by grants from the BBSRC to B.V., A.C., and K.O. F.R. was supported by an MRC capacity award.
Footnotes
Competing interests: BV and KO act as scientific advisers to Intellikine and GlaxoSmithKline, respectively.
References and Notes
- 1.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 2.Corcoran AE, Riddell A, Krooshoop D, Venkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- 3.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, Alt FW. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 4.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 6.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 7.Turner M, Gulbranson-Judge A, Quinn ME, Walters AE, MacLennan IC, Tybulewicz VL. Syk tyrosine kinase is required for the positive selection of immature B cells into the recirculating B cell pool. J Exp Med. 1997;186:2013–2021. doi: 10.1084/jem.186.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweighoffer E, Vanes L, Mathiot A, Nakamura T, Tybulewicz VL. Unexpected requirement for ZAP-70 in pre-B cell development and allelic exclusion. Immunity. 2003;18:523–533. doi: 10.1016/s1074-7613(03)00082-7. [DOI] [PubMed] [Google Scholar]
- 9.Jumaa H, Wollscheid B, Mitterer M, Wienands J, Reth M, Nielsen PJ. Abnormal development and function of B lymphocytes in mice deficient for the signaling adaptor protein SLP-65. Immunity. 1999;11:547–554. doi: 10.1016/s1074-7613(00)80130-2. [DOI] [PubMed] [Google Scholar]
- 10.Pappu R, Cheng AM, Li B, Gong Q, Chiu C, Griffin N, White M, Sleckman BP, Chan AC. Requirement for B cell linker protein (BLNK) in B cell development. Science. 1999;286:1949–1954. doi: 10.1126/science.286.5446.1949. [DOI] [PubMed] [Google Scholar]
- 11.Flemming A, Brummer T, Reth M, Jumaa H. The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat Immunol. 2003;4:38–43. doi: 10.1038/ni862. [DOI] [PubMed] [Google Scholar]
- 12.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 13.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 14.Inabe K, Ishiai M, Scharenberg AM, Freshney N, Downward J, Kurosaki T. Vav3 modulates B cell receptor responses by regulating phosphoinositide 3-kinase activation. J Exp Med. 2002;195:189–200. doi: 10.1084/jem.20011571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aiba Y, Kameyama M, Yamazaki T, Tedder TF, Kurosaki T. Regulation of B-cell development by BCAP and CD19 through their binding to phosphoinositide 3-kinase. Blood. 2008;111:1497–1503. doi: 10.1182/blood-2007-08-109769. [DOI] [PubMed] [Google Scholar]
- 16.Moon KD, Post CB, Durden DL, Zhou Q, De P, Harrison ML, Geahlen RL. Molecular basis for a direct interaction between the Syk protein-tyrosine kinase and phosphoinositide 3-kinase. J Biol Chem. 2005;280:1543–1551. doi: 10.1074/jbc.M407805200. [DOI] [PubMed] [Google Scholar]
- 17.Fruman DA, Snapper SB, Yballe CM, Davidson L, Yu JY, Alt FW, Cantley LC. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85α. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki H, Terauchi Y, Fujiwara M, Aizawa S, Yazaki Y, Kadowaki T, Koyasu S. Xid-like immunodeficiency in mice with disruption of the p85α subunit of phosphoinositide 3-kinase. Science. 1999;283:390–392. doi: 10.1126/science.283.5400.390. [DOI] [PubMed] [Google Scholar]
- 19.Oak JS, Chen J, Peralta RQ, Deane JA, Fruman DA. The p85beta regulatory subunit of phosphoinositide 3-kinase has unique and redundant functions in B cells. Autoimmunity. 2009;42:447–458. doi: 10.1080/08916930902911746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, Smith AJ, Vanhaesebroeck B. Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 21.Clayton E, Bardi G, Bell SE, Chantry D, Downes CP, Gray A, Humphries LA, Rawlings D, Reynolds H, Vigorito E, Turner M. A Crucial Role for the p110δ Subunit of Phosphatidylinositol 3-Kinase in B Cell Development and Activation. Journal of Experimental Medicine. 2002;196:753–763. doi: 10.1084/jem.20020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jou ST, Carpino N, Takahashi Y, Piekorz R, Chao JR, Wang D, Ihle JN. Essential, nonredundant role for the phosphoinositide 3-kinase p110δ in signaling by the B-cell receptor complex. Mol Cell Biol. 2002;22:8580–8591. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fruman DA, Bismuth G. Fine tuning the immune response with PI3K. Immunol Rev. 2009;228:253–272. doi: 10.1111/j.1600-065X.2008.00750.x. [DOI] [PubMed] [Google Scholar]
- 24.Okkenhaug K, Ali K, Vanhaesebroeck B. Antigen receptor signalling: a distinctive role for the p110δ isoform of PI3K. Trends Immunol. 2007;28:80–87. doi: 10.1016/j.it.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tze LE, Schram BR, Lam KP, Hogquist KA, Hippen KL, Liu J, Shinton SA, Otipoby KL, Rodine PR, Vegoe AL, Kraus M, Hardy RR, Schlissel MS, Rajewsky K, Behrens TW. Basal immunoglobulin signaling actively maintains developmental stage in immature B cells. PLoS Biol. 2005;3:e82. doi: 10.1371/journal.pbio.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verkoczy L, Duong B, Skog P, Ait-Azzouzene D, Puri K, Vela JL, Nemazee D. Basal B cell receptor-directed phosphatidylinositol 3-kinase signaling turns off RAGs and promotes B cell-positive selection. J Immunol. 2007;178:6332–6341. doi: 10.4049/jimmunol.178.10.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llorian M, Stamataki Z, Hill S, Turner M, Martensson IL. The PI3K p110delta is required for down-regulation of RAG expression in immature B cells. J Immunol. 2007;178:1981–1985. doi: 10.4049/jimmunol.178.4.1981. [DOI] [PubMed] [Google Scholar]
- 28.Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, DePinho RA, Rickert RC. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graupera M, Guillermet-Guibert J, Foukas LC, Phng LK, Cain RJ, Salpekar A, Pearce W, Meek S, Millan J, Cutillas PR, Smith AJ, Ridley AJ, Ruhrberg C, Gerhardt H, Vanhaesebroeck B. Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature. 2008;453:662–666. doi: 10.1038/nature06892. [DOI] [PubMed] [Google Scholar]
- 32.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 33.Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, Meek S, Smith AJ, Okkenhaug K, Vanhaesebroeck B. The p110beta isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110gamma. Proc Natl Acad Sci U S A. 2008;105:8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daly J, Licence S, Nanou A, Morgan G, Martensson IL. Transcription of productive and nonproductive VDJ-recombined alleles after IgH allelic exclusion. EMBO J. 2007;26:4273–4282. doi: 10.1038/sj.emboj.7601846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagata K, Nakamura T, Kitamura F, Kuramochi S, Taki S, Campbell KS, Karasuyama H. The Ig alpha/Igbeta heterodimer on mu-negative proB cells is competent for transducing signals to induce early B cell differentiation. Immunity. 1997;7:559–570. doi: 10.1016/s1074-7613(00)80377-5. [DOI] [PubMed] [Google Scholar]
- 36.Corcoran AE, Smart FM, Cowling RJ, Crompton T, Owen MJ, Venkitaraman AR. The interleukin-7 receptor alpha chain transmits distinct signals for proliferation and differentiation during B lymphopoiesis. Embo J. 1996;15:1924–1932. [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall AJ, Fleming HE, Wu GE, Paige CJ. Modulation of the IL-7 dose-response threshold during pro-B cell differentiation is dependent on pre-B cell receptor expression. J Immunol. 1998;161:6038–6045. [PubMed] [Google Scholar]
- 38.Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. Critical role for the p110α phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 39.Lin WC, Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc Natl Acad Sci U S A. 1994;91:2733–2737. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grawunder U, Leu TM, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 41.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 42.Janas ML, Hodson D, Stamataki Z, Hill S, Welch K, Gambardella L, Trotman LC, Pandolfi PP, Vigorito E, Turner M. The effect of deleting p110delta on the phenotype and function of PTEN-deficient B cells. J Immunol. 2008;180:739–746. doi: 10.4049/jimmunol.180.2.739. [DOI] [PubMed] [Google Scholar]
- 43.Webb LM, Vigorito E, Wymann MP, Hirsch E, Turner M. Cutting Edge: T Cell Development Requires the Combined Activities of the p110γ and p110δ Catalytic Isoforms of Phosphatidylinositol 3-Kinase. Journal of Immunology. 2005;175:2783–2787. doi: 10.4049/jimmunol.175.5.2783. [DOI] [PubMed] [Google Scholar]
- 44.Beer-Hammer S, Zebedin E, von Holleben M, Alferink J, Reis B, Dresing P, Degrandi D, Scheu S, Hirsch E, Sexl V, Pfeffer K, Nurnberg B, Piekorz RP. The catalytic PI3K isoforms p110gamma and p110delta contribute to B cell development and maintenance, transformation, and proliferation. Journal of leukocyte biology. 2010;87:1083–1095. doi: 10.1189/jlb.0809585. [DOI] [PubMed] [Google Scholar]
- 45.Janas ML, Varano G, Gudmundsson K, Noda M, Nagasawa T, Turner M. Thymic development beyond beta-selection requires phosphatidylinositol 3-kinase activation by CXCR4. J Exp Med. 2010;207:247–261. doi: 10.1084/jem.20091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao JJ, Cheng H, Jia S, Wang L, Gjoerup OV, Mikami A, Roberts TM. The p110alpha isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc Natl Acad Sci U S A. 2006;103:16296–16300. doi: 10.1073/pnas.0607899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bilancio A, Okkenhaug K, Camps M, Emery JL, Ruckle T, Rommel C, Vanhaesebroeck B. Key role of the p110δ isoform of PI3K in B-cell antigen and IL-4 receptor signaling: comparative analysis of genetic and pharmacologic interference with p110δ function in B cells. Blood. 2006;107:642–650. doi: 10.1182/blood-2005-07-3041. [DOI] [PubMed] [Google Scholar]
- 48.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 49.Sadhu C, Masinovsky B, Dick K, Sowell CG, Staunton DE. Essential role of phosphoinositide 3-kinase delta in neutrophil directional movement. J Immunol. 2003;170:2647–2654. doi: 10.4049/jimmunol.170.5.2647. [DOI] [PubMed] [Google Scholar]
- 50.Kodama H, Nose M, Niida S, Nishikawa S. Involvement of the c-kit receptor in the adhesion of hematopoietic stem cells to stromal cells. Exp Hematol. 1994;22:979–984. [PubMed] [Google Scholar]
- 51.Hsu L-Y, Lauring J, Liang H-E, Greenbaum S, Cado D, Zhuang Y, Schlissel MS. A Conserved Transcriptional Enhancer Regulates RAG Gene Expression in Developing B Cells. Immunity. 2003;19:105–117. doi: 10.1016/s1074-7613(03)00181-x. [DOI] [PubMed] [Google Scholar]
- 52.Sayegh CE, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin WC, Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–959. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- 54.Asperti-Boursin F, Real E, Bismuth G, Trautmann A, Donnadieu E. CCR7 ligands control basal T cell motility within lymph node slices in a phosphoinositide 3-kinase-independent manner. J Exp Med. 2007;204:1167–1179. doi: 10.1084/jem.20062079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.