Preface
Ebolaviruses and Marburgvirus are filoviruses that cause haemorhagic fever in primates, with extremely high fatality rates. Studies have focused on elucidating how these viruses enter host cells, with the aim of developing therapeutics. The ebolavirus glycoprotein has been found to play key parts in all steps of entry. Furthermore, recent studies have identified Niemann-Pick C1 (NPC1), a protein that resides deep in the endocytic pathway, as an important host factor in this process.
Introduction
The filovirus family — consisting of the genera Marburgvirus and Ebolavirus — cause haemorhagic fever in primates, including humans. These filamentous, single-stranded, negative-sense RNA viruses are surrounded by a membrane derived from the host cell. There are five recognized species of Ebolavirus and one of Marburgvirus. Among these, Ebola virus (EBOV; formerly known as Zaire ebolavirus) and the Marburg virus Angola variant cause the most severe disease, with case fatality rates reaching ~90%1–3.
Filoviruses are deemed biosecurity and global health risks, and as such are classified as category A priority pathogens. This ‘elite’ ranking is because of the high morbidity and mortality they have caused in humans during sporadic outbreaks, because they are highly infectious and readily spread by person-to-person contact, and because there is a consensus that filoviruses were weaponized during the cold war4. These concerns are compounded by the fact that there are no approved vaccines or drugs to combat any filovirus infection.
Most filoviruses are endemic only in specific regions in Central Africa, although Reston ebolavirus, which has caused disease in monkeys and pigs but not humans, is endemic in the Philippines5. Moreover, a new filovirus, representing a tentative third genus, was recently discovered in Spain6. However, there have been instances of unintentional importation; for example, importation of Marburg virus (from infected monkeys from Uganda) sparked an outbreak with human fatalities in Europe in 1967, and, similarly, importation of infected monkeys from the Philippines caused widespread disease and death in monkeys housed in a facility in Reston Virginia. Hence, there is a pressing need to develop treatments for filovirus infections, whether they arise at endemic sites or are caused by accidental (for example, through importation) or intentional (for example, by bioterrorists) means.
The first step of any virus lifecycle (Fig. 1) is binding to the host cell surface. In the case of filoviruses, the first cells infected are macrophages and dendritic cells, but they can infect most cell types, with the notable exceptions of lymphocytes and other non-adherent cells7. All viruses use attachment factors and/or entry receptors to bind to host cells. Although an attachment factor serves only as a binding moiety, an entry receptor is currently defined as a molecule on the cell surface that actively promotes virus internalization or actively induces virus penetration8 (the process by which the virus breaches a cell membrane barrier and thereby releases its genome into the cytoplasm). Filoviruses engage host cell attachment factors, including DC-SIGN and L-SIGN9–12. However, which molecules serve as entry receptors remains unclear7, 13–15. Several cell surface proteins such as integrins and TYRO3 family members enhance filovirus entry16, 17, but do not function as physical virus receptors18, 19. Recent evidence indicates that T cell immunoglobulin and mucin domain 1 (TIM1) serves as a filovirus receptor on epithelial cells14, but how it promotes entry and which molecules serve as entry receptors on the surface of non-epithelial cells remain to be determined.
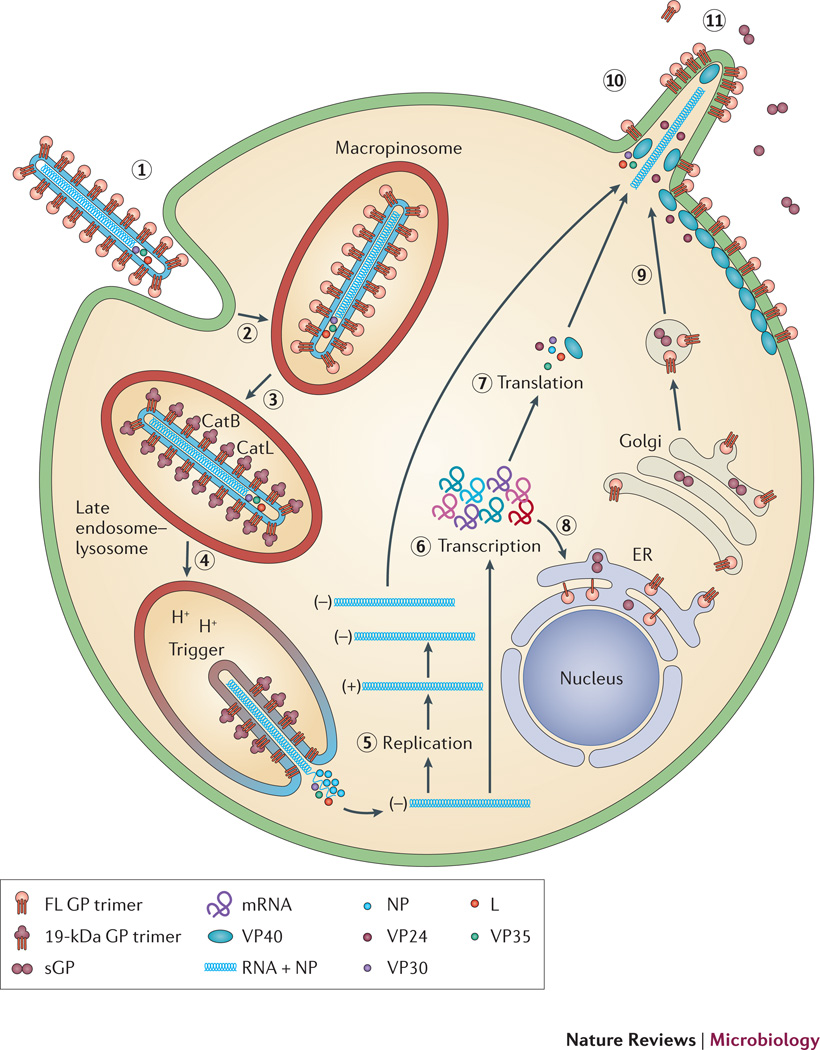
Figure 1. Lifecycle of EBOV.
The virus binds to attachment factors and receptors on the cell surface through the viral spike protein, GP (a). The virus is then internalized into a macropinosome (b) and trafficked to an endosomal compartment containing the Cys proteases cathepsin B and cathepsin L (c). These digest GP to a 19-kDa form, which is triggered to initiate fusion between the viral and endosome membrane (d). After fusion, the viral nucleocapsid is released into the cytoplasm, where the genome is replicated (e) and transcribed (f) with the aid of VP35, VP30, and L, and viral mRNAs are translated (g). mRNAs encoding GP are brought to the ER, where GP is synthesized, modified with N-linked sugars and trimerized (h). GP is further modified in the Golgi and delivered to the plasma membrane in secretory vesicles (i). At the plasma membrane the RNP complex and associated viral proteins assemble with the membrane-associated proteins (matrix proteins VP24 and VP40 and GP) and the virions bud from the cell surface (j). Non-structural forms of the glycoprotein, including soluble GP (sGP), are also secreted (k).
Following binding to the cell surface, filoviruses are internalized by a macropinocytosis-like process20–22 and trafficked to a late endocytic compartment, where penetration occurs. Because they are surrounded by a membrane, filoviruses penetrate into the cytoplasm by membrane fusion with the limiting membrane of a late endosome. After entry, the viral genome is replicated and transcribed, new viral proteins are synthesized, and new virions assemble and bud from the cell surface15, 23, 24 (Fig. 1).
Given its place at the starting line for infection, entry is an ideal target for anti-viral intervention: beat the virus before it gets into the cell. Recent successes in developing antivirals and antibodies that target entry by viral pathogens, including HIV and influenza, have been guided by detailed knowledge of the viral entry mechanisms25, 26. Therefore, it is imperative to learn as much as possible about how filoviruses enter cells.
In this Progress article, we discuss recent insights into filovirus entry, focusing on the viral penetration event. We describe the role of the EBOV glycoprotein (GP) and possible roles of a newly identified host entry factor27, 28, 60, Niemann Pick C1 (NPC1).
EBOV GP mediates virus entry
All stages of EBOV entry (binding to and internalization from the cell surface as well as trafficking to, and fusion with, the limiting membrane of late endosomes) are mediated by trimeric GP spikes15, 29 arrayed around the EBOV particles (Fig. 1). After delivering the virus to a late endosomal compartment, GP must be primed and then triggered to induce the critical membrane fusion event that leads to virus penetration and ensuing replication. Intriguingly, both priming and triggering of EBOV GP (Fig. 2a) are unusual when compared with corresponding processes in other viruses.
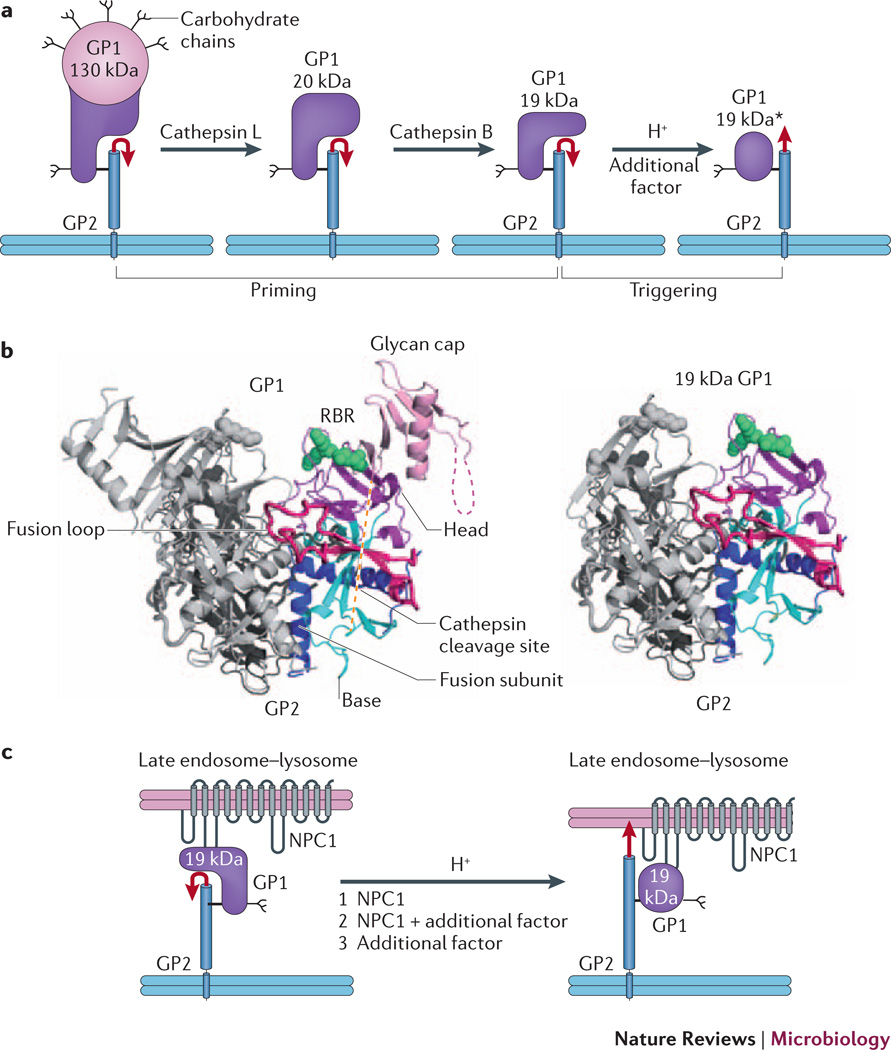
Figure 2. EBOV GP priming and triggering: possible roles of NPC1.
a | Cathepsin L and cathepsin B cleave EBOV GP1, removing the glycan cap and mucin-like domain (pink), thereby generating a 20 kDa and then a 19 kDa form of the glycoprotein. Primed 19 kDa GP requires low pH and the activity of an additional factor to trigger rearrangements that liberate and relocate the fusion loop (red arrow), and thereby initiate the fusion cascade. b | Ribbon diagram of GPΔ (left, PDB 3CSY) and a model for 19 kDa GP (right). The model for cathepsin-primed GP (right), assumes that no conformational changes occur upon cathepsin cleavage. One monomer of each trimer is colored and the other two are in grey. Green spheres depict Lys114, Lys115 and Lys140 (in the RBR), which when mutated impair primed GP binding to NPC1 and GP-mediated infection. c | Possible roles for NPC1 in triggering EBOV fusion. Following delivery to a late endosome–lysosome and priming by cathepsins, primed GP binds to the C-loop of NPC1. In a first model (1), NPC1 is the additional factor depicted in part a that, together with low endosomal pH, triggers the conformational changes that liberate and relocate the fusion loop (red arrow), leading to fusion and therefore penetration of the genome into the cytoplasm to initiate replication. In a second model (2), NPC1 binding to primed GP is necessary, but not sufficient, to trigger GP, and one or more additional factors are still required. In a third scenario (3), NPC1 binds to primed GP, linking the virus to the endosomal membrane, but does not induce conformational changes in GP; in this case an additional factor triggers fusion. If model 3 is correct, NPC1 must have important roles closely upstream of fusion triggering; it may also have an additional upstream role (or roles) if model 1 or 2 applies. Part a is modified with permission from reference30.
Priming EBOV GP to a fusion-competent state
Based on structural information, EBOV GP is categorized as a class I viral fusion protein, and like all fusion proteins in its class, it must be proteolytically primed to set it in a state capable of responding to a fusion trigger (Box 1). Most class I viral fusion proteins are primed either in the Golgi compartment during their biosynthesis or in the extracellular environment on newly budded virus particles released from the surface of an infected cell. For most class I fusion proteins, priming involves removal of only a few, if any, amino acids30, 31.
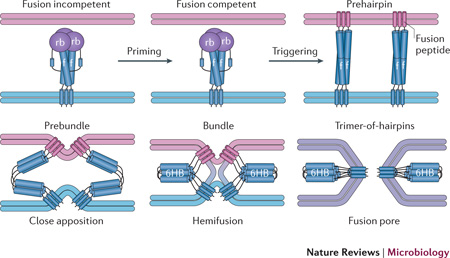
Box 1: Pathway of viral membrane fusion30, 31.
All enveloped viruses penetrate into host cells using a viral membrane fusion protein. The figure depicts a model for membrane fusion by a class I fusion protein; the initial and final protein depictions are based on x-ray structures of several class I fusion proteins. Class I fusion proteins are trimers of three identical units. For most, including EBOV glycoprotein, influenza haemagglutinin and retroviral Envs, each monomeric unit consists of a receptor binding (rb) and a fusion (f) subunit, which are initially present in a single polypeptide chain. Priming by proteolytic cleavage (generally between the receptor binding and fusion subunits) converts the protein from a fusion incompetent to a fusion competent (metastable) form that can respond to a fusion trigger. Triggering exposes and repositions the previously hidden (or tacked-down) fusion peptide (or fusion loop), which then binds hydrophobically to the target membrane. The structure that bridges the viral and target membranes is envisioned to be a trimeric coiled-coil rod, termed a prehairpin, composed of the three (identical) fusion subunits. Subsequent fold-back steps drive the protein through prebundle, bundle and trimer-of-hairpins configurations; the trimer-of-hairpins configuration is a six-helix bundle (6HB) in the case of class I fusion proteins. These structural changes drive the viral (blue) and target (pink) membranes through stages of close apposition and hemifusion, followed by formation of a fusion pore that eventually grows large enough for the viral genome to pass through. Movement of or changes in the receptor-binding subunit are needed to unclamp the fusion subunit, converting it from the fusion-incompetent to the fusion-competent state. Class II and III fusion proteins undergo similar conformational changes (forming prehairpins and trimers-of-hairpins) and induce fusion through similar stages of membrane dynamics. The figure is modified with permission from reference30.
EBOV GP is synthesized as a single polypeptide chain that is cleaved in the Golgi (during biosynthesis) by a furin-like protease into its receptor-binding (GP1) and fusion (GP2) subunits, which remain together through non-covalent interactions and through a disulfide (S-S) bond. Three GP1-S-S-GP2 units come together to form the homotrimer that protrudes from the virion surface15, 29. However, in contrast to similarly organized class I fusion proteins (for example, influenza haemagglutinin), the cleavage site that separates GP1 and GP2 is dispensable for entry32 as well as for infection and pathogenesis33. Therefore, the simple furin cleavage event is not sufficient to prime GP.
Full priming occurs only on fully formed EBOV particles and only after they have been bound, internalized and trafficked to a late endosome in a new target cell (Fig. 1). There, functional priming is mediated by the Cys proteases cathepsin B and cathepsin L34, 35, which cleave within the β13– β14 loop of GP113, 29, 36, a segment that likely crosses over the fusion loop in the native trimer37. Cathepsin priming removes ~60% of the amino acids from GP1, including the mucin-like domain, glycan cap and outermost β-strand of the proposed receptor-binding region (RBR; within the receptor-binding subunit) (Fig. 2b)13, 15, 29, 38–41. Priming seems to occur in two steps, with cathepsin L cleaving GP to a 20 kDa species that is cleaved further by cathepsin B to 19 kDa (Fig. 2a)13, 35, 36. 20 kDa and 19 kDa GP have different biochemical and biological properties. Entry of pseudovirions bearing 20 kDa GP is strongly inhibited by cathepsin B antagonists, whereas entry of pseudovirions bearing 19 kDa GP is not35, 42. Moreover, 19 kDa GP is more sensitive to proteolysis42 and more readily triggered to undergo a conformational change43 than 20 kDa GP.
Two hypotheses have been proposed to explain why GP undergoes such a marked proteolytic priming event. The first is to expose the RBR of GP so that it can interact with an endosomal receptor44. Alternatively, or in addition, priming may potentiate GP for fusion triggering35, 42. As discussed below, both possibilities have received support28, 43, 60.
Triggering EBOV GP
Once primed, all viral fusion proteins must be stimulated by a fusion trigger (Box 1), of which there are four known types: interaction of the receptor-binding subunit of the fusion protein (or a companion viral attachment protein) with a cell surface receptor; sequential interactions (on the cell surface) with a receptor and co-receptor; exposure to low pH in endosomes; and sequential interaction with a cell surface receptor followed by exposure to low pH in endosomes30. Therefore, all known receptor-triggered fusion events occur at the cell surface, whereas all low pH-triggered ones occur in endosomes.
Stimulation by the relevant fusion trigger (for example, receptor binding and/or low pH) elicits a series of marked conformational changes (Box 1). A key early effect is exposure of a hydrophobic fusion peptide (or fusion loop), a protein segment that is vital for fusion and is either buried or tacked down in the native fusion protein. Once liberated and repositioned, the fusion peptide binds to the target membrane, forming a rod-shaped intermediate (a prehairpin), which bridges the viral and target membranes. Next, the fusion protein bends approximately in half, and in so doing pulls the viral membrane and the target membrane (either cell surface or endosome) into close proximity, ultimately leading the two membranes to merge30, 31.
The manner by which primed EBOV GP is triggered does not conform to any known triggering mechanism. Similarly to some other viruses low pH is needed35 (Fig. 2a) to facilitates conformational changes in primed GP43, 45; however, it is not sufficient to trigger primed GP, nor apparently are changes in cation concentrations or further cathepsin digestion42, 43. In vitro, mild treatment, preferentially at low pH, with a reagent that reduces disulfide bonds can trigger primed GP to bind to target membranes through its fusion loop43. Although these observations support the hypothesis that extensive processing potentiates GP for triggering, how primed GP is triggered in vivo and why triggering of primed GP remains sensitive to the Cys protease inhibitor E6435,42 remain unsolved mysteries.
NPC1 is a key EBOV entry factor
Aside from general endocytic machinery and cathepsins, it has remained unclear which host cell factors are crucial for EBOV entry. However, two recent studies identified new players in the process. Intriguingly, both studies revealed a key role for the endosomal membrane protein NPC127, 28.
NPC1 is required for EBOV entry
Carette et al.27 illuminated NPC1 as a crucial EBOV entry factor through a human genome-wide haploid genetic screen27, an approach that had unearthed proteins required for bacterial intoxication and influenza virus infection46. Of the genes identified, all the ones encoding proteins with known functions have roles in the maturation or function of late endosomes or lysosomes. These included cathepsin B, a previously described EBOV entry factor34, 35 (Fig. 2a), and all six subunits of the HOPS complex, a multisubunit machine that assembles components required for fusion between late endocytic organelles, for example between late endosomes and lysosomes to form endolysosomes47–49. The gene that had the greatest number of hits encodes NPC1, a ubiquitously expressed multimembrane-spanning protein that resides primarily in the limiting membrane of late endosomes and lysosomes50, 51. The most recognized role of NPC1 is to aid egress of cholesterol out of late endosomes for redistribution to cellular membranes, including the endoplasmic reticulum and plasma membrane. Consequently, when NPC1 is deficient, cholesterol accumulates in late endosome-lysosomes51–53.
Taking a different approach, Cote et al.28 screened a chemical library for compounds that inhibit infection by pseudovirions bearing EBOV GP28. They showed that inhibitory compound 3.0 and its more potent derivative, compound 3.47, cause cholesterol accumulation in endosomes, a hallmark of Niemann–Pick disease type C (NPC; caused by mutations in the gene encoding NPC1). In a follow-up experiment, NPC1 was the only one of the five tested proteins involved in cholesterol homeostasis to emerge as essential for EBOV entry.
Importantly, both groups found that the role of NPC1 in EBOV entry is independent of its function in cholesterol egress: cells lacking NPC2, an NPC1 partner for cholesterol egress51, 54, or expressing a mutant NPC1 defective in cholesterol egress were still susceptible to EBOV GP-mediated infection.
What is the role of NPC1 in EBOV entry?
NPC1 is clearly involved in EBOV entry, and several lines of evidence suggest that it functions close to, or at the step of, virus–endosome membrane fusion. First, NPC1 is primarily located in late endosomes–lysosomes, where EBOV is thought to enter. Second, EBOV GP particles accumulate in late endosomes and fail to access the cytoplasm in NPC1-null cells. Third, particles bearing primed EBOV GP also require NPC1, suggesting that NPC1 may function after cathepsin priming27, 28 (Fig. 1).
But, what is its precise role? Cote et al.28 showed that a primed trimeric GP ectodomain (GP’ Ecto) bound to a preparation of late endosomes-lysosomes from cells expressing exogenous NPC1, but not from NPC1-null cells. When NPC1-containing late endosomal-lysosomal membranes with bound GP’-Ecto were subsequently solubilized with a detergent, GP’-Ecto could be immunoprecipitated by an antibody to NPC1. Cote et al. further observed a correlation between the potency of compound 3.0 and two of its derivatives for blocking EBOV entry and for blocking GP’-Ecto binding to NPC1-containing late endosome-lysosome membranes. They therefore proposed that NPC1 binds to primed GP. Miller et al.60 have recently provided strong support for this notion. They demonstrated a direct interaction between primed GP and the second lumenal loop—the ‘C loop’—of NPC1 that depends on previously identified critical residues within the RBR in GP113, 38–41. Hence, appealingly, cathepsin priming renders the RBR accessible for binding to NPC1 (Fig. 2b).
Cote et al.28 and Miller et al.60 proposed that, following binding, NPC1 participates in triggering the fusion activity of primed GP. In the simplest model (model 1, Fig. 2c), NPC1 is the necessary and sufficient additional factor that triggers GP at low pH (Fig. 2a) to undergo the requisite fusion-inducing conformational changes (Box 1). An alternate possibility (model 2, Fig. 2c) is that NPC1 participates in, but is not solely responsible for, fusion triggering at low pH. If either of the first two models is correct, EBOV fusion triggering would represent an important exception to the generality that receptor triggering of viral fusion proteins occurs only at the cell surface30. In a third model (Fig. 2c, model 3), NPC1 binds to GP1 but does not actively participate in triggering, which must be mediated by an unknown additional factor, perhaps the reducing potential in endosomes43. If model 3 is correct, then NPC1 probably has essential roles close to, but upstream of, fusion triggering (see below).
Irrespective of whether NPC1 actively participates in fusion triggering (model 1 or 2, Fig. 2c), there are additional ways in which it might support fusion after cathepsin priming. First, binding of NPC1 to primed GP would attach EBOV to the limiting membrane of the late endosome-lysosome50, 51, ensuring that upon fusion the viral genome is delivered directly into the cytoplasm (instead of into small vesicles found within late endosomes-lysosomes48). Second, by binding to primed GP, NPC1 may protect it from further inactivating proteolysis42, 43. And, finally, NPC1 may affect the composition of late endosomes–lysosomes51 (lumen and/or limiting membrane), generating conditions that are optimal for fusion.
NPC1 as an antiviral target
These studies have raised hopes for the development of a therapeutic agent that targets NPC1 to treat EBOV and potentially other filovirus infections. Indeed, compounds that potently inhibit EBOV entry block binding of primed GP to NPC1-containing late endosome-lysosome membranes28. Furthermore, mice heterozygous for NPC1, the late endosomes-lysosomes of which should possess less NPC1 than those of normal mice, are significantly protected from challenges with EBOV and Marburgvirus27. Moreover, NPC1 is required for entry of all five species of EBOV as well as Marburgvirus27, 28. Curiously, given its central role in endosome function and dynamics48, 51, 53, NPC1 does not seem to be required for the entry of ten other viruses tested, including several that enter though late endosomes55. Hence, the collective findings27, 28 suggest that by targeting NPC1 it may be possible to prevent entry and infection by multiple filoviruses, without having dire cell biological consequences.
Another set of observations bear on inhibitor design. U18666A is a small molecule that has been used extensively to mimic the phenotype of cells from patients with NPC52, 56; for example, U18666A causes cholesterol accumulation in late endosomes, similarly to compounds 3.0 and 3.47. Moreover, U18666A blocks EBOV GP-mediated entry and infection27 (J. Shoemaker, C. Scully, G. Olinger, and J.M. W., unpublished observations). Curiously, although U18666A has been shown to interact with purified NPC157, it does not block binding of primed EBOV GP to late endosomal-lysosomal membranes28. This suggests that there may be two classes of agents that interact with NPC1 that can impede filovirus entry and infection. One class, typified by compound 3.47, may bind to the site on NPC1 that engages primed GP, competitively block GP binding. The second class, typified by U18666A, may bind to a different site on NPC1, disabling its role in maintaining the proper composition of late endosome-lysosomes51, 56. Because certain late endosome-lysosome constituents may be needed in addition to NPC1 for EBOV entry (models 2 and 3, Fig. 2c), the second class of compounds could prevent EBOV entry without physically precluding the NPC1–GP interaction. Despite considerations raised above, it may of course be challenging to use either class of compound in a therapeutic setting, given the importance of NPC1 to endosome function and cholesterol homeostasis50–53, 56, 58, 59.
Perspectives
The recent studies by Carette et al.27, Cote et al.28 and Miller et al60 have offered us fresh insights into the cell biology of filovirus entry and have identified a new host protein, NPC1, that may serve as a therapeutic target. However, many questions remain.
First and foremost, further biochemical and cell biological studies are needed to determine the exact roles of NPC1 in EBOV entry. Which residues of the NPC1 C-loop and primed GP engage each other? What is the affinity of the interaction? Is the interaction sufficient to trigger fusion at low pH (Fig. 2c, model 1)? If additional factors are required (Fig. 2c, model 2), what are they and do they work in concert or sequentially with NPC1? Or, if NPC1 is not actively involved in fusion triggering (Fig. 2c, model 3), which of several possible alternate parts does it play, and what is the bona fide fusion trigger?
It will also be important to determine whether compounds typified by 3.47 or U18666A can be used to treat EBOV infections in primates. Finally, structural studies should be carried out on the interface between NPC1 and primed GP to guide further development of small molecules and antibodies for the prevention or treatment of filovirus infections.
Acknowledgements
The authors acknowledge U54 AI057168 for funding work on EBOV entry, Matthew Brecher and Sonia Gregory for help with Fig 2b, and Matthew Brecher, David Castle and Jason Shoemaker for helpful comments on the text.
References
- 1.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan NJ, Martin JE, Graham BS, Nabel GJ. Correlates of protective immunity for Ebola vaccines: implications for regulatory approval by the animal rule. Nat Rev Microbiol. 2009;7:393–400. doi: 10.1038/nrmicro2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn JH, et al. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borio L, et al. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA. 2002;287:2391–2405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- 5.Miranda ME, Miranda NL. Reston ebolavirus in humans and animals in the Philippines: a review. J Infect Dis. 2011;204(Suppl 3):S757–S760. doi: 10.1093/infdis/jir296. [DOI] [PubMed] [Google Scholar]
- 6.Negredo A, et al. Discovery of an ebolavirus-like filovirus in europe. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002304. e1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dube D, et al. Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc Natl Acad Sci U S A. 2010;107:16637–16642. doi: 10.1073/pnas.1008509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backovic M, Rey F. Virus entry: old viruses, new receptors. Curr. Opinion Virol. 2012 doi: 10.1016/j.coviro.2011.12.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuno K, et al. C-type lectins do not act as functional receptors for filovirus entry into cells. Biochem Biophys Res Commun. 2010;403:144–148. doi: 10.1016/j.bbrc.2010.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez CP, et al. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons G, et al. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology. 2003;305:115–123. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- 12.Takada A, et al. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J Virol. 2004;78:2943–2947. doi: 10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dube D, et al. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J Virol. 2009;83:2883–2891. doi: 10.1128/JVI.01956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondratowicz AS, et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci U S A. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JE, Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimojima M, Ikeda Y, Kawaoka Y. The mechanism of Axl-mediated Ebola virus infection. J Infect Dis. 2007;196(Suppl 2):S259–S263. doi: 10.1086/520594. [DOI] [PubMed] [Google Scholar]
- 17.Takada A, et al. Downregulation of beta1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology. 2000;278:20–26. doi: 10.1006/viro.2000.0601. [DOI] [PubMed] [Google Scholar]
- 18.Brindley MA, et al. Tyrosine kinase receptor Axl enhances entry of Zaire ebolavirus without direct interactions with the viral glycoprotein. Virology. 2011;415:83–94. doi: 10.1016/j.virol.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schornberg KL, et al. Alpha5beta1-integrin controls ebolavirus entry by regulating endosomal cathepsins. Proc Natl Acad Sci U S A. 2009;106:8003–8008. doi: 10.1073/pnas.0807578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulherkar N, Raaben M, de la Torre JC, Whelan SP, Chandran K. The Ebola virus glycoprotein mediates entry via a non-classical dynamin-dependent macropinocytic pathway. Virology. 2011;419:72–83. doi: 10.1016/j.virol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nanbo A, et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001121. e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001110. e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bharat TA, et al. Cryo-Electron Tomography of Marburg Virus Particles and Their Morphogenesis within Infected Cells. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001196. e1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez A, Geisbert T, Feldmann H. In: Fields Virology. Knipe D, Howley P, editors. Philadelphia, PA: Lippincott Williams and Wilkins; 2007. pp. 1407–1448. [Google Scholar]
- 25.Teissier E, Penin F, Pecheur E-I. Targeting cell entry of enveloped viruses as an antiviral strategy. Molecules. 2011;16:221–250. doi: 10.3390/molecules16010221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tilton JC, Doms RW. Entry inhibitors in the treatment of HIV-1 infection. Antiviral Res. 2010;85:91–100. doi: 10.1016/j.antiviral.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 27.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cote M, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wool-Lewis RJ, Bates P. Endoproteolytic processing of the ebola virus envelope glycoprotein: cleavage is not required for function. J Virol. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neumann G, et al. Proteolytic processing of the Ebola virus glycoprotein is not critical for Ebola virus replication in nonhuman primates. J Virol. 2007;81:2995–2998. doi: 10.1128/JVI.02486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schornberg K, et al. Role of endosomal cathepsins in entry mediated by the ebola virus glycoprotein. J Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hood CL, et al. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J Virol. 2010;84:2972–2982. doi: 10.1128/JVI.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dias JM, et al. A Shared Structural Solution for Neutralizing Ebolaviruses. Nat Struct Mol Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhn JH, et al. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J Biol Chem. 2006;281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- 39.Manicassamy B, Wang J, Jiang H, Rong L. Comprehensive analysis of ebola virus GP1 in viral entry. J Virol. 2005;79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brindley MA, et al. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J Virol. 2007;81:7702–7709. doi: 10.1128/JVI.02433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mpanju OM, Towner JS, Dover JE, Nichol ST, Wilson CA. Identification of two amino acid residues on Ebola virus glycoprotein 1 critical for cell entry. Virus Res. 2006;121:205–214. doi: 10.1016/j.virusres.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J Virol. 2010;84:163–175. doi: 10.1128/JVI.01832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brecher M, et al. Cathepsin Cleavage Potentiates the Ebola Virus Glycoprotein to Undergo a Subsequent Fusion Relevant Conformational Change. J Virol. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaletsky RL, Simmons G, Bates P. Proteolysis of the Ebola glycoproteins enhances virus binding and infectivity. J Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregory SM, et al. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc Natl Acad Sci U S A. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 47.Epp N, Rethmeier R, Kramer L, Ungermann C. Membrane dynamics and fusion at late endosomes and vacuoles--Rab regulation, multisubunit tethering complexes and SNAREs. Eur J Cell Biol. 2011;90:779–785. doi: 10.1016/j.ejcb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pryor PR, Luzio JP. Delivery of endocytosed membrane proteins to the lysosome. Biochim Biophys Acta. 2009;1793:615–624. doi: 10.1016/j.bbamcr.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 50.Frolov A, et al. Cholesterol overload promotes morphogenesis of a Niemann-Pick C (NPC)-like compartment independent of inhibition of NPC1 or HE1/NPC2 function. J Biol Chem. 2001;276:46414–46421. doi: 10.1074/jbc.M108099200. [DOI] [PubMed] [Google Scholar]
- 51.Lloyd-Evans E, Platt FM. Lipids on trial: the search for the offending metabolite in Niemann-Pick type C disease. Traffic. 2010;11:419–428. doi: 10.1111/j.1600-0854.2010.01032.x. [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi T, et al. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat Cell Biol. 1999;1:113–118. doi: 10.1038/10084. [DOI] [PubMed] [Google Scholar]
- 53.Vitner EB, Platt FM, Futerman AH. Common and uncommon pathogenic cascades in lysosomal storage diseases. J Biol Chem. 2010;285:20423–20427. doi: 10.1074/jbc.R110.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deffieu MS, Pfeffer SR. Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc Natl Acad Sci U S A. 2011;108:18932–18936. doi: 10.1073/pnas.1110439108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lozach P-Y, Huotari J, Helenius A. Late-penetrating viruses. Curr. Opinion Virol. 2011;1:35–43. doi: 10.1016/j.coviro.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Lloyd-Evans E, et al. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat Med. 2008;14:1247–1255. doi: 10.1038/nm.1876. [DOI] [PubMed] [Google Scholar]
- 57.Liu R, Lu P, Chu JW, Sharom FJ. Characterization of fluorescent sterol binding to purified human NPC1. J Biol Chem. 2009;284:1840–1852. doi: 10.1074/jbc.M803741200. [DOI] [PubMed] [Google Scholar]
- 58.Ko DC, Gordon MD, Jin JY, Scott MP. Dynamic movements of organelles containing Niemann-Pick C1 protein: NPC1 involvement in late endocytic events. Mol Biol Cell. 2001;12:601–614. doi: 10.1091/mbc.12.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sobo K, et al. Late endosomal cholesterol accumulation leads to impaired intra-endosomal trafficking. PLoS One. 2007;2:e851. doi: 10.1371/journal.pone.0000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller EH, et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012 doi: 10.1038/emboj.2012.53. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]