Abstract
Changes in gene expression before, during and after five generations of permethrin laboratory selection were monitored in six strains of Aedes aegypti: five F2 – F3 collections from the Yucatán Peninsula of México and one F2 from Iquitos, Perú. Three biological replicate lines were generated for each strain. The response to selection was measured as changes in the lethal and knockdown permethrin concentrations (LC50, KC50) and in the frequency of the Ile1,016 substitution in the voltage gated sodium channel (para) gene. Changes in expression of 290 metabolic detoxification genes were measured using the “Aedes Detox” microarray. Selection simultaneously increased the LC50, KC50 and Ile1,016 frequency. There was an inverse relationship between Ile1,016 frequency and the numbers of differentially transcribed genes. The Iquitos strain lacked the Ile1,016 allele and 51 genes were differentially transcribed following selection as compared to 10–18 genes in the Mexican strains. Very few of the same genes were differentially transcribed among field strains but ten cytochrome P450 genes were upregulated in more than one strain. Laboratory adaptation to permethrin in Ae. aegypti is genetically complex and largely conditioned by geographic origin and preexisting target site insensitivity in the para gene. The lack of uniformity in the genes that responded to artificial selection as well as differences in the direction of their responses challenges the assumption that one or a few genes control permethrin metabolic resistance. Attempts to identify one or a few metabolic genes that are predictably associated with permethrin adaptation may be futile.
Keywords: Aedes aegypti, detoxification, knock-down resistance, artificial selection, permethrin resistance
Introduction
Dengue is the most important mosquito-borne viral disease affecting humans, with Aedes aegypti, the primary mosquito vector, found in nearly 100 tropical countries. Each year, there are an estimated 50–100 million cases of dengue fever (DF) and, depending on the year, 250,000 –500,000 cases of dengue hemorrhagic Fever (DHF) (Gubler 2005). No vaccines or medicines prevent DF, leaving mosquito control as the best strategy to lessen dengue transmission. Unfortunately, most national programs are not equipped to manage the prevention and control aspects of a dengue program and heavy reliance continues to be placed on chemical control (Lloyd 2003).
Adults are commonly treated by space spraying of insecticides with ultra-low volume (ULV), thermal fogging, or aerial application during dengue epidemics. Pyrethroids have become the preferred group of insecticides for ULV spraying. These neurotoxins disrupt the function of the voltage-gated sodium channels (VGSC encoded by the orthologue of para in Drosophila (Loughney et al. 1989)) by preventing the re-polarization phase of action potentials through the nerve cell membrane. Pyrethroid resistance has been reported in Ae. aegypti in many countries, including México, Brazil, Cuba, Thailand, Vietnam and others (da-Cunha et al. 2005; Flores et al. 2009; Jirakanjanakit et al. 2007; Kawada et al. 2009; Martins et al. 2009; Paeporn et al. 2005; Rodriguez et al. 2005). Resistance is conferred by target site insensitivity and/or increased metabolic detoxification. Target site resistance involves structural changes in the VGSC that cause reduced pyrethroid binding. Depending on pyrethroid dose, insects do not display the characteristic knockdown effect and are referred to as knockdown resistant (‘kdr’).
Knockdown resistance arises through nonsynonymous mutations in the VGSC gene and eleven replacement mutations have been identified in Ae. aegypti (Brengues et al. 2003; Saavedra-Rodriguez et al. 2007; Yanola et al. 2010). Two nonsynonymous mutations in domain II, segment 6 on the VGSC gene have been detected in Latin American Ae. aegypti collections. A valine to isoleucine replacement in codon 1,016 (Ile1,016) and a phenylalanine to cysteine replacement in codon 1,534 (Cys1,534) have been definitively associated with permethrin resistance (Harris et al. 2010; Saavedra-Rodriguez et al. 2008; Saavedra-Rodriguez et al. 2007).
Pyrethroids can also be metabolized by two enzyme families, the cytochrome P450 mono-oxygenases (CYP) and the carboxyl/esterases (CCE) (Hemingway et al. 2004). The metabolism of pyrethroids occurs by ester hydrolysis and oxidation at methyl-, methylene-, alkenyl-, or aryl-substituents and ≥ 80 metabolites have been identified from cis- and trans-permethrin (Casida et al. 1983). Identification and isolation of individual detoxification enzymes in mosquitoes has been difficult because of the vast array of potential enzymes many of which have similar substrates. Recently, new mosquito genomic tools and microarray technology have been developed to assist in identification of elevated transcription of individual detoxification genes. Microarrays containing probes for putative detoxification genes have been developed for the malaria vector Anopheles gambiae (David et al. 2005) and also for the dengue vector Ae. aegypti (Strode et al. 2008). Candidate detoxification genes have been identified in pyrethroid resistant strains of An. gambiae (David et al. 2005; Djouaka et al. 2008; Muller et al. 2007) and Ae. aegypti (Marcombe et al. 2009; Strode et al. 2008). These studies support the hypothesis that gene over-expression is not due to transient upregulation caused by the exposure of the adults to permethrin but instead reflect constitutive over-expression of a common subset of detoxification genes in addition to genes that are uniquely expressed in each population (Muller et al. 2008). The challenge in these studies is that susceptible strains with a common genetic background are usually not available to compare, and results are biased by geographical variation or genetic drift in susceptible reference strains.
The objective of the present study was to investigate whether laboratory selection for permethrin resistance in Ae. aegypti derived from Latin America is also associated with changes in detoxification gene expression. The identification of genes associated with permethrin resistance could be a starting point for development of insecticide resistance diagnostic markers in mosquito populations from Latin America. Herein we analyze the response to selection in five mosquito strains from México known to have the Ile1,016 mutation in a range of frequencies and in a strain from Iquitos, Perú without this mutation. We monitored changes in the lethal concentrations (LC50) and knockdown concentrations (KC50) and in the frequency of the Ile1,016 allele. We demonstrate simultaneous increases in LC50, KC50 and Ile1,016 and changes in gene expression as a result of selection. However, microarray analyses demonstrated that laboratory adaptation to permethrin in Ae. aegypti is genetically complex, largely conditioned by target site insensivity in the para gene and dependent upon the geographic origin of the strain.
Results
Bioassays
Six Ae. aegypti lines collected from southern México and one collected from Iquitos, Perú (Table 1) were used in all experiments. The F2 or F3 offspring from field collections were designated FS0 (no prior selection). The KC50, LC50 and Ile1,016 allele frequencies were measured in each of the unselected FS0 lines (Table 1). Low levels of resistance (RR50 < 5) were detected in Iquitos, Lázaro Cárdenas, Solidaridad, and Isla Mujeres unselected FS0 lines. KC50 and LC50 values were similar to those obtained in the New Orleans susceptible line and in Iquitos the Ile1,016 replacement was not detected. Moderate (RR50 = 5 – 10) levels of resistance were detected in the Calderitas and Mérida FS0 lines. High (RR50 >10) levels of resistance were only detected in the Lagunitas FS0. The Ile1,016 allele was detected in all Mexican unselected lines, with frequencies > 0.5 in Lagunitas, Mérida and Lázaro Cárdenas. As in previous studies there was a correlation (R2 = 0.60, P = 0.025) between LC50 and KC50. Furthermore, LC50 and KC50 are strongly correlated (R2 with LC50 = 0.68, P = 0.012; R2 with KC50 = 0.83, P = 0.002) with the Ile1,016 frequency squared (the expected frequency of homozygotes).
Table 1.
Collection site information and adult permethrin resistance prior to artificial selection. LC50 and KC50 are µg of permethrin per 250mL Wheaton Bottle. The LC50 or KC50 resistance ratios are calculated relative to the susceptible New Orleans strain. The initial Ile1,016 allele frequency was calculated from ~50 individuals from each collection site. Pearson's correlation coefficients were calculated between LC50 or KC50 values and the frequency of Ile1,016 squared.
| Country State |
City | Site in City | Coordinates | Generation | LC50 ug/a.i. (95% CI) |
RR LC50 P1/NO |
KC50 ug/a.i. (95% CI) |
RR KC50 P1/NO |
Ile1,016 freq. FS0 |
|---|---|---|---|---|---|---|---|---|---|
| Perú | |||||||||
| Iquitos | Iquitos | F2 | 5.6 (4.3–7.0) | 2.1 | 0.3 (0.26–0.44) | 0.7 | 0.000 | ||
| México | |||||||||
| Quintana Roo | Chetumal | ||||||||
| Calderitas | 18.556269° –88.256181° | F3 | 16.3 (13.2–20) | 6.2 | 1.8 (1.27–2.63) | 3.7 | 0.360 | ||
| Lagunitas | 18.519485° –88.333923° | F3 | 25.4 (22.6–28.6) | 10.2 | 21.7 (18.01–26.1) | 43.4 | 0.854 | ||
| Lázaro Cárdenas | 18.535938° –88.301646° | F3 | 11.0 (7.9–15.2) | 4.2 | 1.6 (0.49–5) | 3.1 | 0.600 | ||
| Solidaridad | 18.528407° –88.302693° | F3 | 6.9 (5.2–8.9) | 2.6 | 1.4 (0.51–3.7) | 2.8 | 0.320 | ||
| Yucatán | |||||||||
| Mérida | 20.948942° –89.640526° | F2 | 22.4 (19.9–25.1) | 8.5 | 5.7 (3.7–8.8) | 11.5 | 0.546 | ||
| United States | |||||||||
| New Orleans | 2.6 | 0.5 | 0.000 | ||||||
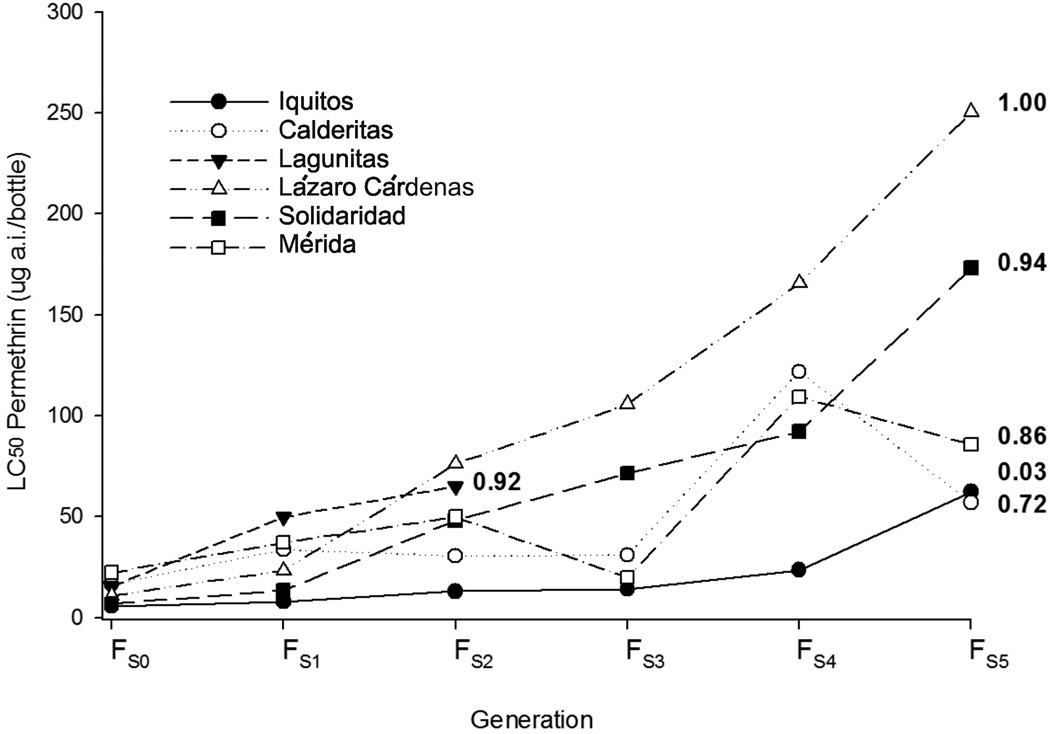
The general response to permethrin selection was an increase in LC50, KC50 and Ile1,016 allele frequencies in all mosquito lines (Figures 2 and 3). However each strain exhibited a distinct response pattern. Table 3 shows the realized heritability (h2) coefficients for KC50 and LC50 calculated during the selection process. During selection in Iquitos, Solidaridad, and Mérida, there was a large h2 for LC50 but not for KC50. Using the experiment-wise error rate the h2 for LC50 was only significant in the Iquitos and Solidaridad collections. The Ile1,016 frequency was almost fixed in Solidaridad FS5 and showed a significant increase in Mérida. In contrast during Lázaro Cárdenas selection, a large h2 was observed for both KC50 and LC50 and Ile1,016 went to fixation. The LC50 h2 during selection in Calderitas was not significant while the KC50 h2 was large and significant and Ile1,016 frequency increased after selection. Selection results for the Lagunitas strain are not shown because all three replicates of this strain died simultaneously in the FS2 due to low oviposition rates.
Figure 2.
LC50 response of mosquito lines to permethrin selection over five generations. The FS5 Ile1,016 allele frequencies were calculated from ~50 individuals and appear in the right side of the graph.
Figure 3.
KC50 response of mosquito lines to permethrin selection over five generations. The FS5 Ile1,016 allele frequencies were calculated from ~50 individuals and appear in the right side of the graph.
Table 3.
Realized heritability (h2) of LC50 or KC50 resulting from five generations of permethrin selection. An ANOVA was performed to test the hypothesis: h2 > 0.
| Selected Line | LC50 h2 |
p* | KC50 h2 |
p* |
|---|---|---|---|---|
| Iquitos | 0.616 | 0.001 | 0.181 | 0.381 |
| Solidaridad | 0.864 | 0.006 | 0.142 | 0.008 |
| Mérida | 0.740 | 0.034 | 0.229 | 0.066 |
| Lázaro Cárdenas | 0.829 | 0.022 | 1.397 | 0.050 |
| Calderitas | 0.241 | 0.573 | 0.789 | 0.014 |
Bonferroni experimentwise rate = 1−(1−0.05)1/5 ≃ 0.01
Microarray validation
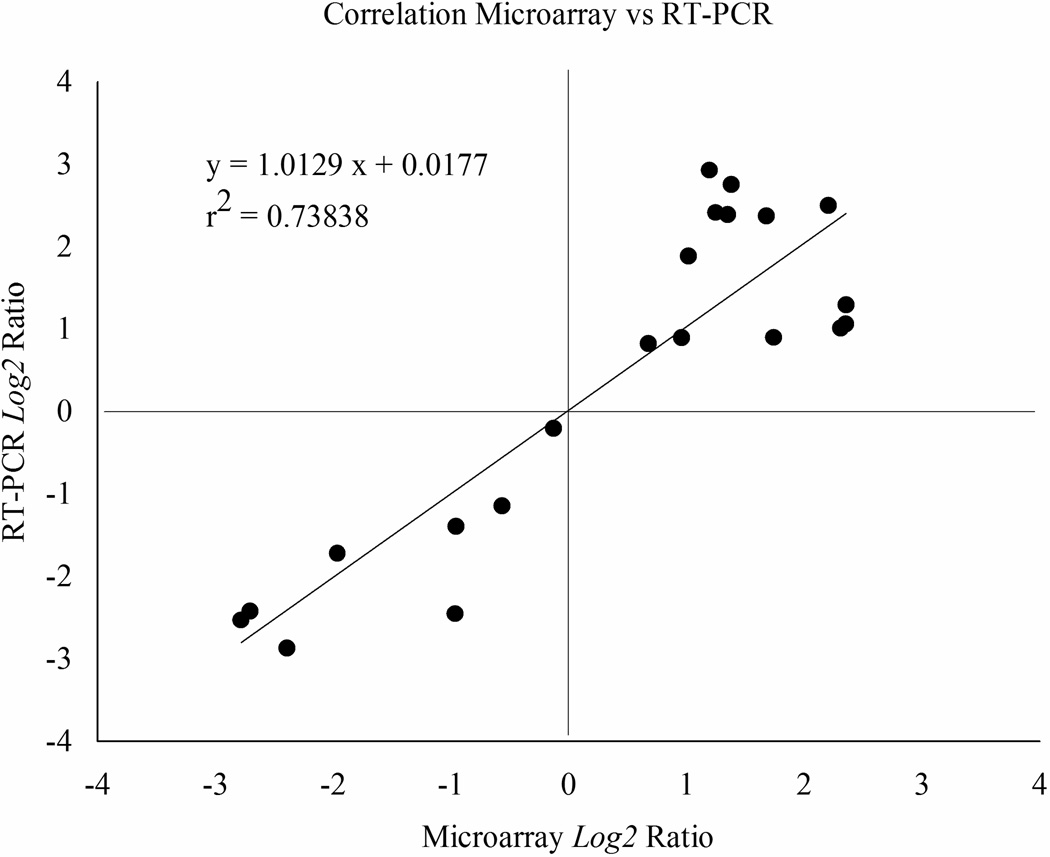
Expression ratios are expressed as M, the log2 of mean transcription ratios, where M = log2 (Cy5/Cy3), Cy5 is the adsorption at 649 nm and Cy3 is the adsorption at 532 nm. Expression ratios from eight genes significantly differentially expressed in the microarray comparisons were correlated with the expression ratios obtained by quantitative-PCR using the same amplified RNA samples (Figure 4). We validated the up-regulation of CYP4J13, CYP325G3, AaeCOE-8, CYP6Nae1 and CYP9J22v1 and the down regulation of AaGSTs1-1. AAEL004388 and AAEL004390 were upregulated in Iquitos FS5 but were down-regulated in Calderitas FS5 and this trend was validated by quantitative-PCR.
Figure 4.
Correlation between microarray and real time expression ratios. Ratios are display in a log2 scale.
Gene expression - FS0 versus New Orleans susceptible strain
Comparisons of transcription patterns among the six FS0 strains relative to New Orleans identified a total of 41 differentially expressed genes. Seventeen of these 41 genes appeared in two or more unselected mosquito lines (Table 4a). Thirteen genes were up-regulated including three epsilon GSTs (3, 4, 6); seven members of the CYP9J family (CYP9J-9, -10, -22, -23, -28, -32), two CYP6 genes (CYP6-Nae1, -Z6) and one peroxinectin (Peroxinectin 4390). CYP6P12, CYP9AE1 and AaGSTs1-2 were downregulated. Only GSTd5 showed inconsistency in expression direction. Table 4b lists the 25 genes that were uniquely differentially transcribed in individual strains. No genes were differentially transcribed in Calderitas or Solidaridad and a single epsilon GSTe7 was upregulated in Lagunitas. Only two CYP9 and two CYP6 genes were up-regulated in Mérida. In contrast, eight genes were differentially transcribed in Lázaro Cárdenas and thirteen genes were differentially transcribed in Iquitos. In total, Solidaridad and Calderitas only exhibited differential transcription of 2–3 genes while Iquitos, Lázaro Cárdenas, Mérida, and Lagunitas exhibited differential transcription at from 10–20 loci.
Table 4.
| a. Genes differentially expressed in two or more unselected FS0 strains relative to the New Orleans strain. Ratio of the average expression of each comparison is displayed on a linear scale. Probability value is shown as a negative log10 scale. | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Activity | Iquitos | Calderitas | Lázaro C | Solidaridad | Mérida | Lagunitas | |||||||
| Gene | Vector Base ID | Ratio | p | Ratio | p | Ratio | p | Ratio | p | Ratio | p | Ratio | p |
| Glutathione transferases | |||||||||||||
| AaGSTe3 | AAEL007947 | 2.06 | 4.29 | 1.99 | 9.80 | 2.93 | 10.98 | ||||||
| AaGSTe4 | AAEL007962 | 2.48 | 13.09 | 3.18 | 10.44 | ||||||||
| AaGSTe6 | AAEL007946 | 2.23 | 9.90 | 2.01 | 9.53 | 2.03 | 10.07 | 3.25 | 9.49 | ||||
| AaGSTs1-2 | AAEL011741 | 0.35 | 4.59 | 0.27 | 14.14 | ||||||||
| GSTd5 | AAEL001071 | 2.04 | 4.23 | 0.46 | 19.18 | ||||||||
| Cytochrome P450 monoxygenases | |||||||||||||
| CYP6Nae1 | AAEL009126 | 2.20 | 5.43 | 2.07 | 6.18 | ||||||||
| CYP6P12V2 | AAEL014891 | 0.37 | 6.82 | 0.45 | 11.83 | 0.48 | 6.99 | ||||||
| CYP6Z6_b | AAEL009123 | 2.08 | 7.32 | 2.25 | 18.07 | 2.22 | 6.27 | ||||||
| CYP9AE1 | AAEL003748 | 0.47 | 4.56 | 0.37 | 4.85 | ||||||||
| CYP9J10 v2 | AAEL014614 | 2.04 | 3.47 | 3.01 | 18.53 | ||||||||
| CYP9J22 v1 | AAEL006802 | 2.28 | 7.86 | 2.64 | 13.31 | 2.27 | 5.70 | ||||||
| CYP9J22 v2 | AAEL014619 | 2.39 | 7.72 | 2.66 | 13.54 | 2.39 | 3.62 | ||||||
| CYP9J23_b | AAEL014615 | 2.35 | 5.54 | 2.33 | 15.37 | ||||||||
| CYP9J28 | AAEL014617 | 3.14 | 6.80 | 2.93 | 15.98 | ||||||||
| CYP9J32 | AAEL008846 | 8.82 | 12.22 | 2.95 | 20.01 | ||||||||
| CYP9J9 v2 | AAEL014605 | 2.06 | 13.54 | 2.00 | 17.62 | ||||||||
| Red/Ox | |||||||||||||
| Peroxinectin 4390 | AAEL004390 | 2.03 | 7.28 | 3.23 | 8.61 | ||||||||
| b. Genes differentially expressed in one of the unselected FS0 strains relative to the susceptible New Orleans strain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Glutathione transferases | |||||||||
| AaGSTd1-1 | AAEL001061 | 4.00 | 11.45 | ||||||
| AaGSTd1-2 | AAEL001061 | 2.38 | 4.50 | ||||||
| AaGSTd1-3 | AAEL001061 | 2.48 | 8.64 | ||||||
| AaGSTe7 | AAEL007948 | 3.25 | 10.39 | ||||||
| AaGSTs1-1 | AAEL011741 | 1.39 | 10.70 | ||||||
| Cytochrome P450 monoxygenases | |||||||||
| CYP4G36 | AAEL004054 | 0.33 | 6.00 | ||||||
| CYP6BB2a | AAEL014893 | 4.66 | 4.77 | ||||||
| CYP6CB1 | AAEL002046 | 2.81 | 24.91 | ||||||
| CYP9J19 v1 | AAEL006810 | 2.57 | 14.59 | ||||||
| CYP9J9 v1 | AAEL006793 | 2.00 | 15.37 | ||||||
| Carboxyl/esterases | |||||||||
| AaeCOE-10 | AAEL001517 | 1.41 | 13.24 | ||||||
| AaeCOE-17 | AAEL004341 | 0.21 | 10.62 | ||||||
| CCae2B | AAEL017553 | 1.28 | 11.07 | ||||||
| CCae5C | AAEL003201 | 0.40 | 9.75 | ||||||
| CCEunk6o | AAEL000545 | 1.38 | 10.21 | ||||||
| Red/Ox | |||||||||
| Peroxinectin 3612 | AAEL003612 | 4.51 | 8.14 | ||||||
| Peroxinectin 4388 | AAEL004388 | 4.51 | 9.44 | ||||||
| Dihydrolipoamide dehydrogenase | AAEL006928 | 0.42 | 5.84 | ||||||
| Aldehyde oxidase 10367 | AAEL010367 | 2.30 | 10.57 | ||||||
| Glutaredoxin 13980 | AAEL013980 | 2.71 | 5.95 | ||||||
| Aldo-keto reductase 15002 | AAEL015002 | 4.08 | 4.85 | ||||||
| Catalase | AAEL013407 | 0.32 | 8.41 | ||||||
| Thioredoxin peroxidase 9051 | AAEL009051 | 2.14 | 3.70 | ||||||
| Others | |||||||||
| 40S ribosomal ptn S17 | AAEL004175 | 2.50 | 11.88 | ||||||
| 60S ribosomal ptn L44 L41 | AAEL003942 | 2.14 | 7.71 | ||||||
Gene expression - FS1 versus FS0
FS1 and FS0 comparisons identified 53 genes that responded to one generation of selection (Table 5). Forty-seven of these genes were detected in the Iquitos strain and 18 were differentially transcribed in Calderitas. The remaining Mexican FS1 lines had 0–5 differentially expressed genes. Twenty of the 53 genes were differentially expressed in two or more lines (Table 5a). Table 5b lists the remaining 33 genes that were uniquely differentially transcribed in Iquitos (28) or Calderitas (5). Transcription patterns in the remaining genes were inconsistent among strains. For example, eight Red/Ox genes were upregulated in Calderitas FS1 but were down regulated in Iquitos FS1. Down regulation of a group of four CCE’s occurred in the Mexican FS1 lines but the same genes were upregulated in the Iquitos FS1 line.
Table 5.
| a. Genes differentially expressed in two or more FS1 strains relative to the unselected FS0. Ratio of the average expression of each comparison is displayed on a linear scale. Probability value is shown as a negative log10 scale. | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Activity | Iquitos | Calderitas | Lázaro C. | Solidaridad | Mérida | Lagunitas | |||||||
| Gene | Vector Base ID | Ratio | p | Ratio | p | Ratio | p | Ratio | p | Ratio | p | Ratio | p |
| Glutathione transferases | |||||||||||||
| GSTd5 | AAEL001071 | 0.37 | 19.11 | 0.46 | 19.18 | ||||||||
| AaGSTe4 | AAEL007962 | 0.41 | 10.30 | 2.48 | 13.09 | ||||||||
| AaGSTs1-2 | AAEL011741 | 20.97 | 14.85 | 2.89 | 14.50 | 0.17 | 14.80 | ||||||
| Cytochrome P450 monooxygenases | |||||||||||||
| CYP6Z6_b | AAEL009123 | 0.43 | 14.01 | 2.25 | 18.07 | ||||||||
| CYP9J9 v2 | AAEL014605 | 2.51 | 11.65 | 2.00 | 17.62 | ||||||||
| Carboxyl/esterases | |||||||||||||
| AaeCOE-2 | AAEL002376 | 2.16 | 14.29 | 0.34 | 9.16 | ||||||||
| AaeCOE-9 | AAEL012509 | 2.81 | 15.15 | 0.44 | 8.52 | 0.44 | 13.24 | ||||||
| AaeCOE-17 | AAEL004341 | 4.38 | 7.19 | 0.46 | 10.02 | 0.46 | 14.26 | ||||||
| AaeCOE-19 | AAEL005112 | 2.25 | 12.17 | 0.32 | 11.28 | ||||||||
| Red/Ox | |||||||||||||
| Aldehyde oxidase 8 | AAEL010382 | 2.14 | 5.93 | 0.45 | 8.30 | 1.00 | |||||||
| Aldo-keto reductase 11 | AAEL015002 | 0.15 | 12.86 | 2.35 | 8.56 | ||||||||
| Aldo-keto reductase 7 | AAEL004118 | 2.04 | 8.88 | 0.43 | 9.28 | ||||||||
| Dual oxidase | AAEL007563 | 0.36 | 10.96 | 3.16 | 8.78 | ||||||||
| Peroxinectin 3612 | AAEL003612 | 0.18 | 16.46 | 4.44 | 10.46 | ||||||||
| Peroxinectin 4386 | AAEL004386 | 0.30 | 11.39 | 3.18 | 9.00 | ||||||||
| Peroxinectin 4388 | AAEL004388 | 0.15 | 17.99 | 3.34 | 7.94 | ||||||||
| Peroxinectin 4390 | AAEL004390 | 0.07 | 21.72 | 4.96 | 12.16 | 2.55 | 14.98 | ||||||
| Peroxinectin 4401 | AAEL004401 | 0.29 | 11.49 | 3.68 | 7.03 | ||||||||
| Thioredoxin peroxidase 5 | AAEL009051 | 0.45 | 7.10 | 2.31 | 10.55 | ||||||||
| HypProt | AAEL004400 | 0.22 | 16.56 | 3.48 | 8.15 | ||||||||
| b. Genes differentially expressed in one of the FS1 strains relative to the unselected FS0 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutathione transferases | |||||||||||||
| AaGSTd1-1 | AAEL001061 | 0.46 | 4.09 | ||||||||||
| AaGSTd6 | AAEL010591 | 0.49 | 10.41 | ||||||||||
| AaGSTe7 | AAEL007948 | 0.34 | 12.21 | ||||||||||
| AaGSTs1-1 | AAEL011741 | 4.63 | 19.91 | ||||||||||
| Cytochrome P450 monooxygenases | |||||||||||||
| CYP305A6 | AAEL002071 | 0.44 | 5.99 | ||||||||||
| CYP306A1 | AAEL004888 | 0.48 | 14.33 | ||||||||||
| CYP325S2 | AAEL000325 | 0.50 | 8.63 | ||||||||||
| CYP4G36 | AAEL004054 | 2.19 | 17.89 | ||||||||||
| CYP4H28 | AAEL003380 | 0.24 | 9.47 | ||||||||||
| CYP6CB2 | AAEL002872 | 0.38 | 16.05 | ||||||||||
| CYP6Z8_b | AAEL009131 | 2.06 | 17.12 | ||||||||||
| CYP6Z9 | AAEL009129 | 2.91 | 13.27 | ||||||||||
| HypProt1055570 | AAGE01055570 | 0.49 | 8.46 | ||||||||||
| HypProt1055570 | AAGE01055570 | 0.50 | 9.58 | ||||||||||
| Carboxyl/esterases | |||||||||||||
| AaeCOE-14 | AAEL012886 | 2.08 | 8.15 | ||||||||||
| AaeCOE-34 | 0.48 | 9.46 | |||||||||||
| CCae2B | AAEL017553 | 3.14 | 13.32 | ||||||||||
| CEunk6o | AAEL000545 | 2.28 | 10.02 | ||||||||||
| Red/Ox | |||||||||||||
| Peroxinectin 5416 | AAEL005416 | 2.38 | 12.02 | ||||||||||
| Heme peroxidase 6014 | AAEL006014 | 2.25 | 6.4 | ||||||||||
| Cu-Zn Superoxide Dismutase | AAEL006271 | 0.48 | 19.05 | ||||||||||
| Ubiquinol-cytochrome c reductase complex 14kd | AAEL007868 | 2.50 | 21.38 | ||||||||||
| Ubiquinol-cytochrome c reductase hinge protein | AAEL010801 | 2.35 | 15.04 | ||||||||||
| Heme peroxidase 13171 | AAEL013171 | 2.08 | 7.43 | ||||||||||
| Aldehyde oxidase 14493 | AAEL014493 | 0.19 | 16.5 | ||||||||||
| Aldoketo reductase 15002 | AAEL015002 | 0.15 | 12.86 | ||||||||||
| Catalase | AAEL013407 | 5.35 | 12.1 | ||||||||||
| Peroxiredoxin 7135 | AAEL007135 | 2.57 | 11.81 | ||||||||||
| Others | |||||||||||||
| 60S ribosomal ptn L44 L41 | AAEL003942 | 0.37 | 15.7 | ||||||||||
| Acidic ribosomal ptn P1 | AAEL005027 | 0.48 | 11.11 | ||||||||||
| ribosomal ptn L34 | AAEL009341 | 0.48 | 13.23 | ||||||||||
| 40S ribosomal ptn S17 | AAEL004175 | 0.38 | 12.4 | ||||||||||
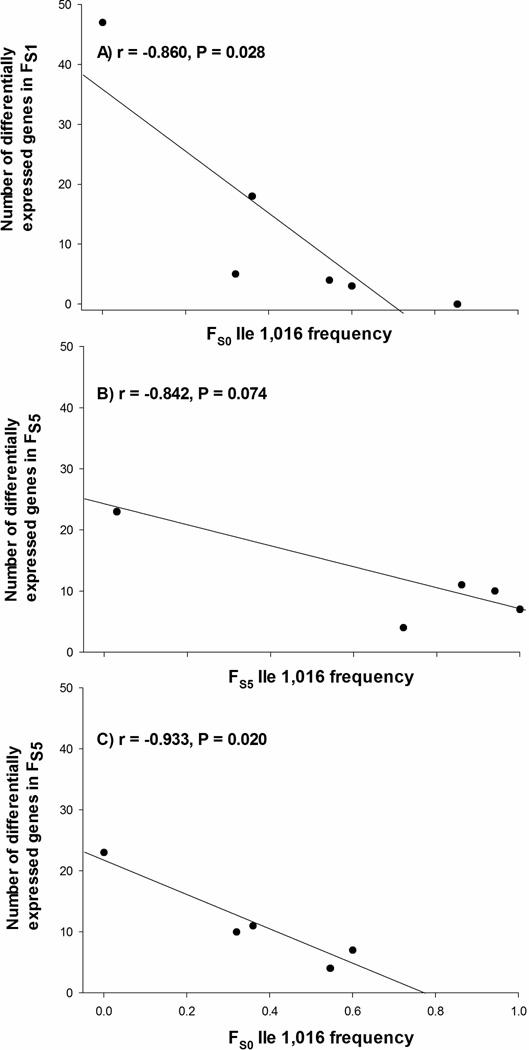
There was a strong negative correlation between numbers of differentially expressed genes in FS0 and the frequency of Ile1,016 in FS0 (Figure 5a) suggesting that there may be less selection on metabolic genes in mosquitoes that are already protected by the Ile1,016 allele. This result was consistent whether we analyzed genes differentially up-regulated (r = −0.87; P= 0.023) or down regulated (r = −0.86; P= 0.029).
Figure 5.
Correlations between Ile1,016 allele frequencies and the numbers of genes differentially transcribed. The Pearson correlation coefficient r is displayed alongside the probability that r = 0. A) FS0 Ile1,016 frequencies versus numbers of genes differentially transcribed in FS1. B) FS5 Ile1,016 frequencies versus numbers of genes differentially transcribed in FS5. C) FS0 Ile1,016 frequencies versus numbers of genes differentially transcribed in FS5
Gene expression - FS5 versus FS1
Direct comparisons between the FS5 and FS1 lines identified 34 genes differentially expressed after four additional generations of selection (Table 6). Lagunitas died in FS2 (Figure 2 and 3). Again, most (25/34) differentially expressed genes occurred in Iquitos and the second most occurred in Calderitas (13). Solidaridad, Lázaro Cárdenas, and Mérida respectively had ten, seven and two genes differentially expressed. Sixteen genes were differentially expressed after four additional generations of selection in two or more of the FS5 strains (Table 6a). Five peroxinectins were upregulated in Iquitos FS5 but down-regulated in the Mexican FS5 lines.
Table 6.
| a. Genes differentially expressed in two or more FS5 strains relative to the unselected FS1. Ratio of the average expression of each comparison is displayed on a linear scale. Probability value is shown as a negative log10 scale. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Activity | Iquitos | Calderitas | Lázaro C. | Solidaridad | Mérida | ||||||
| Gene | Vector base ID | Ratio | p | Ratio | p | Ratio | p | Ratio | p | Ratio | p |
| Glutathione transferases | |||||||||||
| AaGSTs1-1 | AAEL011741 | 0.19 | 17.18 | 0.51 | 15.61 | ||||||
| Cytochrome P450 monoxygenases | |||||||||||
| CYP4J13 | AAEL013555 | 2.64 | 12.18 | 2.38 | 8.8 | 3.20 | 10.73 | ||||
| CYP6Nae1 | AAEL009126 | 2.03 | 6.4 | 2.08 | 4.78 | ||||||
| CYP9J22 v1 | AAEL006802 | 2.55 | 8.37 | 1.95 | 17.79 | ||||||
| CYP9J22 v2 | AAEL014619 | 2.75 | 10.43 | 2.11 | 19.55 | ||||||
| CYP325G3 | AAEL012772 | 2.99 | 10.59 | 2.30 | 5.63 | 2.58 | 8.11 | ||||
| HypProt1055570 | AAGE01055570 | 2.10 | 10.18 | 2.03 | 4.18 | ||||||
| Red/Ox | |||||||||||
| Peroxinectin 3612 | AAEL003612 | 5.13 | 14.79 | 0.17 | 10.68 | 0.39 | 9 | ||||
| Peroxinectin 4386 | AAEL004386 | 3.34 | 11.97 | 0.21 | 8.05 | 0.45 | 5.09 | ||||
| Peroxinectin 4388 | AAEL004388 | 5.10 | 17.27 | 0.15 | 12.4 | 0.42 | 9.17 | 0.44 | 12.31 | ||
| Peroxinectin 4390 | AAEL004390 | 9.25 | 18.22 | 0.26 | 11.47 | ||||||
| Peroxinectin 4401 | AAEL004401 | 2.66 | 6.94 | 0.22 | 7.19 | ||||||
| HypProt4400 | AAEL004400 | 3.76 | 11.37 | 0.19 | 8.39 | 0.40 | 4.41 | ||||
| b. Genes differentially expressed in one of the unselected FS5 lines relative to FS1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutathione transferases | |||||||||||
| AaGSTt4 | AAEL004229 | 0.47 | 14.1 | ||||||||
| AaGSTe2 | AAEL007951 | 2.10 | 6.1 | ||||||||
| AaGSTs1-2 | AAEL011741 | 0.30 | 17.37 | ||||||||
| Cytochrome P450 monoxygenases | |||||||||||
| CYP6Z6_b | AAEL009123 | 4.66 | 18.77 | ||||||||
| CYP6CB1 | AAEL002046 | 4.20 | 11.75 | ||||||||
| CYP9J23_b | AAEL014615 | 2.55 | 10 | ||||||||
| CYP6CB2 | AAEL002046 | 2.13 | 6.92 | ||||||||
| CYP314A1_b | AAEL010946 | 2.04 | 6.67 | ||||||||
| Carboxyl/esterases | |||||||||||
| AaeCOE-8 | AAEL008757 | 2.60 | 9.4 | ||||||||
| AaeCOE-9 | AAEL012509 | 3.16 | 9.37 | ||||||||
| AaeCOE-23 | AAEL005122 | 2.06 | 1.95 | ||||||||
| AaeCOE-25 | AAEL002391 | 0.50 | 1.93 | ||||||||
| AaeCOE-34 | 2.23 | 6.19 | |||||||||
| Ace2 | AAEL012141 | 0.46 | 6.83 | ||||||||
| CCEunk6o | AAEL000545 | 0.40 | 9.97 | ||||||||
| CCae2B | AAEL017553 | 0.36 | 9.97 | ||||||||
| Red/Ox | |||||||||||
| Aldehyde oxidase 10382 | AAEL010382 | 0.45 | 15.23 | ||||||||
| Aldehyde oxidase 14493 | AAEL014493 | 3.84 | 15.47 | ||||||||
| Aldo-keto reductase 15002 | AAEL015002 | 4.69 | 8.26 | ||||||||
| Glutaredoxin 14064 | AAEL014064 | 0.50 | 5.52 | ||||||||
| Other | |||||||||||
| 40S ribosomal ptn S17 | AAEL004175 | 2.43 | 13.47 | ||||||||
| Acidic ribosomal ptn P1 | AAEL005027 | 2.08 | 9.62 | ||||||||
There was again a negative correlation between the number of differentially transcribed genes in FS5 and the frequency of Ile1,016 in FS5 (Figure 5b). This correlation was nonsignificant largely because the frequency of Ile1,016 had approached fixation in the four Mexican strains. Figure 5c shows a strong negative correlation between the frequency of the Ile1,016 allele in FS0 prior to selection and the number of genes differentially expressed in FS5 after 5 generations of selection. This further supports an hypothesis of low amounts of selection acting on metabolic genes in mosquitoes already protected by the Ile1,016 allele.
Gene expression - FS5 versus New Orleans (indirect)
We made an indirect (statistical) comparison of FS5 with New Orleans using the Limma package (http://bioinf.wehi.edu.au/limma/ available on www.bioconductor.org) following (Muller et al. 2007) in each of the strains. This was done to make a comparison of expression ratios (M) among FS5 strains standardized to the uniform genetic background provided by the New Orleans strain. FS5 vs. FS1 M ratios were adjusted to FS1 vs. FS0 M ratios to provide an indirect M ratio of FS5 vs. FS0. This indirect ratio was then adjusted to FS0 vs. New Orleans M ratios to provide an indirect M ratio of FS5 vs. New Orleans. This adjusted all 5 FS5 M ratios according to initial differences in transcription among the five FS0 strains relative to New Orleans.
Twenty four genes were differentially expressed after 5 generations of selection in two or more of the FS5 strains (Table 7a). This included two sigma class GST (AaGSTs1-1, -2) which were independently selected for decreased transcription in Iquitos, Lázaro Cárdenas and Mérida strains. Ten CYPs were independently selected for increased rates of transcription. There was little differential transcription of esterases. The transcription rates of five peroxinectins were greatly increased from 3 – 20 fold (linear scale) through permethrin selection in Iquitos. In contrast, in Calderitas and Mérida, the transcription rates of these peroxinectins decreased from 2–6 fold (linear scale) through permethrin selection.
Table 7.
| a. Genes differentially expressed in two or more FS5 strains relative to New Orleans. Ratio of the average expression of each comparison is displayed on a linear scale. Probability value is shown as a negative log10 scale. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Activity | Vector base ID | Iquitos | Calderitas | Lázaro C. | Solidaridad | Mérida | |||||
| Genes | Ratio | p | Ratio | p | Ratio | p | Ratio | p | Ratio | p | |
| Glutathione S-transferases | |||||||||||
| AaGSTs1-1 | AAEL011741 | 0.20 | 16.08 | 0.34 | 27.37 | 0.39 | 24.89 | ||||
| AaGSTs1-2 | AAEL011741 | 0.10 | 10.87 | 0.31 | 23.81 | ||||||
| Cytochrome P450 mono-oxygenases | |||||||||||
| CYP325G3 | AAEL012772 | 2.85 | 14.24 | 2.08 | 13.11 | 3.03 | 15.05 | ||||
| CYP4J13 | AAEL013555 | 2.14 | 8.02 | 2.25 | 15.15 | 2.89 | 18.82 | ||||
| CYP6CB1 | AAEL002046 | 3.94 | 14.9 | 2.51 | 27.31 | ||||||
| CYP6Nae1 | AAEL009126 | 2.79 | 10.74 | 3.66 | 21.23 | 3.43 | 18.24 | 2.07 | 5.79 | ||
| CYP6Z6_b | AAEL009123 | 4.20 | 18.01 | 2.41 | 7.96 | 2.16 | 20.19 | ||||
| CYP9J22 v1 | AAEL006802 | 3.07 | 14.59 | 2.87 | 12.16 | 5.03 | 27.33 | ||||
| CYP9J22 v2 | AAEL014619 | 3.63 | 16.73 | 3.94 | 27.65 | 5.54 | 28.88 | ||||
| CYP9J23_b | AAEL014615 | 2.20 | 7.69 | 2.51 | 13.36 | 2.06 | 12.76 | 2.85 | 20.3 | ||
| CYP9J28 | AAEL014617 | 2.53 | 4.92 | 2.58 | 16.61 | ||||||
| CYP9J32 | AAEL008846 | 4.76 | 10.21 | 2.75 | 18.46 | ||||||
| CYP9J9 v2 | AAEL014605 | 3.46 | 32.5 | 1.97 | 12.03 | 3.12 | 31.08 | ||||
| Carboxyl/esterases | |||||||||||
| AaeCOE-9 | AAEL012509 | 2.01 | 18.26 | 2.50 | 17.01 | ||||||
| CCae2B | AAEL017553 | 0.29 | 12.77 | 0.49 | 12.2 | ||||||
| Reduction/Oxidation | |||||||||||
| Aldo-keto reductase 4118 | AAEL004118 | 0.42 | 8.47 | 2.60 | 7.81 | ||||||
| Dual oxidase | AAEL007563 | 2.83 | 14.47 | 0.30 | 11.47 | ||||||
| HypProt4400 | AAEL004400 | 4.32 | 14.82 | 0.15 | 21.13 | 0.42 | 16.69 | ||||
| Peroxinectin 3612 | AAEL003612 | 6.02 | 17.12 | 0.16 | 19.13 | 0.47 | 23.74 | ||||
| Peroxinectin 4386 | AAEL004386 | 3.39 | 14.54 | 0.21 | 13.8 | 0.46 | 17.59 | ||||
| Peroxinectin 4388 | AAEL004388 | 8.88 | 22.93 | 0.21 | 15 | 0.46 | 21.27 | ||||
| Peroxinectin 4390 | AAEL004390 | 19.56 | 24.03 | 0.25 | 18.85 | ||||||
| Peroxinectin 4401 | AAEL004401 | 2.45 | 4.91 | 0.28 | 15.34 | 0.51 | 8.53 | ||||
| Thioredoxin peroxidase 4112 | AAEL004112 | 2.39 | 5.57 | 0.48 | 20.73 | ||||||
| b. Genes differentially expressed in one of the FS5 strains relative to New Orleans. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutathione S-transferases | |||||||||||
| AaGSTd1-1 | AAEL001061 | 3.63 | 11.92 | ||||||||
| AaGSTd1-3 | AAEL001061 | 2.35 | 8.01 | ||||||||
| AaGSTe2 | AAEL007951 | 2.04 | 16.52 | ||||||||
| AaGSTe3 | AAEL007947 | 2.50 | 9.16 | ||||||||
| AaGSTe4 | AAEL007962 | 2.41 | 16.97 | ||||||||
| AaGSTe6 | AAEL007946 | 2.10 | 14.71 | ||||||||
| AaGSTe7 | AAEL007948 | 3.53 | 15.75 | ||||||||
| AaGSTt4 | AAEL004229 | 0.40 | 19.63 | ||||||||
| GSTd5 | AAEL001071 | 3.78 | 9.2 | ||||||||
| Cytochrome P450 mono-oxygenases | |||||||||||
| CYP304C1 | AAEL014413 | 0.45 | 10.36 | ||||||||
| CYP305A5 | AAEL002043 | 0.39 | 10.07 | ||||||||
| CYP325S2 | AAEL000325 | 2.06 | 6.02 | ||||||||
| CYP4G36 | AAEL004054 | 0.50 | 2.73 | ||||||||
| CYP4H28 | AAEL003380 | 2.91 | 9.4 | ||||||||
| CYP4K3 | AAEL007798 | 2.23 | 10.34 | ||||||||
| CYP6BB2_a | AAEL014893 | 1.99 | 8.38 | ||||||||
| CYP6CB2 | AAEL002872 | 3.05 | 8.8 | ||||||||
| CYP6P12V1 | AAEL012491 | 0.45 | 7.29 | ||||||||
| CYP6P12V2 | AAEL014891 | 0.35 | 6.74 | 0.50 | 11.82 | ||||||
| CYP6Z9 | AAEL009129 | 0.28 | 9.49 | ||||||||
| CYP9J19 v1 | AAEL006810 | 2.58 | 11.04 | ||||||||
| CYP9J9 v1 | AAEL006793 | 2.16 | 15.83 | ||||||||
| CYP9M9_a | AAEL001807 | 2.04 | 7.55 | ||||||||
| Carboxyl/esterases | |||||||||||
| AaeCOE-2 | AAEL002376 | 2.87 | 12.45 | ||||||||
| AaeCOE-8 | AAEL008757 | 3.51 | 25.63 | ||||||||
| AaeCOE-17 | AAEL004341 | 0.22 | 12.63 | ||||||||
| AaeCOE-19 | AAEL005112 | 3.53 | 11.63 | ||||||||
| CCEjhe1o | AAEL0155590 | 0.50 | 8.1 | ||||||||
| CCEunk6o | AAEL000545 | 0.24 | 16.91 | ||||||||
| CCEae5C | AAEL003201 | 0.34 | 12.78 | ||||||||
| Reduction/Oxidation | |||||||||||
| Aldehyde oxidase 14493 | AAEL014493 | 6.63 | 15.31 | ||||||||
| Aldehyde oxidase 10367 | AAEL010367 | 2.19 | 9.73 | ||||||||
| Aldehyde oxidase 10382 | AAEL010382 | 0.37 | 10.49 | ||||||||
| Aldo-keto reductase 15002 | AAEL015002 | 4.50 | 10.45 | ||||||||
| Aldo-keto reductase 4102 | AAEL004102 | 3.10 | 8.94 | ||||||||
| Catalase | AAEL013407 | 0.23 | 13.32 | ||||||||
| Dihydrolipoamide dehydrogenase | AAEL006928 | 0.31 | 8.88 | ||||||||
| Glutaredoxin 12238 | AAEL012238 | 2.08 | 3.45 | ||||||||
| Glutaredoxin 13980 | AAEL013980 | 3.16 | 7.69 | ||||||||
| Peroxinectin 3933 | AAEL003933 | 0.46 | 6.38 | ||||||||
| Peroxinectin 5416 | AAEL005416 | 0.43 | 10.22 | ||||||||
| Thioredoxin peroxidase 9051 | AAEL009051 | 3.63 | 6.36 | ||||||||
| Thioredoxin peroxidase 10777 | AAEL010777 | 2.91 | 8.36 | ||||||||
| Others | |||||||||||
| 60S ribosomal ptn L44 L41 | AAEL003942 | 2.04 | 6.67 | ||||||||
| Acidic ribosomal ptn P1 | AAEL005027 | 3.12 | 14.03 | ||||||||
| Ribosomal ptn L34 | AAEL009341 | 2.20 | 11.03 | ||||||||
| 40S ribosomal ptn S17 | AAEL004175 | 6.02 | 22.89 | ||||||||
| HypProt | AAGE01055570 | 2.01 | 8.37 | ||||||||
Discussion
Comparative microarray analysis of transcription rates of detoxification genes before, during and after laboratory selection of Ae. aegypti strains with permethrin revealed four important trends. First, artificial selection with permethrin simultaneously increased the LC50 (Figure 2), KC50 (Figure 3) and the frequency of the Ile1,016 allele. Second, there was a consistent inverse relationship between the frequency of the Ile1,016 allele and the numbers of genes that became differentially transcribed, regardless of whether those genes were up- or down-regulated (Figure 5). This relationship was detected for Ile1,016 frequency in FS0 compared to FS1 (Figure 5a), Ile1,016 frequency in FS5 (Figure 5b) or for Ile1,016 frequency in FS0 compared to numbers of genes that become differentially transcribed in FS5 (Figure 5c). Third, very few of the 290 detoxification genes on the detox chip were differentially transcribed in two or more field strains (Table 4–7a). There were 17 genes (5.8%) differentially transcribed in the FS0 versus New Orleans comparisons, 20 genes (6.8%) in the FS1 versus FS0 comparisons, 13 genes (4.4%) in the FS5 versus FS1 comparisons and 24 genes (8.2%) in the FS5 versus New Orleans.
Instead Tables 4–7b list the many genes that were uniquely differentially transcribed in each of the five strains. Fourth, adaptation to permethrin did not cause a uniform up- or down regulation of detoxification genes. An intuitive a priori hypothesis is that upregulation of a detoxification gene should accompany selection of insecticide resistance. For example, this pattern was seen in Table 7 with some of the CYP genes. However, none of these CYP genes appeared in all five strains and in the case of the peroxinectins, selection caused an increase in Iquitos but a simultaneous decrease in the Calderitas and Mérida strains.
The observation that artificial selection with permethrin simultaneously increased the LC50 and KC50 along with the frequency of the Ile1,016 allele is not surprising. Table 3 indicates a large and significant h2 for LC50 for all strains except for Calderitas. This suggests that there is a large amount of additive genetic variance for permethrin adaptive genes in the five field strains. In Figure 2, the LC50 in Calderitas and Mérida appears to increase up until generation 4 when it suddenly decreases. This might have been associated with lethal or deleterious recessive alleles linked to the para locus that approach fixation as the Ile1,016 allele increased in frequency. However similar patterns were not noted in Lázaro Cárdenas or Solidaridad. The h2 for KC50 was not expected to be as large as LC50 because Ile1,016 allele is mostly recessive in its expression (Saavedra-Rodriguez et al. 2008; Saavedra-Rodriguez et al. 2007). This also would explain why h2 in Lázaro Cárdenas was greater than one. As Ile1,016 approached fixation in Lázaro Cárdenas there was a nonlinear increase in Ile1,016 homozygotes; the response to selection (R) was greater than the selection differential (S) and since h2 = R/S, h2>1. h2 was small in Iquitos where Ile1,016 was effectively absent. Note however that Ile1,016 was present albeit in very low frequency in Iquitos FS5 (Figure 3). Eventually selection increased its frequency to 0.03. This predicts fewer than 9×10−4 Ile1,016 homozygotes in the FS5, too few for selection to act upon especially when fewer than 500 individuals survived to pass on genes in the next generation. But (Saavedra-Rodriguez et al. 2008; Saavedra-Rodriguez et al. 2007) showed that Ile1,016 heterozygotes have a higher recovery rate following knockdown and this might account for the early initial appearance of Ile1,016 in Iquitos FS5.
The observation of a consistent inverse relationship between Ile1,016 frequency and the numbers of genes that became differentially transcribed through selection is one of the most interesting outcomes of this experiment. The most parsimonious explanation for this observation is that Ile1,016 confers a much larger selective advantage to both homozygous and heterozygous mosquitoes than the metabolic detoxification genes represented on the Ae. aegypti detox chip. Mosquitoes lacking or with a low frequency of Ile1,016 (e.g. Iquitos, Calderitas) can only very slowly evolve permethrin resistance by accumulating slight advantages conferred by the many detoxification genes. In contrast, these same genes would confer only slight additional survival advantages to the Mexican mosquitoes with a high frequency of Ile1,016.
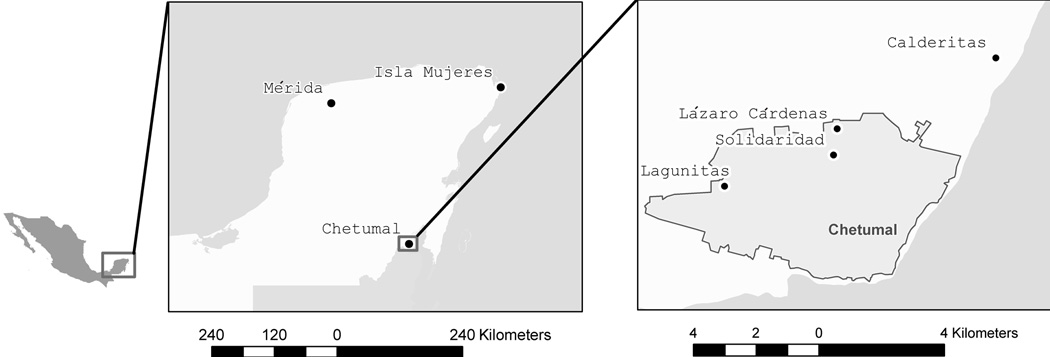
Few of the 290 detoxification genes on the detox chip were differentially transcribed in two or more of the field strains. This outcome can be largely attributed to the pre-existing high frequency of Ile1,016 in the Mexican strains. It would be interesting to compare the relatively long list of genes differentially transcribed in Iquitos with other Ae. aegypti field strains in which para substitutions are in low frequency. The observation that many were uniquely transcribed in individual strains suggests that permethrin adaptation in Ae. aegypti is genetically complex. It is clear that this adaptation is largely conditioned by target site insensitivity in the para gene but also probably reflects the frequency of various metabolic genes in different populations. This phenomenon occurs among geographically proximate collections; Lázaro Cárdenas, Solidaridad, and Lagunitas are located within the city of Chetumal.
A potential criticism of our experimental design is that it did not test whether differentially transcribed genes were a result of adaptation to the laboratory rather than to permethrin. To do so would have required analyzing microarrays on each of five lines (maintained without selection) in the FSO, FS1 and FS5 generations and this was not done. However, we have applied the same experimental design to identify genes responsive to temephos (an organophosphate insecticide) in four of the same collection sites (Saavedra-Rodriguez - unpublished). If differential transcription was a result of laboratory adaptation or reflected a xenobiotic metabolism response to insecticide exposure then the same sets of genes should have been differentially expressed in the present study and in the temephos study. This was examined by sorting all of the genes in Table 7 with all of the genes with significant differential expression between the FS5 vs. NO in the temephos experiment to identify genes with differential expression in both permethrin and temephos experiments (Table 8). Of the 70 differentially transcribed genes in the current experiment (Table 7) and the 80 differentially transcribed genes in the temephos experiment, there were 15 genes with differential expression in both experiments. Those highlighted in grey in Table 8 were differentially transcribed in the same direction in the same collection in both experiments and could therefore represent laboratory adaptation. Patterns consistent with lab adaptation were seen in three genes in Mérida and six genes in Iquitos. No comparisons fit these criteria for Calderitas and only one fit in Lázaro Cárdenas. Of the 27 comparisons in Table 8, 10 were consistent with a pattern expected for lab adaptation. But none appeared in all four collections. In general then there was little or no consistent evidence of laboratory adaptation for the majority of the 70 differentially transcribed genes in the current experiment.
Table 8.
Genes with differential expression in both permethrin and temephos experiments. Those highlighted in grey were differentially transcribed in the same direction in the same collection in both experiments and could therefore represent laboratory adaptation.
| Gene | Experiment | Collection | |||
|---|---|---|---|---|---|
| Iquitos | Calderitas | Lázaro C. | Mérida | ||
| AaGSTs1-1 | Permethrin | 0.20 | - | 0.34 | 0.39 |
| Temephos | - | - | 2.58 | 0.23 | |
| AaGSTs1-1 | Permethrin | 0.10 | - | 0.31 | - |
| Temephos | 0.44 | - | 2.14 | - | |
| AaGSTd1-1 | Permethrin | 3.63 | - | - | - |
| Temephos | 2.39 | - | - | - | |
| CYP304C1 | Permethrin | 0.45 | - | - | - |
| Temephos | - | 0.49 | - | - | |
| CYP6BB2_a | Permethrin | - | - | 1.99 | - |
| Temephos | - | - | 2.43 | - | |
| CYP6Z9 | Permethrin | 0.28 | - | - | - |
| Temephos | 0.39 | - | - | - | |
| CYP9J22 v1 | Permethrin | - | - | 3.07 | 5.03 |
| Temephos | - | - | 0.49 | 4.59 | |
| CYP9J22 v2 | Permethrin | - | - | 3.63 | 5.54 |
| Temephos | - | - | 0.44 | 4.08 | |
| CYP9J32 | Permethrin | 4.76 | - | 2.75 | - |
| Temephos | 2.48 | 3.73 | - | - | |
| CYP9J9 v2 | Permethrin | - | - | 3.46 | 3.12 |
| Temephos | - | - | 0.46 | - | |
| Aldehyde oxidase 14493 | Permethrin | 6.63 | - | - | - |
| Temephos | - | - | 0.45 | - | |
| Catalase | Permethrin | 0.23 | - | - | - |
| Temephos | 0.41 | - | - | - | |
| Thioredoxin peroxidase 4112 | Permethrin | 2.39 | - | - | - |
| Temephos | - | - | 2.46 | 0.40 | |
| Dihydrolipoamide dehydrogenase | Permethrin | 0.31 | - | - | - |
| Temephos | - | - | - | - | |
| AaeCOE-17 | Permethrin | 0.22 | - | - | - |
| Temephos | 0.36 | - | - | - | |
A unique outcome of this selection experiment is that many genes exhibited differential expression. Toxicity studies in vertebrates have demonstrated that pyrethroids generate reactive oxygen species (ROS) and cause oxidative stress (Kale et al. 1999; Sayeed et al. 2003). Several studies have demonstrated that pyrethroid intoxication alters the antioxidant system in erythrocytes and causes pyrethroid-induced lipid peroxidation (LPO) (Fetoui et al. 2008; Fetoui et al. 2010). The increased oxidative stress results in an increase in the activity of antioxidant enzymes such as superoxide dismutase (SOD) and catalase (CAT) (Kale et al. 1999). While no similar studies have been made insects, it is interesting that peroxinectins were prominent among the oxidative stress genes listed in Table 7. Peroxinectins are cell adhesive ligands synthesized in invertebrate hemocytes and stored in secretory granules in an inactive form (Schmidt et al. 2010). Peroxinectin is released in response to elicitors, such as bacterial endotoxins (lipopolysaccharide (LPS) or beta-1,3-glucans) and are subsequently activated outside the cells to mediate hemocyte attachment and spreading (Johansson and Soderhall 1989). Peroxinectins are multifunctional molecules involved in encapsulation, opsoninization, and can act as peroxidases (Johansson et al. 1995). In the latter respect, peroxinectins are similar to a family of vertebrate peroxidases that includes myeloperoxidase (Johansson et al. 1995). Peroxinectin is the first protein discovered to have combined adhesive/peroxidase functions. It is possible that peroxinectins have a protective function in insects. If exposure to pyrethroids causes insect tissues to produce ROS, LPOs, and elevate oxidative stress that damage membranes; the peroxidase activity of peroxinectin may protect cells against this damage.
An intuitive a priori hypothesis is that upregulation of a detoxification gene should accompany insecticide selection. For example, with its expression mostly restricted to the indirect flight muscle and neurons, Flanagan and Smythe (2011) suggested that AaGSTs1-1 may serve a protective role in highly aerobic tissues or tissues sensitive to oxidative damage. Thus its down regulation in this study seems counterintuitive. The observation that permethrin laboratory adaptation did not cause a uniform up- or downregulation of detoxification genes suggests the possibility that some detoxification genes may be disadvantageous in mosquitoes with Ile1,016. This might also explain why selection increased transcription of the five peroxinectins in Iquitos strain while decreasing transcription in the Calderitas and Mérida strains. Perhaps, in the absence of pyrethroids or in the presence of Ile1,016, peroxinectin expression is selected against. Peroxinectins were only one of a few mechanisms that the Iquitos strain had for pyrethroid protection. Prior to selection of Ile1,016, expression of peroxinectins was 2–6 fold greater in Calderitas and Mérida but as Ile1,016 increased in frequency increased peroxinectin activity was selected against.
Uniform upregulation was seen with some of the CYP genes in Table 7 even though no gene was significant in all five strains. Enzymes of the CYP6 family are known to be generally involved in detoxification of xenobiotics in insects. In An. gambiae, fourteen CYP6 family genes are clustered on chromosome III. CYP6Z1 was shown to metabolize DDT, carbaryl and xanthotoxin, while CYP6Z2 was capable of metabolizing carbaryl (Chiu et al. 2008). Our results show that CYP6Z6 was upregulated in the Calderitas, Mérida and Lagunitas FS0 strains. After selection, this gene was upregulated in Calderitas, Mérida and Iquitos. CYP6Z6 has a 63% and 59% identity with An. gambiae CYP6Z2 and CYP6Z1, respectively, suggesting that it might have a role in detoxification of permethrin. CYP6Z6 was also over-transcribed in larval and adult Ae. aegypti in a permethrin resistant strain from Martinique (Marcombe et al, 2009). Larvae exposed to sublethal concentrations of the herbicide glyphosate and benzo[a]pyrene significantly induced CYP6Z6 (Riaz et al. 2009). Paralogues of CYP6Z6, -8 and -9 also were induced by fluoranthene (Poupardin et al. 2008) and were over transcribed in a permethrin resistant strain from Northern Thailand (Strode et al, 2008). CYP6 gene transcription was also analyzed in different Ae. aegypti mosquito tissues, showing that genes in the subfamilies CYP6Z, CYP6M and CYP6N are preferentially transcribed in the alimentary canal, with the exception of CYP6Z6 which is preferentially transcribed in head and anterior midgut (Poupardin et al. 2010).
A second CYP6 member that responded to selection was CYP6Nae1 (AAEL009126, new VectorBase ID = CYP6N6). This gene was upregulated in the unselected strains from Mérida and Lagunitas, however, after five generations of permethrin selection, CYP6Nae1 was significantly up regulated in all Mexican strains. CYP6Nae1 has not been associated with insecticide detoxification in Ae. aegypti, however, paralogues CYP6N12 (AAEL009124, 87% nucleotide similarity) and CYP6N11 (AAEL009138, 58% nucleotide similarity) were induced in larvae by the herbicide glyphosate (Riaz et al, 2009). In a microarray analysis, CYP6N12 was induced in larvae exposed to fluoranthene, however, RT-PCR showed that this gene is also induced by permethrin, temephos and copper (Poupardin et al, 2008).
CYP4J13 was upregulated in larvae from a Isla Mujeres (México) permethrin resistant strain and in adults from a Northern Thailand resistant strain (Strode, et al 2008) and a paralogue of this gene, CYP4J15 (AAEL013556, 48% nucleotide similarity) was upregulated in larvae in a permethrin resistant strain from Martinique. On the other hand, CYP325G3 has not been previously identified in Ae. aegypti insecticide resistant strains, although, members of this family were upregulated in a permethrin resistant strain of An. gambiae (David et al. 2005).
CYP9J family genes are commonly up regulated in insecticide resistant strains. Our results indicate that genes CYP9J-9, -10, -22, -23, -28 and -32 are upregulated before or after selection in most field strains. Except for CYP9J32, most of the CYP9J genes occur on chromosome III in two clusters at 4 cM on the p arm and at 50 cM in the q arm (Strode et al., 2007; Saavedra-Rodriguez et al., 2008). Genes of these clusters were also identified as QTL involved in permethrin resistance in mosquitoes from México (Saavedra-Rodriguez et al. 2008). CYP9J-9, -22, and -23 were also upregulated in a permethrin resistant strain from Martinique (Marcombe et al, 2008). Five CYP9J genes were upregulated in a Northern Thailand resistant strain and seven were upregulated in a Mexican permethrin resistant strain (CYP9J-8, -10, -19, -24, -27, -28, and -32) (Strode et al, 2008). It is possible that CYP9J genes in the 4 cM and 50 cM clusters were increased solely by selection for Ile1,016 located at 31 cM on chromosome III. In other words, selecting Ile1,016 could have swept (increased) other alleles on chromosome III that did not necessarily confer resistance. If this were true then we would have expected no increase in CYP9J genes in Iquitos in which Ile1,016 does not occur. However Table 7 shows that CYP9J23 and CYP9J28, both in the 50cM cluster, increased 2.5 fold in Iquitos. Still the possibility of a selective sweep cannot be excluded for CYP9J22 and CYP9J9 (both in the 4cM cluster) which increased 3–5 fold in three of the Mexican strains. Furthermore while there are 12 CYP9J genes in the 4 cM cluster and 7 in the 50 cM cluster, only 7 increased in expression during selection.
A common goal of the current study and of the many microarray studies cited above is to identify the principal genes conditioning pyrethroid resistance. However, the lack of uniformity in the genes that responded to artificial and natural selection as well as in the direction of their responses (e.g. peroxinectins) poses a critical question regarding the assumption that one or a few genes control pyrethroid metabolic resistance. Table 4 compares transcription patterns in six field strains with transcription in the standard susceptible New Orleans strain and basically shows that all collections have very different transcription patterns. Even the proximate collections of Lagunitas, Lázaro Cárdenas, and Solidaridad (Figure 1) exhibited distinct transcription profiles. This result might be a result of genetic drift arising when subpopulations are established by one or a few of the adults that survived (i.e. founder effects) control efforts by permethrin and other tactics. However, Tables 6 and 7 show that application of a uniform artificial selection regime did not cause collections to converge in their transcription profiles. These results collectively suggest that Ae. aegypti populations may have a multitude of genes that can respond to pyrethroid selection. If so, attempts to identify one or a few metabolic genes that are predictably associated with pyrethroid adaptation may prove futile.
Figure 1.
Map of collections sites in México
Experimental procedures
Collection sites and colony rearing conditions
The Mexican lines were collected as larvae from the states of Yucatán (Mérida) and from Quintana Roo (Lázaro Cárdenas, Solidaridad, Calderitas and Lagunitas) in the city of Chetumal (Figure 1). Eggs of the Iquitos strain were from a laboratory strain collected from and maintained in Iquitos by Dr. Amy Morrison. F1 or F2 offspring were reared to adults, bloodfed, and eggs were collected and shipped to Colorado State University where additional F2 or F3 generations were reared to generate sufficient larvae and adults for bioassay and for initiation of three replicate lines for artificial selection with permethrin. Each replicate line was initiated with 100 males and 100 females placed in a 30 cm3 cage (BugDorm-1, Mega View Science, Co). Mosquito larvae were reared in plastic containers holding 2 L of water maintained at 30°C and provided with brewer’s yeast. Pupae were placed in 30 cm3 cages and eclosed adults were provided with a 10% (w/v) sucrose solution. Adults were fed on an artificial membrane feeder containing defibrinated sheep blood every three days. Cages were housed in growth chambers held at 14:10 photoperiod, and 28°C with 85% relative humidity.
Bioassays and permethrin selection
FS0 adults were bioassayed to estimate the LC50 and KC50 for permethrin (47.6% cis – 50.4 % trans; Chem Service, West Chester, PA). The insides of 250 ml Wheaton bottles were coated with 1 ml of acetone containing five different concentrations of permethrin. Twenty-five 3–4 day old adults were gently aspirated into each bottle. The number of mosquitoes unable to fly was scored every 10 minutes for up to one hour. After exposure, all mosquitoes were gently transferred into a 400 ml cardboard carton (Huhtamaki, USA) and mortality was scored after 24 hours. Each bioassay was performed in triplicate to have ~75 mosquitoes per concentration. LC50 and KC50 were estimated using a logistic regression model in R version 2.11.1 (http://cran.r-project.org/) (Source Code in Supplement 1). Confidence limits were calculated using the IRMA quick calculator software (http://sourceforge.net/projects/irmaproj/files/).
Adult permethrin selection was performed in triplicate for each strain. Replicates were maintained during five generations of permethrin selection. The first selection consisted in exposing 250–700 3–4 day old females and males from each line to the previously estimated LC50. Permethrin exposure time was for one hour and ~100 adults were aspirated into each impregnated bottle. Knocked down and alive mosquitoes were transferred to a 400 ml cardboard carton (Huhtamaki, USA) and mortality was registered at 24 hours after exposure. Survivors for each selection were transferred to insect rearing cages and blood fed. At the beginning of each of the subsequent four generations of selection, a bioassay was performed to estimate the LC50 following the previous generation of selection. From each replicate 300 – 700 adults were then selected using the new LC50. For some mosquito lines in certain generations, a lower permethrin concentration was used for selection, depending upon the damage inflicted by permethrin exposure (i.e. multiple appendage loss or blood feeding appetite loss). Selected line names were designated Fs1, Fs2, Fs3, Fs4 or Fs5 indicating the generation of selection.
Realized Heritability
Realized heritability (h2) is calculated as the ratio of the response to selection (R) to the selection differential (S) and is correlated with the amount of additive genetic variance for a trait. A low h2 predicts no additive genetic variance for a trait and a poor or very slow response to artificial selection while a high h2 predicts a large additive genetic variance at one or a few loci that condition a trait and predicts a rapid response to artificial selection. When estimating h2 for insecticide resistance, R is estimated as the mean difference between the LC50 in offspring of surviving parents as compared to the LC50 in the parents and S is the proportion of surviving parents.
Realized heritability was calculated using the method of Tanaka and Noppun (1989). The LC50 or KC50 were estimated in R version 2.11.1 (Supplement 1) and transformed with the natural logarithm. The proportion surviving (pt) was the weighted average of the survival rate among the three replicates in generation t. The standard deviation of susceptibility (σt) was the reciprocal of the slope from the logistic regression analysis in generation t. The intensity of selection (it) was obtained using pt in Appendix-Table A in (Falconer 1989). St was σt multiplied by it. Rt was the change in ln(LC50) or ln(KC50) from generation t to t+1. The cumulative response to selection R was the sum of Rt = 1,…5 while the cumulative selection differential S was the sum of St = 1,…5. Following Tanaka and Noppun (1989), significance of the regression of R on S was tested by ANOVA in R 2.11.1 where the slope (h2 = ΔR/ΔS) was treated as a continuous variable with one degree of freedom.
Isoleucine 1,016 allele frequency
Fifty mosquitoes from the unselected FS0, and the selected FS1 and FS5 lines were genotyped. DNA was isolated from individual mosquitoes by salt extraction (Black and DuTeau 1997) and suspended in 200 µl TE buffer (10 mM Tris-HCl, 1 mM EDTA pH 8.0). The frequency of the Ile1,016 allele was determined by melting curve PCR following (Saavedra-Rodriguez et al. 2007).
Differential expression profiles
The DNA microarray ‘Aedes Detox Chip’ v.2 (Strode et al. 2008), was used to follow changes in the expression of detoxification genes. This microarray contains 318 70-mer probes representing 290 detoxification genes including 183 CYP, 28 GST, 44 CCE and 35 additional enzymes potentially involved in response to oxidative stress in Ae. aegypti. The three biological replicates from each mosquito line were processed separately. Direct comparisons between three different points of selection were: 1) ‘Unselected- FS0’ relative to the New Orleans susceptible reference strain; 2) Permethrin selected-FS1 relative to FS0, and 3) FS5 relative to FS1. RNA isolations, cDNA synthesis and labeling reactions were performed independently for each biological replicate. Total RNA was extracted from batches of thirty 3 day old adults (15 females and 15 males) using the RNeasy ®Midi Kit (Qiagen) according to manufacturer’s instructions for total RNA isolation from animal tissues. Total RNA quantity and quality was assessed using a Nanodrop® spectrophotometer. cDNA synthesis, labeling reaction and hybridization to the array were performed as in Strode et al. (2008).
Spot finding, signal quantification and spot superimposition for both dye channels were performed using the Axon Instruments Genepix Personal 4100A laser scanner and Genepix 5.1 software (Axon Instruments, Molecular Devices, Union City, CA, USA). Spots that did not satisfy the criteria described by Strode et al. (2008) were excluded from analysis. Normalization and statistical analyses were performed on R using the limma package http://bioinf.wehi.edu.au/limma/ available on www.bioconductor.org following (Muller et al. 2007). Results are expressed as M, the log2 of mean transcription ratios where M = log2(Cy5/Cy3). An arbitrary threshold of M = 1 (i.e. two-fold) was used to identify differentially expressed genes. The probability threshold was set at 3.00 (i.e. −log10 (0.001)).
Quantitative PCR for microarray validation
Transcription profiles of eight differentially expressed genes in the FS5 strains were validated by real-time quantitative PCR using the same RNA samples as were used for microarray experiments. Four µg of total amplified RNA (RiboAmpTM RNA amplification kit) were used for cDNA synthesis with Superscript Reverse Transcriptase III (Invitrogen) and oligo-(dT)15–18 primer (Invitrogen). Resulting cDNAs were diluted 100 times for real-time quantitative PCR reactions. Primer pairs used for quantitative PCR (Table 2) were optimized and tested by melting curve analysis and agarose gel electrophoresis to insure that they produced a unique amplification product. Real-time quantitative 20 µL PCR were performed in triplicate on a CFX-96 system (BioRad) using iQ SYBR Green Supermix (BioRad), 0.3µM of each primer and 5 µL of diluted cDNAs. For each gene analyzed, a cDNA dilution scale from 1:1,000,000 times was performed to assess efficiency of PCR. Data analysis was performed according to the ΔCt method taking into account PCR efficiency (Pfaffl 2001) and using the gene encoding the ribosomal protein L8 (Vector Base ID AAEL000987) for normalization. Results were expressed as average M obtained in the three replicates.
Table 2.
Real time PCR primers used for microarray validation
| Gene | Vector Base ID |
5'-->3' forward sequence | 5'-->3' reverse sequence | cDNA size (bp) |
|---|---|---|---|---|
| Glutathione transferases | ||||
| AaGSTe-2 | AAEL007951 | GCCCGATGATGACGTGAAG | TGGCTTGCTTAACCAGTTCTTTC | 246 |
| AaGSTs1-1 | AAEL011741 | GCCCGATGATGACGTGAAG | TCCAGGATGGCGACAAAGTA | 159 |
| Cytochrome P450 monooxygenases | ||||
| CYP4J13 | AAEL013555 | TATGCGTATGTGCCGTTTAGT | ATAGGGCGTATTTCTGTCC | 62 |
| CYP325G3 | AAEL012772 | GCACGTTGGATATGATTTGT | TCTGTCCGATAGTATTTGGTTAGT | 178 |
| CYP6Nae1 | AAEL009126 | GGATTTCCAATACTTCCACGA | TTCCACTTGGCACCCTCC | 96 |
| CYP9J22 v1 | AAEL006802 | CAGAGGCTGTACGAGGAGATAGT | CGAAGGGACTCCGACACTA | 116 |
| CYP9J32 | AAEL008846 | CGATTTCATCCAGACCATTTA | CGTCTTCTTTGTTCGCTCC | 176 |
| Carboxyl/esterases | ||||
| AaeCOE-8 | AAEL008757 | CGGAGTCTTTCTTGAAGGGTA | TTCGGAGTGTTTCATTTCGTAG | 111 |
| Red/Ox & other | ||||
| Peroxinectin 4388 | AAEL004388 | CAACCTCCACTGGGATGACG | ATGTTCTGAAGTACGGCGATTA | 72 |
| Peroxinectin 4390 | AAEL004390 | TCAGCAGCCAATGGAACAAA | GGTCGCAGGGCAAAGTCA | 132 |
| 60S ribosomal protein L8 | AAEL000987 | TGAAGGGAACCGTCAAGCAA | CATCAGACCGATTGGCAGAAC | 200 |
Supplementary Material
Acknowledgements
Larvae were collected by collaborators at the Universidad Autónoma de Nuevo León and Universidad Autónoma de Yucatán. The Iquitos, Perú collection was provided by Amy Morrison (UC Davis). This work was funded in part by NIH-NIAID U01 AI088647.
References
- Black WC, DuTeau NM. RAPD-PCR and SSCP analysis for insect population genetic studies. In: Crampton JM, Beard CB, Louis C, editors. Molecular Biology of Insect Disease Vectors: A Methods Manual. London: Chapman & Hall; 1997. pp. 362–363. [Google Scholar]
- Brengues C, Hawkes NJ, Chandre F, McCarroll L, Duchon S, Guillet P, Manguin S, Morgan JC, Hemingway J. Pyrethroid and DDT cross-resistance in Aedes aegypti is correlated with novel mutations in the voltage-gated sodium channel gene. Medical and Veterinary Entomology. 2003;17:87–94. doi: 10.1046/j.1365-2915.2003.00412.x. [DOI] [PubMed] [Google Scholar]
- Casida JE, Gammon DW, Glickman AH, Lawrence LJ. Mechanisms of Selective Action of Pyrethroid Insecticides. Annual Review of Pharmacology and Toxicology. 1983;23:413–438. doi: 10.1146/annurev.pa.23.040183.002213. [DOI] [PubMed] [Google Scholar]
- Chiu TL, Wen ZM, Rupasinghe SG, Schuler MA. Comparative molecular modeling of Anopheles gambiae CYP6Z1, a mosquito P450 capable of metabolizing DDT. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8855–8860. doi: 10.1073/pnas.0709249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da-Cunha MP, Lima JBP, Brogdon WG, Moya GE, Valle D. Monitoring of resistance to the pyrethroid cypermethrin in Brazilian Aedes aegypti (Diptera : Culicidae) populations collected between 2001 and 2003. Memorias Do Instituto Oswaldo Cruz. 2005;100:441–444. doi: 10.1590/s0074-02762005000400017. [DOI] [PubMed] [Google Scholar]
- David JP, Strode C, Vontas J, Nikou D, Vaughan A, Pignatelli PM, Louis C, Hemingway J, Ranson H. The Anopheles gambiae detoxification chip: A highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouaka RF, Bakare AA, Coulibaly ON, Akogbeto MC, Ranson H, Hemingway J, Strode C. Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. Bmc Genomics. 2008;9 doi: 10.1186/1471-2164-9-538. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS. Introduction to Quantitative Genetics, Third Edition. Harlow, Essex, UK/New York: Longmans Green/John Wiley & Sons; 1989. [Google Scholar]
- Fetoui H, Garoui EM, Makni-Ayadi F, Zeghal N. Oxidative stress induced by lambda-cyhalothrin (LTC) in rat erythrocytes and brain: Attenuation by vitamin C. Environmental Toxicology and Pharmacology. 2008;26:225–231. doi: 10.1016/j.etap.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Fetoui H, Makni M, Garoui EM, Zeghal N. Toxic effects of lambda-cyhalothrin, a synthetic pyrethroid pesticide, on the rat kidney: Involvement of oxidative stress and protective role of ascorbic acid. Experimental and Toxicologic Pathology. 2010;62:593–599. doi: 10.1016/j.etp.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Flanagan JU, Smythe ML. Sigma-class glutathione transferases. Drug Metabolism Reviews. 2011;43:194–214. doi: 10.3109/03602532.2011.560157. [DOI] [PubMed] [Google Scholar]
- Flores AE, Solis GR, Salas IF, Ramos FJS, Garcia GP. Resistance to Permethrin in Aedes aegypti (L.) in Northern México. Southwestern Entomologist. 2009;34:167–177. [Google Scholar]
- Gubler D. The emergence of epidemic dengue fever and dengue hemorrhagic fever in the Americas: a case of failed public health policy. Revista Panamericana De Salud Publica-Pan American Journal of Public Health. 2005;17:221–224. doi: 10.1590/s1020-49892005000400001. [DOI] [PubMed] [Google Scholar]
- Harris AF, Rajatileka S, Ranson H. Pyrethroid Resistance in Aedes aegypti from Grand Cayman. American Journal of Tropical Medicine and Hygiene. 2010;83:277–284. doi: 10.4269/ajtmh.2010.09-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemingway J, Hawkes NJ, McCarroll L, Ranson H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochemistry and Molecular Biology. 2004;34:653–665. doi: 10.1016/j.ibmb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Jirakanjanakit N, Rongnoparut P, Saengtharatip S, Chareonviriyaphap T, Duchn S, Bellec C, Yoksan S. Insecticide susceptible/resistance status in Aedes (Stegomyia) aegypti and Aedes (Stegomyia) albopictus (Diptera : Culicidae) in Thailand during 2003–2005. Journal of Economic Entomology. 2007;100:545–550. doi: 10.1603/0022-0493(2007)100[545:irsias]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Johansson MW, Lind MI, Holmblad T, Thornqvist PO, Soderhall K. Peroxinectin, a Novel Cell-Adhesion Protein from Crayfish Blood. Biochemical and Biophysical Research Communications. 1995;216:1079–1087. doi: 10.1006/bbrc.1995.2731. [DOI] [PubMed] [Google Scholar]
- Johansson MW, Soderhall K. Cellular-Immunity in Crustaceans and the Propo System. Parasitology Today. 1989;5:171–176. doi: 10.1016/0169-4758(89)90139-7. [DOI] [PubMed] [Google Scholar]
- Kale M, Rathore N, John S, Bhatnagar D. Lipid peroxidative damage on pyrethroid exposure and alterations in antioxidant status in rat erythrocytes: a possible involvement of reactive oxygen species. Toxicology Letters. 1999;105:197–205. doi: 10.1016/s0378-4274(98)00399-3. [DOI] [PubMed] [Google Scholar]
- Kawada H, Higa Y, Komagata O, Kasai S, Tomita T, Nguyen TY, Luu LL, Sanchez RAP, Takagi M. Widespread Distribution of a Newly Found Point Mutation in Voltage-Gated Sodium Channel in Pyrethroid-Resistant Aedes aegypti Populations in Vietnam. Plos Neglected Tropical Diseases. 2009;3 doi: 10.1371/journal.pntd.0000527. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd LS. Best Practices for Dengue Prevention and Control in the Americas Vol. Strategic Report 7. City: U.S. Agency for International Development; 2003. Best Practices for Dengue Prevention and Control in the Americas; p. 106. [Google Scholar]
- Loughney K, Kreber R, Ganetzky B. Molecular Analysis of the Para Locus, a Sodium-Channel Gene in Drosophila. Cell. 1989;58:1143–1154. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- Marcombe S, Poupardin R, Darriet F, Reynaud S, Bonnet J, Strode C, Brengues C, Yebakima A, Ranson H, Corbel V, David JP. Exploring the molecular basis of insecticide resistance in the dengue vector Aedes aegypti: a case study in Martinique Island (French West Indies) Bmc Genomics. 2009;10 doi: 10.1186/1471-2164-10-494. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins AJ, Lima JBP, Peixoto AA, Valle D. Frequency of Val1016Ile mutation in the voltage-gated sodium channel gene of Aedes aegypti Brazilian populations. Tropical Medicine & International Health. 2009;14:1351–1355. doi: 10.1111/j.1365-3156.2009.02378.x. [DOI] [PubMed] [Google Scholar]
- Muller P, Donnelly MJ, Ranson H. Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. Bmc Genomics. 2007;8 doi: 10.1186/1471-2164-8-36. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Warr E, Stevenson BJ, Pignatelli PM, Morgan JC, Steven A, Yawson AE, Mitchell SN, Ranson H, Hemingway J, Paine MJI, Donnelly MJ. Field-Caught Permethrin-Resistant Anopheles gambiae Overexpress CYP6P3, a P450 That Metabolises Pyrethroids. Plos Genetics. 2008;4 doi: 10.1371/journal.pgen.1000286. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeporn P, Supaphathom K, Sathantriphop SPM, Sangkitporn S. Insecticide susceptibility of Aedes aegypti in Tsunami-affected Areas in Thailand. Dengue Bulletin. 2005;29:210–213. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29 doi: 10.1093/nar/29.9.e45. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupardin R, Reynaud S, Strode C, Ranson H, Vontas J, David JP. Cross-induction of detoxification genes by environmental xenobiotics and insecticides in the mosquito Aedes aegypti: Impact on larval tolerance to chemical insecticides. Insect Biochemistry and Molecular Biology. 2008;38:540–551. doi: 10.1016/j.ibmb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Poupardin R, Riaz MA, Vontas J, David JP, Reynaud S. Transcription profiling of eleven cytochrome P450s potentially involved in xenobiotic metabolism in the mosquito Aedes aegypti. Insect Molecular Biology. 2010;19:185–193. doi: 10.1111/j.1365-2583.2009.00967.x. [DOI] [PubMed] [Google Scholar]
- Riaz MA, Poupardin R, Reynaud S, Strode C, Ranson H, David JP. Impact of glyphosate and benzo[a]pyrene on the tolerance of mosquito larvae to chemical insecticides. Role of detoxification genes in response to xenobiotics. Aquatic Toxicology. 2009;93:61–69. doi: 10.1016/j.aquatox.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Rodriguez MM, Bisset JA, De Armas Y, Ramos F. Pyrethroid insecticide-resistant strain of Aedes aegypti from Cuba induced by deltamethrin selection. Journal of the American Mosquito Control Association. 2005;21:437–445. doi: 10.2987/8756-971X(2006)21[437:PISOAA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Saavedra-Rodriguez K, Strode C, Suarez AF, Salas IF, Ranson H, Hemingway J, Black WC. Quantitative Trait Loci Mapping of Genome Regions Controlling Permethrin Resistance in the Mosquito Aedes aegypti. Genetics. 2008;180:1137–1152. doi: 10.1534/genetics.108.087924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra-Rodriguez K, Urdaneta-Marquez L, Rajatileka S, Moulton M, Flores AE, Fernandez-Salas I, Bisset J, Rodriguez M, Mccall PJ, Donnelly MJ, Ranson H, Hemingway J, Black WC. A mutation in the voltage-gated sodium channel gene associated with pyrethroid resistance in Latin American Aedes aegypti. Insect Molecular Biology. 2007;16:785–798. doi: 10.1111/j.1365-2583.2007.00774.x. [DOI] [PubMed] [Google Scholar]
- Sayeed I, Parvez S, Pandey S, Bin-Hafeez B, Haque R, Raisuddin S. Oxidative stress biomarkers of exposure to deltamethrin in freshwater fish, Channa punctatus Bloch. Ecotoxicology and Environmental Safety. 2003;56:295–301. doi: 10.1016/s0147-6513(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Soderhall K, Theopold U, Faye I. Role of Adhesion in Arthropod Immune Recognition. Annual Review of Entomology. 2010;55:485–504. doi: 10.1146/annurev.ento.54.110807.090618. [DOI] [PubMed] [Google Scholar]
- Strode C, Wondji CS, David JP, Hawkes NJ, Lumjuan N, Nelson DR, Drane DR, Karunaratne SHPP, Hemingway J, Black WC, Ranson H. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochemistry and Molecular Biology. 2008;38:113–123. doi: 10.1016/j.ibmb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Noppun V. Heritability Estimates of Phenthoate Resistance in the Diamond-Back Moth. Entomologia Experimentalis Et Applicata. 1989;52:39–47. [Google Scholar]
- Yanola J, Somboon P, Walton C, Nachaiwieng W, Prapanthadara LA. A novel F1552/C1552 point mutation in the Aedes aegypti voltage-gated sodium channel gene associated with permethrin resistance. Pesticide Biochemistry and Physiology. 2010;96:127–131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.