Abstract
Ffh is the signal sequence recognition and targeting subunit of the prokaryotic signal recognition particle (SRP). Previous structural studies of the NG GTPase domain of Ffh demonstrated magnesium-dependent and magnesium-independent binding conformations for GDP and GMPPNP that are believed to reflect novel mechanisms for exchange and activation in this member of the GTPase superfamily. The current study of the NG GTPase bound to Mg2+ GDP reveals two new binding conformations—in the first the magnesium interactions are similar to those seen previously, however, the protein undergoes a conformational change that brings a conserved aspartate into its second coordination sphere. In the second, the protein conformation is similar to that seen previously, but the magnesium coordination sphere is disrupted so that only five oxygen ligands are present. The loss of the coordinating water molecule, at the position that would be occupied by the oxygen of the γ-phosphate of GTP, is consistent with that position being privileged for exchange during phosphate release. The available structures of the GDP-bound protein provide a series of structural snapshots that illuminate steps along the pathway of GDP release following GTP hydrolysis.
Keywords: signal recognition particle (SRP), SRP54, Ffh, FtsY, GTPase, crystallography, magnesium coordination
INTRODUCTION
The prokaryotic GTPase Ffh is one component of the Signal Recognition Particle (SRP) cotranslational targeting pathway in bacteria.1 SRP, which recognizes the hydrophobic signal peptide as it emerges from the ribosome, is targeted to the plasma membrane by interaction between the two-domain “NG” GTPase of Ffh and the structurally homologous “NG” GTPase of its receptor, FtsY.2–5 The two proteins are members of and structurally similar to the superfamily of GTPases, which includes small G proteins such as ras, and the heterotrimeric G proteins.6,7 However their mechanism of regulation is likely to be distinct from the other GTPases8 –11 and recent structural and biochemical studies have only begun to dissect the molecular logic of the SRP GTPase system.8,12,13
A basic question, for example, is how the proteins are regulated by and regulate GTP binding and hydrolysis. A series of crystal structures of the NG GTPase domain of Ffh and FtsY from the prokaryotes T. aquaticus, E. coli, and the archae A. ambivalens,8, 14 –18 suggest a number of novel features relevant to understanding SRP-mediated targeting. These include an open active site which appears to be regulated by interaction between the N and G domains of the GTPase,15,17 a stable “empty-state” which, in the structures of the T. aquaticus protein, is maintained by a network of interactions between highly conserved side-chains of the characteristic GTPase sequence motifs,14 and the lack of a requirement for an exchange factor for nucleotide release.19 A non-canonical binding mode for the non-hydrolyzable GTP analog GMPPNP suggests that, in contrast to the “classic” model of the GTPase switch, GTP binding does not “activate” the protein. Instead, the GTP-docked protein may itself be “activated” by interaction with its membrane receptor.8
The active site of the NG GTPase is defined by highly conserved sequence motifs, termed motifs I–IV, characteristic of all members of the GTPase superfamily, as well as conserved elements unique to the SRP family of GT-Pases.20 Previous structures of the GDP complex of the NG domain from T. aquaticus17 revealed that the elements that recognize the guanine base—motif IV, the so-called “closing loop,” and a buried lysine residue— disordered in the absence of nucleotide, become organized on binding as part of a network of interactions that links the conformation of the binding pocket to the relative position of the N domain. In contrast, the elements of the active site that interact with the β- and γ-phosphate groups—sequence motifs II and III—which are ordered in the apo protein, become, in the presence of Mg2+GDP, somewhat disordered, as many of their stabilizing interactions are disrupted. The structure of a magnesium-free GDP complex revealed a novel conformation of the bound nucleotide that allowed the protein to recover the stabilizing interactions characteristic of the apo state.17 These structures were interpreted as steps along a putative pathway for magnesium and GDP release, with the first, disordered, conformation being representative of the state following release of inorganic phosphate, and the second representative of the state immediately prior to GDP release and recovery of the stable apo conformation.17
In the course of studies aimed at obtaining a structure of the Ffh NG in its GTP-bound conformation, we obtained crystals which, when solved, revealed instead a new GDP-bound form. These crystals contain two Mg2+GDP-bound monomers in the asymmetric unit that exhibit two new distinct nucleotide binding conformers. One reveals a conformational change of motif II that suggests a functional role for a highly conserved aspartate side-chain; the second reveals a pentacoordinate magnesium ion that provides insight into the role of the Mg2+ ion in the mechanism of phosphate and nucleotide exchange. These structures, with the previous structures of the apo and GDP-bound protein, can be interpreted as a series of snapshots along the pathway of nucleotide release and recovery of the quiescent apo state. That a substantial rearrangement of the conformation of active site motifs II and III must occur in the catalytically competent state of the GTPase has been shown.8 However, particularly because there appear to be a number of novel mechanistic features of the SRP GTPases, fleshing out our understanding of the interactions with the products of hydrolysis—inorganic phosphate, magnesium ion, and GDP—is likely to provide valuable insights into both the functional and structural basis for specific elements of the protein structure and the biological logic of this distinct family of GTPases.
RESULTS AND DISCUSSION
Overview of the Structures
The crystal structure of the Mg2+GDP complex of Ffh NG reported here has been refined at 2.1 Å resolution to an Rcryst of 0.194 and an Rfree of 0.236. The two non-crystallographically related monomers in the asymmetric unit were refined without NCS restraints, and we refer to the two independent monomers as monomers A and B, below. Crystallization was carried out at room temperature, using a mother liquor that contained Mg2+GTP at 2 mM. However, in both binding sites in the asymmetric unit, the bound species is clearly identifiable as GDP, and the position and distinct coordination of the magnesium ion in each site is unambiguous (see below). The electron density maps are overall quite good, and the main-chain conformation is everywhere well defined. All residues are within most-favored and allowed regions of the Ramachandran plot.21 Crystallographic and refinement statistics are reported in Table I.
TABLE I.
Statistics
| Data collection | |
| Space group | P21 |
| Unit cell | a = 55.4 Å, b = 113.6 Å, c = 61.4 Å, β = 111.8° |
| Resolution range | 20.0–2.1Å |
| Rsyma | 0.045 (0.159)b |
| Completeness | 99.5 (99.9) |
| Redundancy | 3.2 (3.2) |
| Average I/σ(I) | 24.1 (6.1) |
| Refinement | |
| Number of reflections (Test set) | 38,000 (2,023) |
| Rcrystc | 0.194 |
| Rfree | 0.236 |
| Number of protein atoms | 4620 |
| Number of heterogen atoms | 351 |
| Average temperature factor (A2) | |
| (Å2) | |
| Protein | 26.1 |
| MgGDP | 20.5 |
| Water | 31.0 |
| RMS Bonds (Å) | 0.018 |
| RMS Angles (°) | 1.669 |
Rsym = Σ|Ih − 〈Ih〉|/ΣIh, where 〈Ih〉 is the average over symmetry equivalents.
Values in parentheses are the high resolution shell.
Rcryst = Σ|Fo − Fc|/ΣFo. Rfree is calculated for 5% of the reflections omitted from the refinement.
An overview of the structure of monomer A is presented in Figure 1. Overall, the structures of the two monomers in the asymmetric unit are very similar to each other, having an RMS deviation over Cα atoms of 0.53 Å, and they are similar to the previously reported structure of the Mg2+GDP complex (PDB id 1ng1),17 with RMS deviations of 0.78 and 0.84 Å for monomers A and B, respectively. Significantly, when the three structures are superimposed over the G domains alone, the dynamic elements of the nucleotide binding sites previously identified in the comparison of the GDP and apo crystal structures—the guanine recognition motif IV, the “closing loop,” and the α4 helix at the N/G domain interface—all overlap (< 0.2 Å shift). However, the helical N-domain undergoes a slight twist between the different structures such that the distal ends of the domain deviate by as much as 1.5 Å. This shift is distinct from that between the apo and the Mg2+GDP bound structures (as in that case the relative position of the α4 helix, and the N/G interface itself, is shifted by ~ 0.9 Å), but it suggests that the position of the N domain is less restricted than was previously described.17,18
Fig. 1.
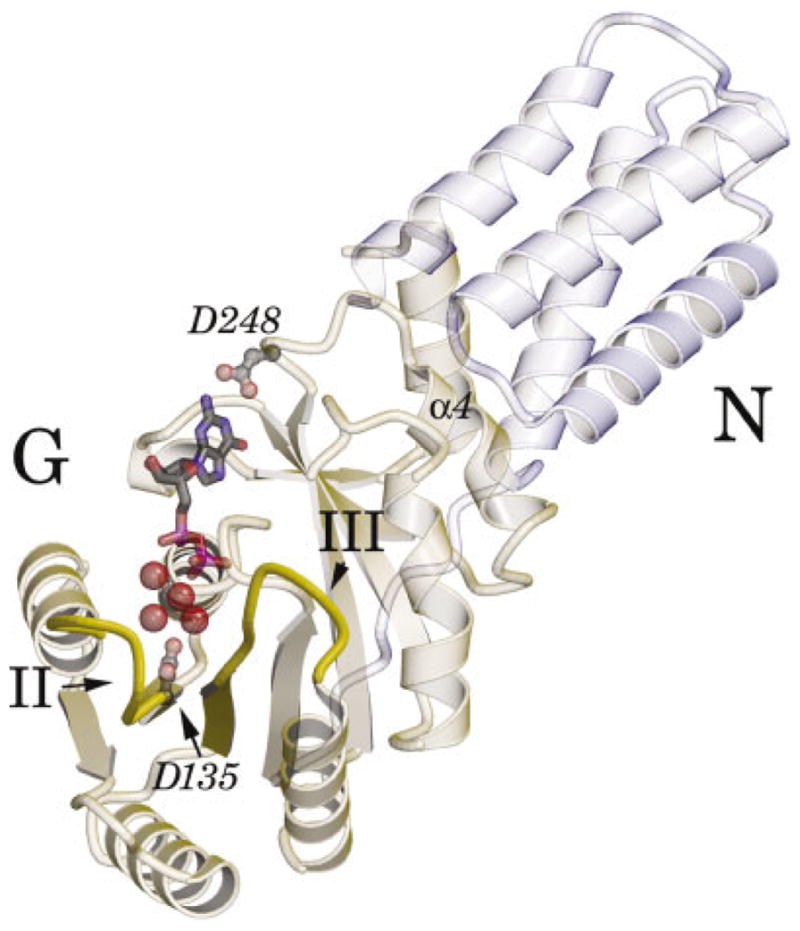
Ribbon diagram of the nucleotide bound protein. The ribbon diagram of monomer A of the Mg23GDP complex of the Ffh NG domain is oriented to view into the active site. The 3-helical N domain is in light blue, and the G domain is in light green. The motif I P-loop at the center of the G domain interacts with the phosphate groups of GDP (ball-and-stick). Motif II to the left, and motif III to the center right (indicated), interact with the bound magnesium ion through intervening water molecules. The hydrated magnesium ion is shown as a CPK representation. The carboxylate group of Asp248 hydrogen-bonds the guanine base; the carboxylate of Asp135 contributes to the second coordination sphere of the magnesium in monomer A, but is usually found in a different conformation (see text).
The GDP binding interactions of monomers A and B are similar to those previously observed for the SRP GTP-ase.17 In particular, Asp248 hydrogen bonds the guanine base (Fig. 1), the “closing loop” packs against it, and a buried water molecule bridges the N1 nitrogen of the ligand to the buried side-chain of Lys117. In each site, an oxygen of the β-phosphate coordinates the bound magnesium ion (see Fig. 3), as is characteristic of Mg2+GDP binding in other members of the GTPase superfamily.22 In contrast to this common structure, which positions the base and the α- and β-phosphates of GDP, there is striking variation between monomers A and B in the adjacent active site pocket and sequence motifs II and III (see Fig. 1). These differences occur under identical solution conditions, and so are due to differences in the crystal packing environment of each monomer. What they reveal, however, are a set of conformational states of the active site loops in the SRP GTPase, and behavior of the active site water molecules that may give insight into the mechanism by which the products of GTP hydrolysis are released. These include a novel conformation of motif II in monomer A, a pentacoordinate state of the magnesium ion in monomer B, and shifts in the positions of water molecules that suggest an uncoupling of the protein sequence motifs from the bound ligand during the process of nucleotide release. They are discussed in turn below.
Fig. 3.
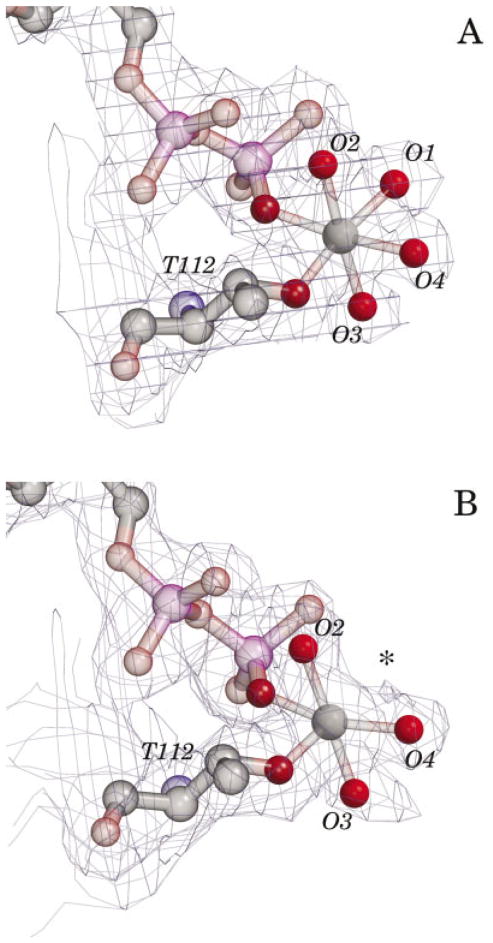
Difference in magnesium ion hydration in monomers A and B. The 2Fo-Fc electron density maps of the hydrated magnesium ions in the active sites of monomers A and B are shown, contoured at 1.0 3. In monomer A, each of the coordinating water molecules is clearly defined. In contrast, although three coordinating waters are well defined in monomer B, and the temperature factors of the magnesium ions and coordinating waters are similar, no coordinating water at position 1 (asterisk) is visible in the electron density map. As this position is the site of interaction with the 3-phosphate oxygen of bound GTP, facile exchange may be functionally significant. The magnesium omit difference map allowed for an identical interpretation. Thr112 and the phosphate groups of the bound GDP are shown in each figure. The map cover radius was 1.1 Å, including the missing water position.
Disorder and Rearrangement of Motif II
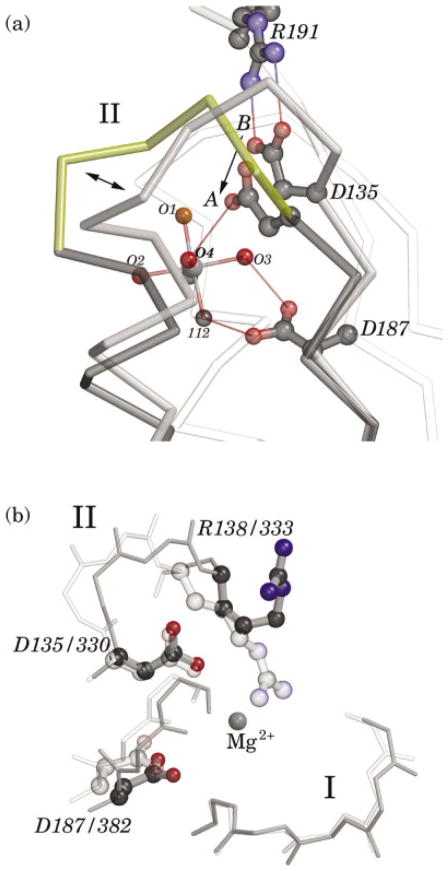
The residues of sequence motif II are highly conserved in the SRP GTPases, but are unique to this family and distinct from other GTPases. Their position relative to the GTPase active site, corresponding to the region of “switch 1” in other GTPases, constitutes them as likely to be involved in the conformational changes that signal GTP binding and hydrolysis.6,22 The sequence of motif II in the T. aquaticus Ffh is DTQRPAA, with Asp135, Arg138, and Ala141 being universally conserved in the SRP GTPases.23 In the structure of the apo Ffh NG domain the side-chains of motif II are involved in a number of interactions that stabilize the conformations of motifs II and III in the empty state.14 In the first structure of an Mg2+GDP complex of Ffh NG,17 it was found that these interactions, in particular a salt bridge between Asp135 and Arg191 of motif III, were broken, resulting in disorder, but not rearrangement, of the loop that comprises the motif.14 These interactions are also absent in the crystal structure reported here in monomer A but there has been substantial further rearrangement [Fig. 2(a), and see Fig. 4]. Most significant is that the carboxylate group of the highly conserved Asp135 side-chain has moved by ~ 3 Å towards the magnesium ion, so that it contributes to its second coordination sphere by hydrogen-bonding one of the coordinating water molecules, O4 [Fig. 2(a)]. The conformational change along the polypeptide backbone of residue 135 is actually fairly slight—a small shift in φ/ψ angles is leveraged along the side-chain—and the key conformational change of the backbone occurs further along the motif, between residues Pro139 and Ala140. There, a peptide flip both facilitates movement of Asp135, and has two additional consequences [Fig. 2(a)]. The adjacent helix α1a shifts significantly, ~ 1 Å approximately along its axis, and the α-carbon of the conserved motif II arginine residue, Arg138, is shifted 3.8 Å towards the active site pocket. Its side-chain, though disordered in this structure, is clearly reoriented towards the active site.
Fig. 2.
Conserved Asp 135 contributes to the magnesium binding site. a: The localized shift in the position of Asp135 is shown following alignment of monomers A and B. The positions of the magnesium ion and most of the rest of the protein are essentially invariant, and the background includes the their overlap (white). The conformation in monomer B (grey), in which the aspartate side-chain forms a salt bridge with Arg191, is similar to that seen in previous structures of the Ffh NG domain. In monomer A (yellow), however, the side-chain is shifted so that the terminal oxygen is moved by 3 3.1 Å towards the magnesium site (single arrow), where it can contribute to the second coordination sphere of the bound magnesium ion. Large shifts at Pro139 and Ala140 (double arrow) accompany a peptide flip at the proline and movement of the alanine from an 3L conformation to an 3R. This conformation of the main-chain is very well defined in the electron density map. The motif II loop is poorly ordered in the previous structure of an Mg23GDP complex of Ffh NG, and similar localized disorder has been noted in a comparison of structures of the T. aquaticus FtsY (C. Reyes & R. Stroud, personal communication). b: An overlay of the positions of the motif I P-loop, motif II Asp135, and Arg138, and motif III Asp187 in the structures of the T. aquaticus Ffh NG Mg23GDP complex reported here (darker), and the apo E. coli FtsY NG15 (lighter). The structures have been aligned by superposition of their P-loops, and the corresponding residue numbering in the two sequences is shown. The position of the Mg23 ion in the Ffh NG structure is indicated; no magnesium was bound in the apo FtsY structure, and Arg333 extends towards that position in the apo FtsY. The Arg138 side-chain is shown for Ffh NG, but is very poorly ordered in this structure.
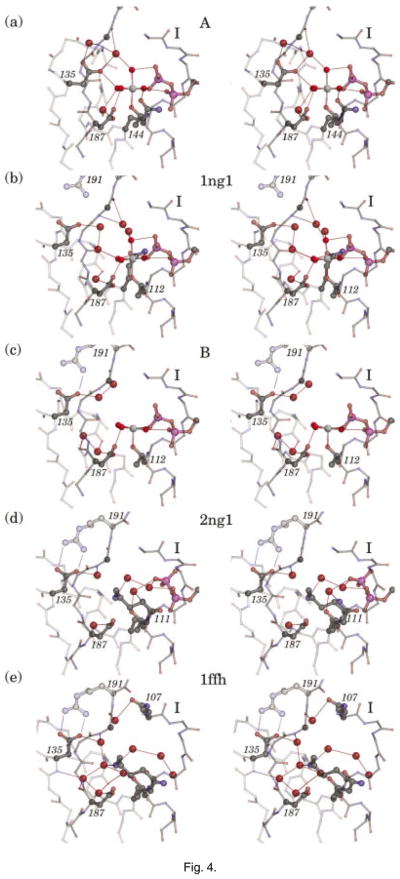
Fig. 4.
Snapshots of the active-site interactions in GDP complexes of Ffh NG. A series of stereo diagrams generated from structures of the apo and GDP complexes of Ffh NG superimposed over their P-loop regions. The resolutions of the structures shown range from 2.0 –2.3 Å and the structures are of similar quality. The backbone of the P-loop (right front), motif II (back), and motif III (left front) are indicated. a: Monomer A (this manuscript), a Mg23GDP complex with motif II/III rearranged and Asp135 shifted towards the active site. b: PDB id 1ng1, a Mg23GDP complex, with motif II/III disordered only. c: Monomer B (this manuscript), an Mg23GDP complex, in which the magnesium ion is five-coordinate. d: PDB id 2ng1, a magnesium-free GDP complex in which the 3-phosphate of the GDP is flipped away from the P-loop and the protein adopts the “apo” conformation. e: PDB id 1ffh, a structure of the apo protein in which the water interactions in the active site is well defined. In (a) and (b) an extensive network of water hydrogen-bonding interactions links the magnesium site to the backbone of motif III. Note the hydrogen bonds to the carbonyl and amide nitrogens of Gly190 (C3 position indicated by a ball along the main-chain), and note the reestablishment of hydrogen-bonding between Asp135 and Arg191 as the apo structure is recovered in (d) and (e). In this series of figures the side-chains of all residues that interact with the waters are shown (Gln107, Lys111, Thr112, Asp135, Gln144, and Asp187).
Interestingly, the partial collapse of motif II towards the active site yields a conformation that is closely reminiscent of that seen in two structures of the apo NG domain from other species, E. coli FtsY and A. ambivalens Ffh.15,16 Indeed, the conformation of motif II observed in monomer A here is almost identical to that first observed in the structure of the E. coli FtsY [Fig. 2(b)]. Monomer A and FtsY NG (1ftsy) superimpose over their G domains with an overall RMS deviation of 1.52 Å for 158 α-carbons and good overlap over the motif I P-loop. However, in contrast to the comparison previously made with the apo Ffh NG (1ffh.pdb),17 the region of motif II also overlaps remarkably well, with the alpha carbon atoms at the conserved aspartate (i.e., Asp135) being only 0.6 Å apart, and their side-chains oriented identically towards the magnesium binding site. While the α-carbons of the conserved arginine (i.e., Arg138) are separated by ~ 1.5 Å in such an alignment, the residue is reoriented in monomer A such that the two side-chains are directed in essentially the same direction towards the active site. The conformation of the motif II loop in the A. ambivalens Ffh NG structure is similar.
The occurrence of similar conformations of motif II in structures of the different SRP GTPases suggests that the rearrangement seen here is functionally significant, and it provides further evidence for the role of conserved residues of motif II in the SRP GTPases. Asp187 of motif III is universally conserved in the superfamily of GTPases, and is believed to contribute electrostatic stabilization of the bound magnesium ion.24 –26 The structure observed in monomer A confirms the notion that conserved Asp135 of motif II participates in magnesium binding as well;16 its actual chemical contribution to the reaction, or its regulation, remains to be elucidated. The reorientation of the Arg138 side-chain is also significant, because arginine has been implicated as a key player in the chemistry of GTP hydrolysis.27–29 The position of the side-chain is consistent with the catalytic role previously suggested from inspection of the apo protein structures.14,15 Taken with a previous modeling study,8 this identifies two conserved arginines (the other being Arg191 of motif III) as turning towards the active site of the SRP GTPase during different stages of GTP binding and hydrolysis, and suggests that regulation of the accessible conformational space of the two motifs, as opposed to external provision of an “arginine finger,”27 may provide the mechanism for regulation of GTP hydrolysis in the targeting complex.
Interestingly, a structural rearrangement of motif II over exactly the region identified here (and similarly, in a context of structures with little overall change) has been observed in a series of crystal structures of the apo and nucleotide-bound FtsY from T. aquaticus (C. Reyes & R.M. Stroud, personal communication). This provides further support for the notion that the conformational substates of motif II trapped in different crystal structures reflect a conserved propensity for conformational change (i.e., that its function is manifested by shifts between conformational states separated by relatively low energy barriers). We can propose, therefore, that the conformation observed in our crystal structure (and others) is representative of a step in the pathway following GTP hydrolysis, a point to which we return below.
Differences in the Magnesium Coordination
The position of the bound magnesium ion and its coordinating waters are well defined in both monomers in the asymmetric unit (Fig. 3). The magnesium ion is almost always observed to be six-coordinated in the structures of GTPases.8,22,30 –32 Four of the coordinating groups are invariant, with one of the coordinating oxygens being supplied by a conserved side-chain hydroxyl (from Thr112 of motif I in Ffh NG) and another by the β-phosphate of the bound nucleotide.17 In addition, two water molecules, corresponding here to O2 and O3, are always present, and these have second sphere interactions with the nucleotide α-phosphate (O2) and the carboxylate of the conserved aspartate, Asp187 of motif III in Ffh NG (O3). The remaining two oxygen ligands, corresponding to O1 and O4 here, are, in the structures of various GTPases, supplied by different groups in different states, and so can be considered to be dynamic. O1 occupies the position which, when GTP is bound, is occupied by an oxygen of the γ-phosphate group,24,30,33,34 and O4 occupies a position that is replaced by the hydroxyl of a conserved threonine in the GTP-bound state of some GTPases (e.g., in ras35). In the Mg2+GDP complex seen in monomer A the magnesium is also six-coordinated [Fig. 3(a)], with the addition, in this structure, of a new hydrogen bond between O4 and the carboxylate group of the shifted Asp135 of motif II (discussed above).
What is striking, however, is that in monomer B the magnesium exhibits a well defined five-coordinate configuration [Fig. 3(b)]. There is little evidence in the difference map for a coordinating water at position O1; although a small ‘nub’ of electron density in the 2Fo-Fc map contoured at 0.5 σ may represent a minor population of fully-coordinated molecules, we can place an upper limit of 20% occupancy at that position based on occupancy refinement trials. Importantly, the temperature factors of the magnesium ion and its coordinating water molecules are fairly similar to those in monomer A (in monomer A they average 19.6 Å2, σ 1.9 Å2 and in B, 24.6 Å2, σ 2.7 Å2), and are similar to the temperature factors of the coordinating oxygens of Thr112 and the bound GDP (which average 17.3 Å2, σ 1.4 Å2). During refinement the coordination bond lengths were restrained to 2.08 Å (and the bond angles restrained to 90° and 180°).36 There are no deviations from the standard magnesium coordination geometry in the structure, with the exception of the bond trans to the vacant position O1 in monomer B. This bond length shortens to 2.03 Å, which is consistent with the behavior of pentacoordinate magnesium ion37 and it has the consequence that the magnesium ion appears to shift slightly out of the plane defined by the other coordinating atoms.
A five-coordinated magnesium ion has been observed previously, in a study of GDP complexes of the heterotrimeric G protein Giα1.38 In that case, however, a low occupancy water molecule was found at a distance of 2.7 α from the magnesium ion. Here, we see no evidence for ordered water in the close neighborhood of the magnesium ion, as the nearest water molecules are 4.8 –5.4 Å away. Interestingly, however, there is no apparent steric hindrance for occupancy of a water at position O1. It appears, instead, that because the active-site region of monomer B has extensive hydrogen bonding and packing interactions across a crystal contact with monomer A, there are no neighboring groups free to contribute stabilizing interactions with a coordinating water at position O1. The absence of these interactions may be a key factor in the observed coordination geometry. Although the lowest energy configuration for magnesium is hexacoordinate, in computational studies the pentacoordinate state (with one water molecule in the second coordination shell) is less than 5 kcal/mol higher in energy.39 What the structure here implies is that the particular geometry of interactions between the protein and GDP, each providing a number of negatively charged groups to both the first and second coordination spheres of the bound magnesium ion, destabilizes binding at the coordination position O1 such that without additional interactions water freely exchanges. Assuming 10% occupancy at position O1, we can make a “back of the envelope” estimate of the free energy difference: 1.36 kcal/mol at 297 K for binding of water at position O1, a ~6 kcal/mol destabilization relative to the calculated hexacoordinate state. This is a significant effect, but it may well have been obscured in previous crystal structures because other stabilizing interactions (e.g., hydrogen bonds ~5 kcal/mol) are available to the coordinating water.
The conformation observed in monomer B is, therefore, energetically feasible, and it must occur transiently during the course of GTP hydrolysis and release because in the corresponding structures of the GTP-bound state of other GTPases, the water at position O1 is replaced by an oxygen atom of the γ-phosphate. This provides a functional significance for the destabilization we infer from the coordination structure in monomer B: the interactions of the protein-magnesium complex, which comprise residues of motif I and motif III, facilitate exchange of the oxygen ligand at coordination position O1. It would be interesting to see whether behavior consistent with this notion is exhibited by structures of the stabilized GTPase/GAP complexes,28,40,41 in which the is thought to act as a mimic of the planar transitional state during hydrolysis of the phosphodiester bond. However, direct comparison at this point is proscribed by the varying refinement protocols used among the limited dataset of structures (and in some cases, the anomalous coordination bond lengths reported). An interesting parallel is found in the high-resolution ternary substrates complex structures of the hypoxanthine phosphoribosyltransferase (HPRT) of Trypanosoma cruzi, with 5-phosphoribosylopyrophosphate (PRPP), purine analog 7-hydroxypyrazolo-pyrimidine (HPP) and two divalent metal ions bound.42 The four ions in the dimeric structure show standard coordination bond lengths, except in the direction of an important catalytic interaction. In these structures the position of bound PRPP, HPP, and Mg2+ (or Mn2+) are highly constrained within a sequestered binding site, and the distance between the metal ion and the O1 oxygen of PRPP is too long to be a true metal ligand, 2.5–2.6 Å, while the distance to the trans water molecule is shorter than the average. Completion of the coordination sphere of this divalent ion was proposed to contribute a driving force in bond breakage between the ribose and leaving pyrophosphate group.
Reassembly of the ‘apo’ Conformation
If we suppose that the two structures reported here represent accessible and functionally relevant conformations of the Ffh NG GTPase, and also consider the previous structures of the Mg2+GDP, GDP and apo proteins, we can assemble a tentative map of the conformational changes undergone during the end stage following GTP hydrolysis as the protein releases Pi, Mg2+, and GDP, and recovers its stable apo structure. We focus here on the motif II–III interaction, where we see several new conformers of the “active site network” (Fig. 4). We take structure A as that closest to the hydrolysis step; its significance is suggested by the observation of similar conformations in the structures of E. coli FtsY and A. ambivalens Ffh. Its conformation is the most distant from the structure of the apo protein and it suggests a functional role for highly conserved Asp135 of motif II in its interaction with the O4 coordinating water of the bound magnesium ion, as well. In this state, there is a network of water-mediated hydrogen bond interactions that links the sequence motifs II and III, particularly the central Gly190,8 to the bound magnesium ion and motif I [Fig. 4(a)]. The structure of the Mg2+GDP complex reported previously in another crystal form (1ng1) can be taken as the subsequent step, as the motif II backbone rearranges to its apo conformation, but remains disordered.17 A water molecule moves in to maintain the interaction with the O4 coordinating position, and the Gly190-H2O-Mg2+ bridge is also maintained [Fig. 4(b)]. Note that in both of these structures, the position of Arg191 is poorly defined, and it does not contribute stabilizing salt-bridge interactions. Monomer B can be taken to reveal the next stage in the transition, in which the water network is disrupted, and the Arg191/Asp135 salt bridge begins to form and stabilize the motif II/III region [Fig. 4(c)]. As discussed above, not only is the magnesium ion predominantly five-coordinate, but there are no hydrogen bonds bridging across the active site. The structure of the magnesium-free GDP complex (2ng1) can be taken as subsequent to release of the magnesium ion, freeing the β-phosphate to rotate away from the P-loop, and allowing apo-like side-chain interactions to re-form [Fig. 4(d)]. Finally, the structure of the apo protein (1ffh) reveals the re-establishment of a fairly extensive network of water-mediated hydrogen bonds, and the presence of the Arg191/Asp135 salt bridge that stabilizes the conformations of motifs II and III [Fig. 4(e)].
Interestingly, throughout this transition there are three water positions that are maintained. The first, the water hydrogen-bonded to the side-chain of Asp187 buried in the active site pocket, is present throughout. The second, which interacts with the backbone amide and carbonyl of Gly190, is present in the Mg2+GDP structures, but can be considered to shift to a second position during the transition (represented by structure B) to the apo form. There, the water hydrogen-bonds the backbone amide of Gly190 and the side-chain oxygen of Asp135. In the first instance, the water bridges Gly190 and the waters coordinating the magnesium ion; in the second, the water bridges Gly190 and Asp135 and waters bound in the motif I P-loop. The protein can be thought of as releasing a set of interacting waters, and recapturing them in a different position as it assumes a new conformational state.
That different conformations of the protein are trapped in different crystal forms is not in itself surprising, but to the extent that they suggest the conformational space available to specific polypeptide regions that are likely to reflect the functional dynamics of the protein, the set of different structures can become particularly informative. Only limited regions of the Ffh NG protein structure have been observed to occupy distinct conformational states in its different crystal structures.8,14,17,43 Whether the structural details of the observed substates of motif II correspond exactly to a reaction pathway cannot yet be established. However, they do provide insight into an underlying structural dynamic, and they provide confirmation of a common structural state in the two SRP GTPases, Ffh and FtsY, in which the conserved Asp135 of motif II interacts with the bound magnesium ion. Similarly, the distinct coordination of the magnesium ion observed in monomer B, presumably an intermediate trapped by a particular packing interaction, provides insight into destabilization of the O1 coordination position, privileged for facile exchange, that may well contribute to the mechanism of hydrolysis and exchange in the SRP GTPase and in other GTPases as well. The scheme presented in Figure 4 is of course at this point speculative, but it allows us to rationalize the series of conformational states observed in these multiple crystal forms. It highlights both the value of multiple crystal forms, which trap different conformers of the same protein binding state that may, indeed, represent different functional states, and the value of the non-crystallographic symmetry present in the crystal structure reported here, which captured two different conformations of the same species under identical solution conditions. These structures allow us to at least tentatively begin to sketch out the “movie” that describes how the protein works, and provide the basis for consideration of the structural dynamics of the GTPase and the mechanism by which water and side-chain interactions manipulate the functional cycle.
MATERIALS AND METHODS
Crystallization and Data Collection
The NG domain of T. aquaticus Ffh14 was concentrated using a Centricon 30 (Amicon) to 32 mg/ml in 2 mM MgCl2, and GTP was added from a 100 mM stock to make 2 mM final concentration. Crystals were grown in 4 M Na Formate (corresponding Hampton Crystal Screen I, condition #33). Sitting drop (1.2 μl) vapor diffusion crystallization experiments were setup at room temperature; after two days the drops were streaked using a nylon fiber from a seed stock of crystal clusters grown under the same conditions. A single chunky 100 μ crystal was cryoprotected by addition of two-drop volumes of a mother liquor containing 20% ethylene glycol and harvested in a 200 micron loop for data collection. Data were measured at 100 K at DND-CAT beamline 5-ID-B of the Advanced Photon Source (APS) using a 165 mm MARCCD detector, with a 20 second exposure time for each 1° oscillation. A total of 150° rotation was measured, the detector distance was 120 mm, and the wavelength was 0.964 Å. The data were indexed and integrated with DENZO and merged and scaled with SCALEPACK44 (Table I). As noted in the text, the GTP in the crystallization solution was hydrolyzed to GDP, and this was the species bound in the crystal.
Structure Solution and Refinement
The space group was found to be P21 with two monomers in the asymmetric unit. The structure was solved by molecular replacement with the program AmoRe45 using the structure of the apo Ffh (1ffh.pdb), with waters removed, as the starting model. The two monomers in the asymmetric unit were further positioned by rigid body refinement, and then refined using simulated annealing and positional minimization protocols in XPLOR.46 The model was inspected with O47 and rounds of manual rebuilding and placement of water molecules alternated with positional and isotropic temperature factor refinement with problematic regions of the structure omitted. Subsequently, the incomplete model was refined further using the maximum likelihood target in REFMAC.48 We found it necessary to use a WEIGH MATR value of 0.25 (half the default) at this resolution to limit distortion the protein model. The resulting maps allowed rapid resolution of ambiguous regions in the structure, and yielded a final Rcryst of 0.194 and Rfree of 0.236. The average temperature factors of the two monomers in the asymmetric unit are similar (26.4 Å2 and 25.7 Å2, respectively). Despite the difference in coordination, the two active-site magnesium ions have similar temperature factors as well (20.0 Å2 and 24.7 Å2, respectively). The pentacoordinate magnesium ion in monomer B was refined with six coordinating groups, but with the occupancy of oxygen O1 set to 0.0. (Setting the occupancy of O1 to 1.0 during refinement, which yields a large negative difference peak, did not affect the deviation in geometry we describe.) The coordination bond lengths were restrained to 2.08 Å (and the bond angles restrained to 90° and 180°)36 during refinement. Except for the bond trans to position O1 in monomer B (2.03Å), there are no large deviations from standard geometry. The bond lengths of the six coordinating groups in monomer A average 2.08 Å, σ 0.015 Å, and the four remaining bonds in monomer B average 2.07 Å, σ 0.008 Å. Refinement statistics are summarized in Table I.
Analysis
The final model consists of residues 1–295 in each monomer, each with bound Mg2+GDP. All residues are within the allowed regions of the Ramachandran plot (98.1% in the favored regions) as defined by MOLPRO-BITY.21 We used LSQMAN49 to determine root mean squared deviations between atomic positions in the compared structures.
Coordinates
Atomic coordinates and structure factors have been deposited with the ePDB and given PDB ID code 1o87.
Acknowledgments
We thank Yi Fan for technical assistance, and Savita Padmanabhan for guidance during data reduction. This work was partly supported by a faculty development grant 76296-549401 from the Howard Hughes Medical Institute, by support from the R.H. Lurie Cancer Center to the Structural Biology Facility at Northwestern University, and by grant GM58500 from the NIH. The DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) at the Advanced Photon Source is supported by the U.S. National Science Foundation through Grant DMR-9304725 and the State of Illinois through the Department of Commerce and the Board of Higher Education Grant IBHE HECA NWU 96. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research under Contract No. W-31-102-Eng-38.
References
- 1.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 2.Jagath JR, Rodnina MV, Wintermeyer W. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. J Mol Biol. 2000;295:745–753. doi: 10.1006/jmbi.1999.3427. [DOI] [PubMed] [Google Scholar]
- 3.Shepotinovskaya IV, Freymann DM. Conformational change of the N-domain on formation of the complex between the GTPase domains of Thermus aquaticus Ffh and FtsY. Biochim Biophys Acta. 2002;1597:107–114. doi: 10.1016/s0167-4838(02)00287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. Embo J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powers T, Walter P. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- 6.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 7.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 8.Padmanabhan S, Freymann DM. The conformation of bound GMPPNP suggests a mechanism for gating the active site of the SRP GTPase. Structure. 2001;9:859–867. doi: 10.1016/s0969-2126(01)00641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rapiejko PJ, Gilmore R. Empty site forms of the SRP54 and SR alpha GTPases mediate targeting of ribosome-nascent chain complexes to the endoplasmic reticulum. Cell. 1997;89:703–713. doi: 10.1016/s0092-8674(00)80253-6. [DOI] [PubMed] [Google Scholar]
- 10.Song W, Raden D, Mandon EC, Gilmore R. Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 2000;100:333–343. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- 11.Miller JD, Wilhelm H, Gierasch L, Gilmore R, Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 1993;366:351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y, Qi HY, Hyndman JB, Ulbrandt ND, Teplyakov A, Tomasevic N, Bernstein HD. Evidence for a novel GTPase priming step in the SRP protein targeting pathway. Embo J. 2001;20:6724–6734. doi: 10.1093/emboj/20.23.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan SO, Walter P. Induced nucleotide specificity in a GTPase. Proc Natl Acad Sci U S A. 2003 doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 15.Montoya G, Svensson C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–369. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- 16.Montoya G, Kaat KT, Moll R, Schäfer G, Sinning I. The crystal structure of the conserved GTPase of SRP54 from the archeon Acidianus ambivalens and its comparison with related structures suggests a model for the SRP-SRP receptor complex. Structure. 2000;8:515–525. doi: 10.1016/s0969-2126(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 17.Freymann DM, Keenan RJ, Stroud RM, Walter P. Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP. Nat Struct Biol. 1999;6:793–801. doi: 10.1038/11572. [DOI] [PubMed] [Google Scholar]
- 18.Ramirez UD, Minasov G, Focia PJ, Stroud RM, Walter P, Kuhn P, Freymann DM. Structural basis for mobility in the 1. 1 A crystal structure of the NG domain of Thermus aquaticus Ffh. J Mol Biol. 2002;320:783–799. doi: 10.1016/s0022-2836(02)00476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moser C, Mol O, Goody RS, Sinning I. The signal recognition particle receptor of Escherichia coli (FtsY) has a nucleotide exchange factor built into the GTPase domain. Proc Natl Acad Sci. 1997;94:11339–11344. doi: 10.1073/pnas.94.21.11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 21.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 22.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 23.Zwieb C, Samuelsson T. SRPDB (signal recognition particle database) Nucleic Acids Res. 2000;28:171–172. doi: 10.1093/nar/28.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prive GG, Milburn MV, Tong L, de Vos AM, Yamaizumi Z, Nishimura S, Kim SH. X-ray crystal structures of transforming p21 ras mutants suggest a transition-state stabilization mechanism for GTP hydrolysis. Proc Natl Acad Sci USA. 1992;89:3649–3653. doi: 10.1073/pnas.89.8.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Via A, Ferre F, Brannetti B, Valencia A, Helmer-Citterich M. Three-dimensional view of the surface motif associated with the P-loop structure: cis and trans cases of convergent evolution. J Mol Biol. 2000;303:455–465. doi: 10.1006/jmbi.2000.4151. [DOI] [PubMed] [Google Scholar]
- 27.Scheffzek K, Ahmadian MR, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 28.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 29.Rittinger K, Walker PA, Eccleston JF, Nurmahomed K, Owen D, Laue E, Gamblin SJ, Smerdon SJ. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 30.Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR. Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 31.Jagath JR, Rodnina MV, Lentzen G, Wintermeyer W. Interaction of guanine nucleotides with the signal recognition particle from Escherchia coli. Biochemistry. 1998;37:15408–15413. doi: 10.1021/bi981523a. [DOI] [PubMed] [Google Scholar]
- 32.Zhang B, Zhang Y, Wang Z, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions or Rho family GTP-binding proteins. J Biol Chem. 2000;275:25299–25307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- 33.Kjeldgaard M, Nissen P, Thirup S, Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993;1:35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- 34.Berchtold H, Reshetnikova L, Reiser CO, Schirmer NK, Sprinzl M, Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- 35.Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protoon-cogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 36.Harding MM. The geometry of metal-ligand interactions relevant to proteins. Acta Crystallogr D. 1999;55:1432–1443. doi: 10.1107/s0907444999007374. [DOI] [PubMed] [Google Scholar]
- 37.Glusker JP. Structural sspects of metal liganding to functional groups of proteins. Adv Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 38.Coleman DE, Sprang SR. Crystal structures of the G protein Gi alpha 1 complexed with GDP and Mg2+: a crystallographic titration experiment. Biochemistry. 1998;37:14376–14385. doi: 10.1021/bi9810306. [DOI] [PubMed] [Google Scholar]
- 39.Bock CW, Kaufman A, Glusker JP. Coordination of water to magnesium cations. Inorg Chem. 1994;33:419–427. [Google Scholar]
- 40.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AIF4 —activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 41.Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Structure at 1. 65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 42.Focia PJ, Craig SP, III, Eakin AE. Approaching the transition state in the crystal structure of a phosphoribosyltransferase. Biochemistry. 1998;37:17120–17127. doi: 10.1021/bi9821465. [DOI] [PubMed] [Google Scholar]
- 43.Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z. Oscillation data reduction program. In: Sawyer L, Isaacs NW, Bailey S, editors. Data collection and processing. Warrington: SERC Daresbury Laboratory; 1993. pp. 55–62. [Google Scholar]
- 45.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 46.Brunger AT, Krukowski A, Erickson JW. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr A. 1990;46(Pt 7):585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- 47.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 49.Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Meth Enzymol. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]