Abstract
Ergosterol is the predominant sterol of fungi and green algae. Although the biosynthetic pathway for sterol synthesis in fungi is well established and is known to use C24-methylation-C24 (28)-reduction (Δ24(28)-olefin pathway) steps, little is known about the sterol pathway in green algae. Previous work has raised the possibility that these algae might use a novel pathway because the green alga Chlamydomonas reinhardtii was shown to possess a mevalonate-independent methylerythritol 4-phosphate not present in fungi. Here, we report that C. reinhardtii synthesizes the protosterol cycloartenol and converts it to ergosterol (C24β-methyl) and 7-dehydroporiferasterol (C24β-ethyl) through a highly conserved sterol C24- methylation-C25-reduction (Δ25(27)-olefin) pathway that is distinct from the well-described acetate-mevalonate pathway to fungal lanosterol and its conversion to ergosterol by the Δ24 (28)-olefin pathway. We isolated and characterized 23 sterols by a combination of GC-MS and proton nuclear magnetic resonance spectroscopy analysis from a set of mutant, wild-type, and 25-thialanosterol-treated cells. The structure and stereochemistry of the final C24-alkyl sterol side chains possessed different combinations of 24β-methyl/ethyl groups and Δ22(23)E and Δ25 (27)-double bond constructions. When incubated with [methyl-2H3]methionine, cells incorporated three (into ergosterol) or five (into 7-dehydroporiferasterol) deuterium atoms into the newly biosynthesized 24β-alkyl sterols, consistent only with a Δ25 (27)-olefin pathway. Thus, our findings demonstrate that two separate isoprenoid-24-alkyl sterol pathways evolved in fungi and green algae, both of which converge to yield a common membrane insert ergosterol.
Keywords: green algae, sterol evolution, cycloartenol, membranes, sterol C24-methyl transferase, sterol C25-reductase
The defining feature of eukaryote membranes, other than animals that possess the C27 cholesterol, is the presence of C28- to C30-steroidal compounds of varied side-chain constructions characterized by a C24-alkyl group. The addition of C1 to C3 side chains is derived by transmethylation reactions requiring S-adenosyl-L-methionine as the methyl donor and catalyzed by the sterol C24-methyltransferase (24-SMT) family of enzymes (1, 2). Using differences in the C24-alkyl group size, stereochemistry, and complexity in further transalkylations at C22, C23, C27, and C28 as taxonomic traits, and linking them to steroidogenesis, we are able to reason the grouping of 24-alkyl sterol- containing organisms into more or less primitive and advanced forms of life (3). Fossil steranes identified from their diagenetic remains in sedimentary rocks confirm the evolution of 24-alkyl sterol diversity in Eukarya noted in the chemotaxonomy studies and further suggest an ancient origin (< 2.7 billion years ago) of the sterol frame (4–6).
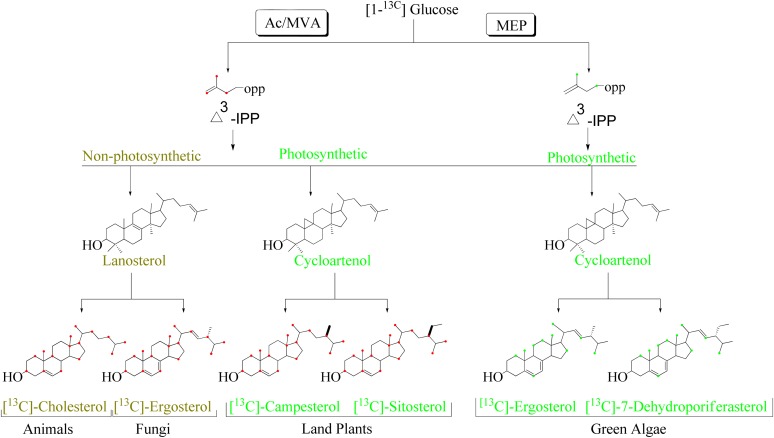
In accordance with the shifting geneome-metabolome congruence in sterol evolution, three general pathways of isoprenoid-sterol biosynthesis are often considered: one in animals that yields cholesterol (C24-H), one in fungi that yields ergosterol (C24β-methyl), and one in land plants that yields stigmasterol (C24α-ethyl) (7–9). These compounds can be assembled modularly in three stages: In module I, synthesis of the basic C5-unit, isopentenyl diphosphate (Δ3-IPP) from glucose, can originate from two independent and nonhomologous metabolic pathways, namely the acetate-mevalonate or the mevalonate-independent 2-C-methyl-D-erythritol 4-phosphate pathways (Fig. 1) (1, 10–12). In module II, Δ3-IPP is converted to the protosterols, lanosterol (nonphotosynthetic lineage), or cycloartenol (photosynthetic lineage) (2, 3, 13, 14). In module III, protosterols are converted to Δ5-sterols via a C24-reduction or coupled C24-alkylation-reduction pathway that yields the exquisite 24-alkyl sterol patterns observed throughout nature (1, 7).
Fig. 1.
Pattern of 13C-incorporation into isopentenyl diphosphate and sterols from incubation of [1-13C]glucose. Ac/MVA, acetate-mevalonate pathway; MEP, mevalonate-independent methylerythritol 4-phosphate pathway.
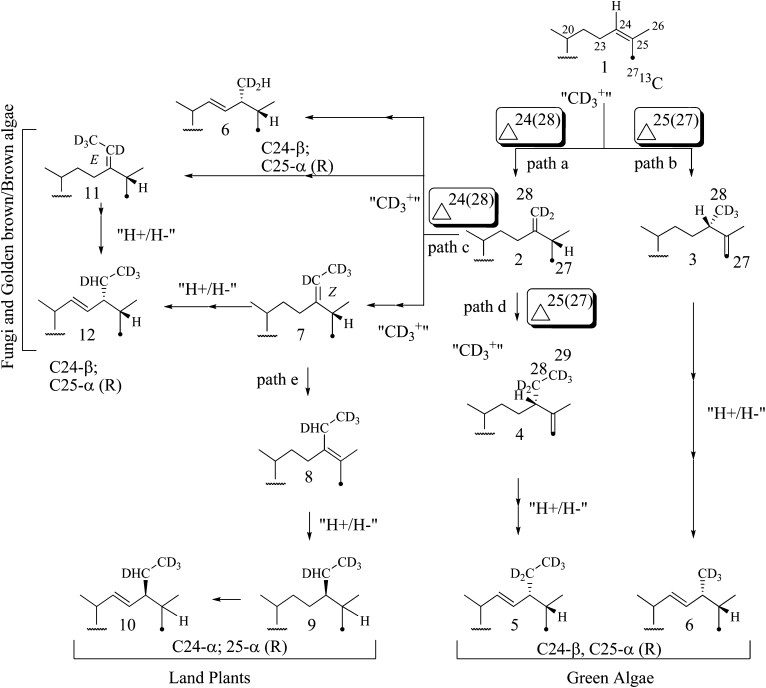
A striking finding to emerge from in vivo isotopically labeling, sterol biosynthesis inhibitor-treatments, and cell-free 24-SMT enzyme studies among phylogenetically diverse algae that include the green, brown, and golden brown algae (15–18) that is not necessarily obvious by current bioinformatic analyses (19) is that ergosterol biosynthesis in the green algae may use a different set of enzymatic reactions in module III from fungi. A crucial difference between ergosterol biosynthesis in these organisms involves the sterol C24-methyl transferase activities and side-chain reductase specificities involved in generating the final ergosterol side chain. Thus, green algae express a 24-SMT that recognizes cycloartenol (16) and a sterol C25(27)-reductase (25-SR) of unknown substrate preference that operate in tandem to introduce stereoselectively the methyl cation at C24 and the hydride ion at C25 from S-adenosyl-L-methionine and NADPH, respectively (Fig. 2). Alternatively, fungi synthesize a 24-SMT that prefers substrates zymosterol or lanosterol, depending on the organism, and a sterol C24(28)-reductase the prefers ergosta-5,7,24(28)-trienol as substrate (1). In both sterol C-methylation-reduction pathways, the resulting ergosterol contains the same stereochemistry at C24 and C25 (1).
Fig. 2.
Sterol C24-alkylation-reduction pathways to 24-alkyl sterol products. The incorporation sites at C24 from incubation with [methyl-2H3]methionine are shown; 13C27 label is shown as a dot.
Therefore, it was surprising that several investigations on sterol biosynthesis in the green alga Chlamydomonas reinhardtii reported that 24-alkyl(idene) sterol biosynthesis followed the fungal Δ24(28)-olefin pathway (20, 21). To deduce generalities for a Δ25(27)-olefin pathway in the synthesis of algal ergosterol and its 24-ethyl homolog, we have examined the ability of C. reinhardtii to synthesize sterol intermediates in the presence and absence of an inhibitor of the sterol C24-methylation reaction and after genetic manipulation to induce intermediates to accumulate in the cell. In a painstaking analysis of the minor and trace compounds of mutant and treated cells, 23 different sterols were detected, many of which contained a Δ25(27) bond consistent with a Δ25(27)-olefin pathway to ergosterol. Moreover, the sterol profiles of these cells failed to show C24(28)-ethylidene derivatives required in the synthesis of 24β-ethyl(idene) sterols synthesized in golden brown or brown algae or which can serve as precursor of land plant sitosterol (22, 23). Analysis of isotopically labeled ergosterol and 7-dehydroporiferasterol isolated from C. reinhardtii grown in the presence of [methyl-2H3]methionine provided further verification for the Δ25(27)-olefin pathway to Δ5,7-C24β-alkyl sterols, which ultimately become the architectural components of algal cell membranes.
MATERIALS AND METHODS
C. reinhardtii strains and culture conditions
C. reinhardtii wild-type strains 21gr (mt+ CC-1690 and 6145c; CC-1691) and ergosterol mutants KD7 and KD21 (24), obtained from the Chlamydomonas Genetics Center, Duke University (Durham, NC), were grown at 23°C on a 13:11 h light:dark cycle with aeration in medium I or medium II of Sager and Granick (25) as previously described (26). The KD7PY mutant was a product of a cross between 6145c and KD7 and was selected due its its ability to grow on agar plates in medium I containing nystatin (2 mM). For the inhibitor studies, cells (1 × 106/ml) were cultured for 3 days in medium I containing 1 µM 25-thialanosterol iodide salt. Cell number was determined using a hemocytometer.
Source and analysis of sterols
Sterol analysis was performed as described previously (27). Briefly, algal cells at approximately 1 × 107 cells/ml were harvested by centrifugation and saponified in 10% aqueous methanolic KOH (10% w/v) at reflux for 30 min to give hexane-soluble neutral lipids. The neutral lipids were routinely examined by GC-MS (30 m HP-5 capillary column coupled to a HP 6890 gas chromatograph interfaced to a 5973 mass spectrometer at 70 eV; GC flow rate of He was set at 1.2 ml/min, injector port was 250°C, and the initial temperature was set at 170°C, held for 1 min, and increased at 20°C/min to 280°C) and HPLC equipped with a photodiode array detector used to provide UV spectra relevant to double bond character in the molecule. In several cases, sterols isolated from the nonsaponifiable lipid fraction and purified by HPLC (analytical Phenomenex Luna column, ODS-100A, eluted with methanol at 20°C at 1 ml/min or analytical TOSOHAAS TSK gel column, ODS-120A with acetonitrile/isopropanol [65/35, v/v] at 35°C at 1 ml/min) were examined by proton nuclear magnetic resonance spectroscopy (1HNMR) (spectra measured in deuterochloroform solutions on a Varian Unity Inova 500 MHz spectrometer with the chemical shifts referenced to chloroform resonating at 7.265 ppm and reported as δ in ppm, ppm) to confirm structure and stereochemistry of the side-chain C24-alkyl group. Authentic reference specimens for comparative GC-MS and 1HNMR analyses are taken from our sterol collection reported in references 27–31 and from literature values (32). Sterols are referenced to the retention time of cholesterol in capillary GC at 13.8 min (old column) or 14.5 min (new column) and in HPLC at 16.5 min (Luna column) or 26.8 min (TSK gel) affording the relative retention times to cholesterol in GC as the RRTc or in HPLC as the αc values.
Feeding of [methyl-2H3]methionine to C. reinhardtii
L-[methyl-2H3]methionine (98 atom % of 2H) (Sigma, St. Louis) was administered to wild-type C. reinhardtii cultures inoculated with 1 × 107 cells/ml at 1 mg/ml. After 3 days inoculation under light, the cells were harvested by centrifugation, and the total sterols from the cultures were examined by GC-MS.
RESULTS AND DISCUSSION
Total sterols of wild-type strain
When this work was undertaken, little information was available on the sterol composition of Chlamydomonas. Several recent reports documented that this alga synthesizes two major sterol products, ergosterol and 7-dehydroporiferasterol, and variably a minor compound, ergost-7-enol (20, 21, 33–35). Moreover, [3-3H]squalene-2,3-oxide incubated in a microsomal enzyme preparation converts exclusively to cycloartenol (36). On the other hand, information is lacking about the types and amount of C4 methyl- or C24(28)-ethylidene intermediates involved in the sequence of chemicals to ergosterol or its 24-ethyl homolog from which a reliable sterol biosynthesis pathway could be constructed.
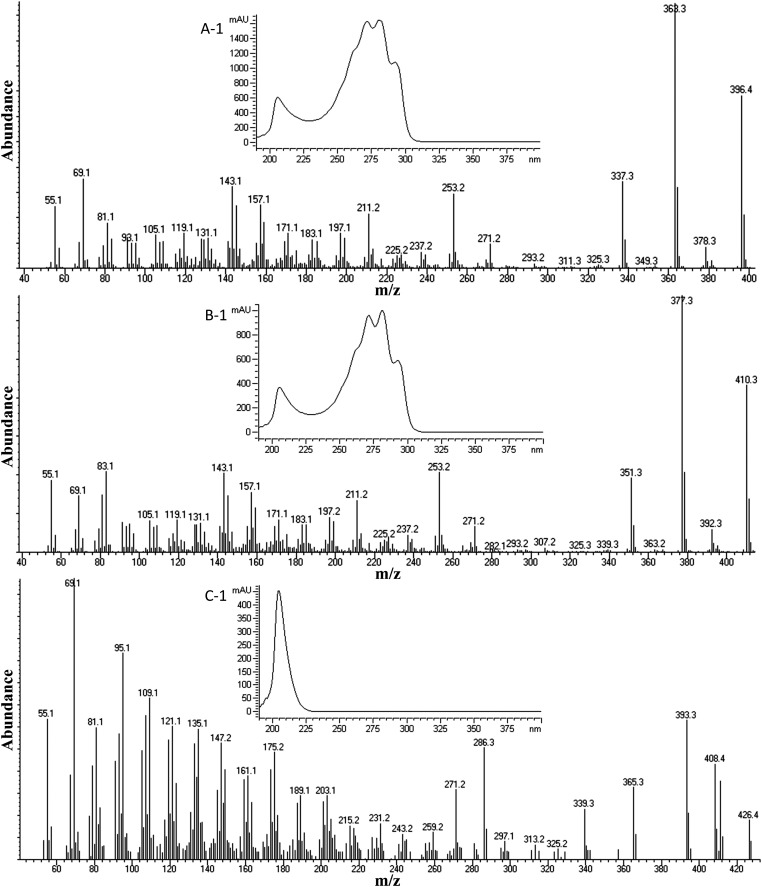
In the present study, the total sterol fraction of C. reinhardtii was analyzed by GC-MS, and 11 sterols were detected (Fig. 3), including ergosterol, 7-dehydroporiferasterol, and cycloar tenol, which possessed mass and UV spectra similar to authentic specimens (Fig. 4). The major sterols in the sterol composition, ergosterol and 7-dehydroporiferasterol, were purified by HPLC, and 1HNMR analysis confirmed their structure and C24β-methyl/ethyl group stereochemistry (supplementary Table I). Minor compounds detected in the GC chromatogram were identified according to GC retention times and mass spectra relative to standards corresponding to cycloartenol, 4α,14α-dimethylergosta-8,25(27)-dienol, 4α,14α-dimethylergosta-8,24(28)-dienol (obtusifoliol) ergosta-7,25(27)-dienol, ergosta-8,25(27)-dienol, ergost-7-enol, porifersta-7,25(27)-dienol, porifersta-8,25(27)-dienol, and poriferst-7-enol (Tables 1 and 2). The finding of a set of C4-methyl intermediates, including cycloartenol, in the sterol composition of wild-type cells was significant (Fig. 4C) because it confirmed the “photosynthetic lineage” of sterol biosynthesis in this alga. Moreover, the natural occurrence of Δ25(27)-sterols in cells was consistent with ergosterol formation proceeding from a Δ25(27)-olefin pathway.
Fig. 3.
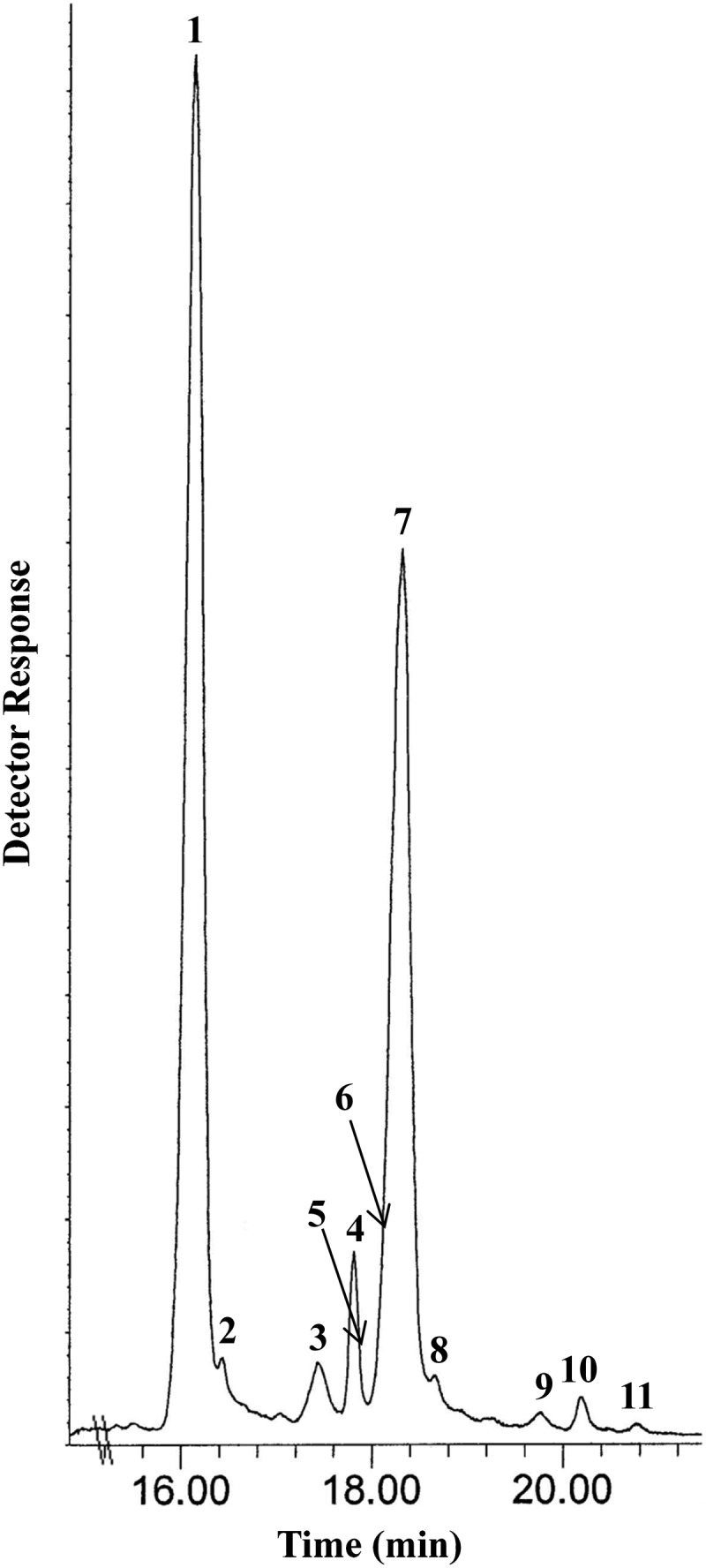
Gas-liquid chromatograph separation of sterols isolated from wild-type C. reinhardtii. Peaks in the total ion current (TIC) chromatogram correspond to peak 1, ergosta-5,7,22-trienol (ergosterol); peak 2, ergosta-8,25(27)-dienol; peak 3, ergosta-7,25(27)-dienol; peak 4, ergost-7-enol; peak 5, 4α,14α- dimethylergosta-8,25(27)-dienol; peak 6, 4α,14α-dimethylergosta-8,24(28)- dienol; peak 7, porifersta-5,7,22-trienol (7-dehydroporifersterol); peak 8, porifersta-8,25(27)-dienol; peak 9, porifersta-7,25(27)-dienol; peak 10, poriferst-7-enol; and peak 11, cycloart-24(25)-enol (cycloartenol).
Fig. 4.
Mass spectra and UV spectra (inset) for select sterols from control C. reinhardtii cells analyzed by GC-MS shown in Figure 1. A: GC peak 1. B: Peak 7. C: Peak 11.
TABLE 1.
Chromatographic and spectral properties of sterols from Chlamydomonas reinhardtii
| Systematic Name | Structurea | GC (RRTc) | UV (λmax) | MW (M ) |
| Cycloart-24(25)-enol | 1 | 1.43 | EA | 426 |
| 24β-Methyl cycloart-25(27)-enol | 2 | 1.57 | EA | 440 |
| Cycloart-24(28)-enol | 3 | 1.59 | EA | 440 |
| 4α, 14α-Dimethyl ergosta-8,25(27)-dienol | 4 | 1.23 | EA | 426 |
| 4α, 14α-Dimethyl ergosta-8,24(28)-dienol | 5 | 1.25 | EA | 426 |
| Ergosta-8,25(27)-dienol | 6 | 1.15 | EA | 398 |
| 4α-,14α-Dimethyl porifersta-8,25(27)-dienol | 7 | 1.40 | EA | 440 |
| Ergosta-7,25(27)-dienol | 8 | 1.17 | EA | 398 |
| Porifersta-8,25(27)-dienol | 9 | 1.29 | EA | 412 |
| Ergost-7-enol | 10 | 1.23 | EA | 400 |
| Porifersta-7,25(27)-dienol | 11 | 1.34 | EA | 412 |
| Ergosta-5,7-dienol | 12 | 1.2 | 282 | 398 |
| Poriferst-7-enol | 13 | 1.39 | EA | 414 |
| Ergosta-5,7,22-trienol | 14 | 1.10 | 282 | 396 |
| Poriferata-5,7-dienol | 15 | 1.35 | 282 | 412 |
| Porifersta-5,7,22-trienol | 16 | 1.28 | 282 | 410 |
| Lanosta-8,24-dienol b | 17 | 1.33 | EA | 426 |
| 4α-,14α-Dimethyl cholesta-8,24-dienol | 18 | 1.15 | EA | 412 |
| 14α-Methyl cholesta-8,24-dienol | 19 | 1.07 | EA | 398 |
| Cholesta-7,24-dienol | 20 | 1.12 | EA | 384 |
| Ergosta-5,7,25(27)-trienol | 21 | 1.18 | 282 | 396 |
| Ergosta-5,7,22,25(27)-tetraenol | 22 | 1.12 | 282 | 394 |
| Porifersta-5,7,25(27)-trienol | 23 | 1.33 | 282 | 410 |
| Porifersta-5,7,22,25(27)-tetraenol | 24 | 1.26 | 282 | 408 |
Structures of sterols are shown in Fig. 7.
Lanosterol was not detected in the cells and is given for reference purpose only.
TABLE 2.
Sterol composition of Chlamydomonas reinhardtii cells
| Sterola | WTb | KD7c | KD21c | 25-TLd | KD7PYc |
| 1 | 0.3 | 0.7 | 2.5 | 1.1 | |
| 2 | 0.3 | ||||
| 3 | 0.3 | ||||
| 4 | tre | tr | tr | 0.9 | |
| 5 | tr | tr | tr | tr | tr |
| 6 | 2.7 | ||||
| 7 | tr | ||||
| 8 | 2.6 | 14.9 | 8.1 | 21.4 | |
| 9 | 1.6 | ||||
| 10 | 3.5 | 3 | 0.5 | ||
| 11 | 0.5 | 6.4 | 10.6 | ||
| 12 | 21.3 | ||||
| 13 | 0.9 | 4.6 | |||
| 14 | 50.8 | 36.6 | |||
| 15 | 70.4 | ||||
| 16 | 37.2 | 27.5 | 1.5 | ||
| 17 | |||||
| 18 | 15.5 | ||||
| 19 | 0.5 | ||||
| 20 | 8.8 | ||||
| 21 | 22.6 | 21.5 | |||
| 22 | 19.4 | 12.7 | |||
| 23 | 1.7 | 2.9 | |||
| 24 | 35 | 26.8 |
Structures of sterols are shown in Fig. 7.
WT, wild-type cells.
Mutant cell lines.
25TL, 25-thialanosterol salt treated cells.
tr, trace amount of sterol at less than 0.3%; blank refers to no sterol detected in cells.
Induced accumulation of sterol intermediates
To generate a more robust sterol profile from which a sterol biosynthesis pathway for C. reinhardtii could be established, we next examined the sterol composition of cells engineered to produce modified sterol compositions using mutant strains generated previously by Bard et al. (24), one mutant created by us from the Bard strains, and mutants after inhibitor treatment of wild-type cells. The sterol composition of KD7 was examined first because this mutant strain was reported to accumulate six unconventional C28 and C29 Δ25(27)-sterol products, specifically a C28-7,25(27)-diene, C28-5,7,25(27)-triene, and C28-5,7,22,25(27)-tetraene and the corresponding C29-ethyl homologs (37); two of them, ergosta-7,25(27)-dienol and porifersta-7,25(27)-dienol, were detected in our analysis of the sterol composition of wild-type cells (Table 2), suggesting that the Δ25(27)-sterol pathway might be operational in C. reinhardtii. In our investigation of the sterols from KD7, we detected eight sterols, including the six reported by Bard et al. (24) and two minor C4-methyl sterols that were detected in the wild-type cells, 4α,14α-dimethylergosta-8,25(27)-dienol and obtusifoliol (Table 2). In similar fashion, we investigated the sterol composition of KD21 reported previously to synthesize C28-7-ene, C28-5,7-diene, C29-7-ene, and C29-5,7-diene sterols, and in our studies we detected the same major sterols as well as two minor sterols obtusifoliol and cycloartenol, which were detected in wild-type cells (Table 2). Using HPLC, we purified four sterols from KD7 and two sterols from KD21. The structures of these compounds were confirmed by 1HNMR (supplementary Table I). Based on spectra of reference materials (31, 37, 38), the sterols from the different cell types possessed one or more functional groups of a Δ22E double bond, a C24β-methyl/ethyl stereochemistry, and Δ25(27)-double bond in the sterol side chain. In addition, the combination of UV, MS, and 1HNMR analyses shows sterols from KD7 and KD21 to be populated by Δ7 and Δ5,7 nuclei.
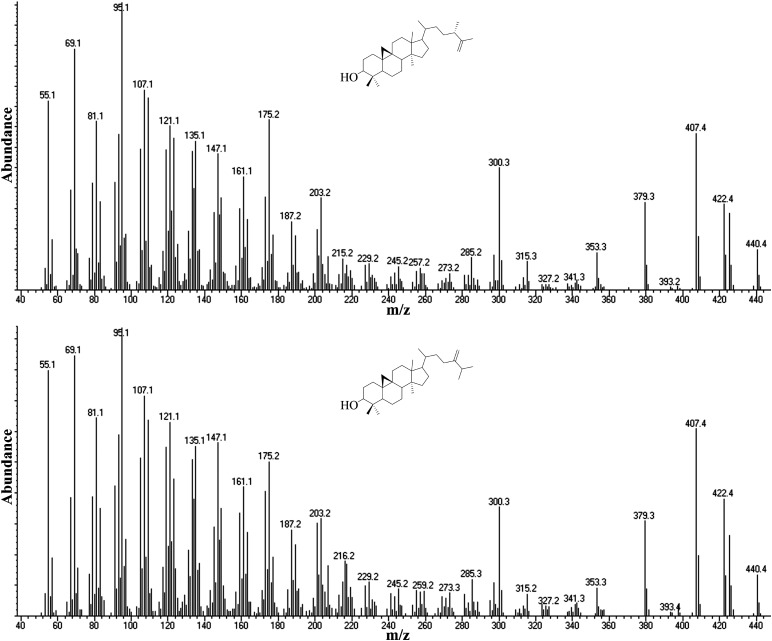
Analysis of a strain (KD7PY) derived by selection on nystatin of cells produced in a cross of KD7 and wild-type C. reinhardtii yielded 13 cometabolites, of which three were new C4-sterol intermediates not detected in wild-type, KD7, or KD21 cells. KD7PY cells accumulated trace amounts of cyclolaudenol and 24(28)-methylenecycloartanol along with minor amounts of 4α,14α-dimethylporifersta-8,25(27)-dienol (Table 2). The mass spectra of the structural isomers cyclolaudenol and 24(28)-methylenecycloartanol are almost identical to each other and, depending on the nature of the GC column, they coelute as we reported previously (13) (Fig. 5). Indeed, Bard et al. (24) also noted the coelution of Δ25(27)- and Δ24(28)-sterols in their investigation of KD7 sterols, making structure identification equivocal based on GC-MS analysis. Our use of capillary GC column allows us to separate these structural isomers such that the Δ25(27)-olefin elutes before the Δ24(28)-olefin by a retention factor Δ25(27)/Δ24(28) of approximately 0.99 (Table 1); this chromato graphic technique was developed from studies on 24-SMT action (37, 38).
Fig. 5.
Mass spectra of cyclolaudenol and 24(28)-methylenecycloartanol.
A final incubation of C. reinhardtii with 25-thialanosterol salt, designed to block sterol C24-methyltransferase activity (27, 39), led to growth inhibition after progressive addition of 25-thialanosterol to the medium (1–10 µM). At 1 µM, 10 sterols were detected by GC-MS analysis (Table 2); three of them were previously unidentified in the other cells studied and were determined to be 4α,14α-dimethylcholesta-8,24-dienol (31-norlanosterol), 14α-methylcholesta-8,24-dienol (14α-methylzymosterol), and cholesta-7,24-dienol, all sterols lacking a C24-alkyl group in the side chain.
Incorporation of [methyl-2H3]methionine
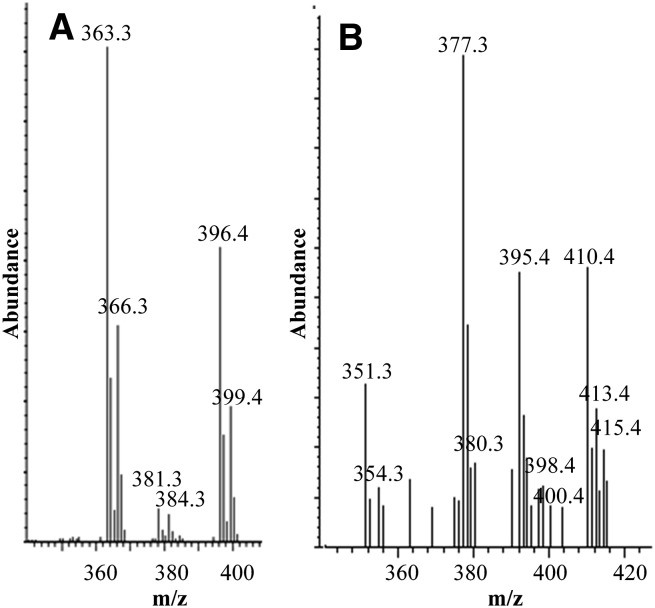
The wild-type C. reinhardtii strain contains two major sterols, ergosterol and 7-dehydroporiferasterol. The corresponding mass spectra for these 24β-alkyl sterols in the high mass end revealed ions at M+, M+-CH3, M+-H2O, and M+-CH3-H2O. Relevant ions for ergosterol appeared at m/- 396, 381, 378, and 363 amu and for 7-dehydroporiferasterol at m/z 410, 395, 392, and 377 amu (Fig. 3). When the alga was administered [methyl-2H3]methionine, three deuterium atoms were incorporated into the side chain of ergosterol (M+ m/z 399), and three or five atoms were present in the 7-dehydroporiferasterol (M+ m/z 413 and 415) (Fig. 6). These experiments with [methyl-2H3]methionine established that the C24β-methyl/ethyl group in C. reinhardtii sterols is introduced via a Δ25(27)-olefin intermediate, analogous to 24β-methyl sterols in other green algae (Chlorella and Trebouxia; 22, 40) and in the nonphotosynthetic, Chlorella-like alga Prototheca (13, 41). Our results rule out the Δ24(28)-olefin pathway used in fungal ergosterol biosynthesis (39, 42) because, in the fungal pathway, the methyl groups in 24-methyl and 24-ethyl sterols incorporate two and four deuterium atoms, respectively (Fig. 2).
Fig. 6.
Mass spectra of GC peaks corresponding to ergosterol (A) and 7-dehydroporiferasterol (B) from cells administered [methyl-2H3]methionine.
General consideration for ergosterol biosynthesis in C. reinhardtii
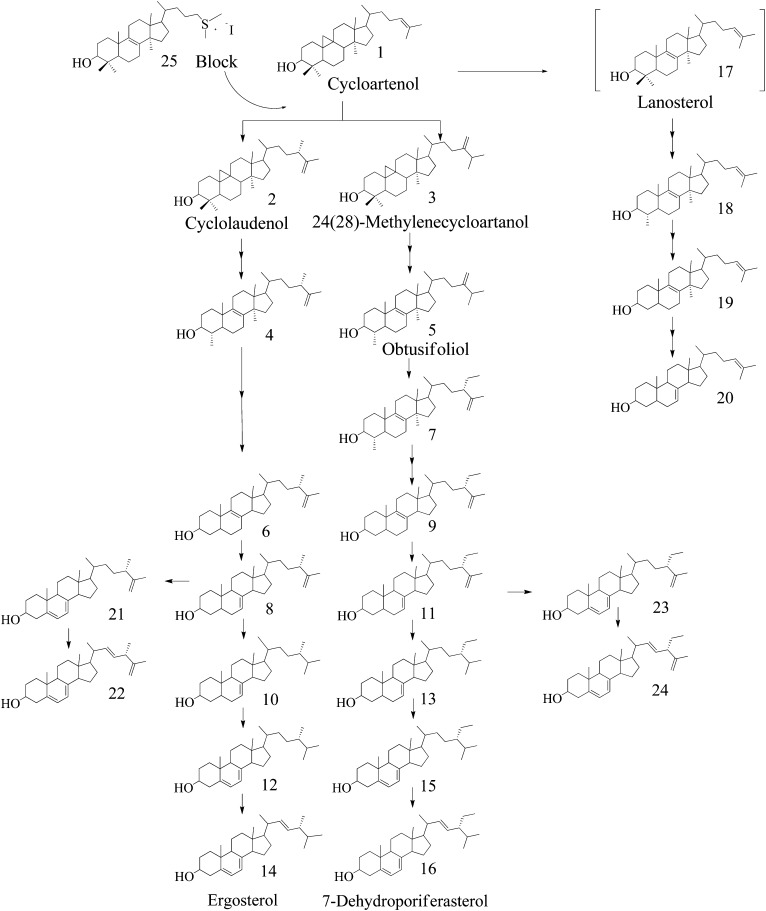
In all, 23 different sterols have been detected in C. reinhardtii vegetative cells, many of them Δ25(27)-sterols. In assembling the intermediates from the different cell types and treatments into a committed ergosterol pathway for the wild-type strain, several compounds were considered to be by-products. Thus, on the basis of our present understanding of biosynthetic relationships that posit that cycloartenol is a key branch point intermediate of phytosterol metabolism because it can serve as the Δ24(25)-protosterol precursor of Δ24(28)-sterols as well as Δ25(27)-sterols, the following pathway is suggested: cycloartenol → cyclolaudenol → ergosta-8,25(27)-dienol → ergost-7-enol → ergosta-5,7-dienol → C28 ergosterol (Fig. 7). The proposed cycloartenol-ergosterol sequence eliminates the typical intermediates in fungal ergosterol biosynthesis of zymosterol and fecosterol [ergosta-8,24(28)-dienol], which convert to ergost-7-enol through the action of a sterol 8-7 isomerase and sterol 24(28)-reductase enzyme (supplementary Fig. I1); a parallel pathway exists in the formation of C29 sterols, which requires the intermediacy of a Δ24(28)-derivative. In this case, the C1-transfer product 24(28)-methylenecycloartanol converts to obtusifoliol, which can serve as substrate for the second C1-transfer reaction catalyzed by 24-SMT to form 4α,14α-dimethylporifersta-8,25(27)-dienol. The latter sterol then converts to 7-dehydroporiferasterol (Fig. 7). Accumulation of 14α-methylzymosterol and 4α,14α-dimethylporifersta-8,25(27)-dienol, intermediates in mutant or treated cells, is consistent with complete demethylation at C4 occurring before C14-demethylation in the biosynthesis of 24-alkyl sterols in C. reinhardti. Although 24(28)-methylenelophenol is considered a substrate for Δ24(28)-ethylidene products and a branch point intermediate in the biosynthesis of 24-methyl and 24-ethyl sterols in plants (7, 43), the proposed removal of both C4 methyl groups before the elimination of the C14 methyl group rules out this compound as intermediate to 7-dehydroporiferasterol in C. reinhardtii.
Fig. 7.
Proposed sterol biosynthesis pathway from cycloartenol to ergosterol and 7-dehydroporiferasterol in C. reinhardtii; inhibition of sterol C24-methyltransferase action from incubation with 25-thialanosterol salt is shown. In brackets is shown lanosterol, which was not detected in cells but is presumed to be an intermediate to cholesta-7,24-dienol. Systematic naming of sterols are reported in Tables 1 and 2.
In defining evolutionary characters, data from several sou rces (1, 2), including natural product profiling, bioinformatic analyses, and isotopic labeling studies (15, 19), point toward the existence of a mevalonate-independent pathway to sterols in Chlorophyta, including green algae, and diatoms (44–46). Alternatively, other algal groups associated with the Streptophyta operate the acetate-mevalonate pathway to sterols, such as the Klebsormidales Spirogyra aligned with land plants (45) in addition to all fungal and animal systems (1, 40). Our work confirms that the chemistry of the C24-alkylation-reduction reactions provides an independent set of characters. Thus, the cycloartenol -Δ25(27)-olefin pathway to ergosterol/7-dehydroporiferasterol defines sterol biosynthesis in green algae, and the cycloartenol-Δ24(28)-olefin pathway to 24β-methyl/ethyl sterols or to 24α-methyl sterols defines sterol biosynthesis in brown and golden brown algae or in diatoms, respectively (2, 47). In dinoflagellates and choanflagellates, respectively, the lanosterol -Δ24(28) pathway to uncommon C30-phytosterols (gorgosterol) or to ergosterol defines the sterol pathway (18, 32, 48).
In support of the phyla-specific differences reported in module III, the cloned 24-SMT from the ascomyetous fungus Paracoccidiodes brasiliensis (49) recognizes lanosterol and cycloartenol (as expected for conformational reasons [50]) and converts them to a single Δ24(28) product. Only the lanosterol-based product, however, is further converted to ergosterol because fungi lack enzymes required to open the 9β,19-cyclopropane ring system (1, 2). Future studies in progress to determine whether the C. reinhardtii 24-SMT can catalyze lanosterol and cycloartenol to the same set of Δ25(27)- and Δ24 (28)-C1 transfer products should further illuminate the evolution of these pathways.
Although the information is incomplete, three competing phylogenies constructed from these characters reasonably describe the separate evolutions of ergosterol bio syn thesis. We surmise that in green algae the ergosterol pathway starts with (i) the mevalonate-independent methylerythritol 4-phosphate pathway to Δ3-IPP, followed by (ii) the isoprenoid pathway to cycloartenol, which becomes (iii) the Δ25(27)-sterol pathway. In contrast, the three modules acting successively in ergosterol biosynthesis in fungi are: (i) the acetate-mevalonate pathway to Δ3-IPP, (ii) the isoprenoid pathway to lanosterol, and (iii) the Δ24(28)-olefin pathway. Thus, there is considerable diversity with respect to pathways and to the formation of ergosterol, a likely result of convergent evolution in the biosynthesis of membrane inserts.
It is possible that green algae and fungi had a common ancestor and that the fungi lost the ability to make cycloartenol as a result of mutational divergence in which the cycloartenol synthase underwent “channel switching” in the catalytic reaction path to form lanosterol (1). In such a model in which the acetate-mevalonate pathway was common to algae and fungi, in module II, cycloartenol formation would precede lanosterol for thermodynamic reasons (3), and in module III, the sterol C24-methyltransferase reductases were recruited into a patchwork assembly of enzymatic reactions that recognize the same sterol template for catalysis. The presence of lanosterol synthases in some algae and land plants (36, 51–53) and the Δ25(27)-olefin pathway as the source of C30 sterols of elongated side chains extending from C27 in dinoflagellates (32) is consistent with this proposal.
In similar fashion, channel switching in sterol C24-methyl transferase activities may have been an evolutionary event that redirected the reaction path from a single product to one or more 24-alkyl(idene) product(s) that therefore is partially responsible for the chemical traits of phylogenetic significance in ergosterol of algae or fungi and sitosterol of higher plants. Although these catalysts may have evolved by duplication and functional divergence (54), the way in which the enzymatic reactions within modules and the modules themselves have been organized to yield end products remains enigmatic. A combination of the organic/enzymatic approach and amino acid sequence alignments and X-ray structures of the relevant enzymes in more organisms across Domains may lead to a better understanding of sterol evolution.
Note added in proof
The original article appeared online with three tables in text. Table 1 was subsequently moved to supplementary data online and the other tables renumbered.
Supplementary Material
Footnotes
This work was supported by the National Science Foundation Grant MCB-0929212 (W.D.N.) and by National Institutes of Health Grant GM-25661 (W.J.S.). Its contents are solely the responsibilities of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Nes W. D. 2011. Biosynthesis of cholesterol and other sterols. Chem. Rev. 111: 6423–6451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodwin T. W. 1981. Biosynthesis of plants sterols and other triterpenoids. Biosynthesis of isoprenoids, vol. I Porter J. W., Spurgeon S. L., Wiley, New York: 443–480 [Google Scholar]
- 3.Nes W. D, Nes W. R. 1980. Lipids in evolution. Plenum Press, New York, 244 [Google Scholar]
- 4.Brocks J. J., Logan G. A., Buick R., Summons R. E. 1999. Archean molecular fossils and the early rise of eukaryotes. Science. 285: 1033–1036 [DOI] [PubMed] [Google Scholar]
- 5.Love G. D., Grosjean E., Stalvies C., Fike D. A., Grotzinger J. P., Bradley A. S., Kelly A. E., Bhatia M., Meredith W., Snape C. E., et al. 2009. Fossil steroids record the appearance of demospongia during the Cryogenian period. Nature. 457: 718–721 [DOI] [PubMed] [Google Scholar]
- 6.Volkman J. K. 2005. Sterols and other triterpenoids: source specificity and evolution of biosynthetic pathways. Org. Geochem. 36: 139–159 [Google Scholar]
- 7.Benveniste P. 2004. Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 55: 429–457 [DOI] [PubMed] [Google Scholar]
- 8.Porter F. D., Herman G. E. 2011. Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 52: 6–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alcazar-fuoli L., Mellado E., Garcia-effron G., Lopez J. F., Grimalt J. O., Cuenca-estrella J. M., Rodriguez-tudela J. L. 2008. Egosterol biosynthesis pathway in Aspergillus fumigatus. Steroids. 73: 339–347 [DOI] [PubMed] [Google Scholar]
- 10.Disch A., Schwender A. J., Muller C., Lichtenthaler H. K., Rohmer M. 1988. Distribution of the mevalonate and glycerlaldehyde phosphate/pyruvate pathways for isopreniod biosynthesis in unicellular alga and the cyanobacterium Synechocystis PCC 6714. Biochem. J. 333: 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohmer M. 1999. The discovery of the mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 16: 565–574 [DOI] [PubMed] [Google Scholar]
- 12.Hunter W. N. 2007. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 282: 21573–21577 [DOI] [PubMed] [Google Scholar]
- 13.Nes W. D., Norton R. A., Crumley F. G., Madigan S. J., Katz E. R. 1990. Sterol phylogenesis and algal evolution. Proc. Natl. Acad. Sci. USA. 87: 7565–7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips D. R., Rasbery J. M., Bartell B., Matsuda S. P. T. 2006. Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9: 305–314 [DOI] [PubMed] [Google Scholar]
- 15.Nes W. R., McKean M. R., 1977. Biochemistry of steroids and other isopentenoids. University Park Press, Baltimore, MD [Google Scholar]
- 16.Mangla A. T., Nes W. D. 2000. Sterol C-methyl transferase from Prototheca wickerhamii: mechanism, sterol specificity and inhibition. Bioorg. Med. Chem. 8: 925–936 [DOI] [PubMed] [Google Scholar]
- 17.Wojciechowski Z. A., Goad L. J., Goodwin T. W. 1973. S-Adenosyl-methionine-cycloartenol methyltransferase activity in cell-free system from Trebouxia sp. and Scenedesmus obliqus. Biochem. J. 136: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giner J-L., Djerassi C. 1991. Biosynthetic studies of marine lipids. 33. Biosynthesis of dinosterol, peridinosterol and gorgosterol: Unusual pattern of bioalkylation in dinoflagellate sterols. J. Org. Chem. 56: 2357–2363 [Google Scholar]
- 19.Desmond E., Gribaldo S. 2009. Phylogenomics of sterol synthesis: Insights into the origin, evolution and diversity of a key eukaryotic feature. Genome Biol. Evol. 1: 364–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brumfield K. M., Moroney J. V., Moore T. S., Simms T. A., Donze D. 2010. Functional characterization of the Chlamydomonas reinhardtii ERG3 ortholog, a gene involved in the biosynthesis of ergosterol. PLoS ONE. 5: e8659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salimova E., Boschetti A., Eichenberger W., Lutova L. 1999. Sterol mutants of Chlamydomonas reinhardtii: characterisation of three strains deficient in C24(28)-reductase. Plant Physiol. Biochem. 37: 241–249 [Google Scholar]
- 22.Goad L. J., Lenton J. B., Knapp F. F. 1974. Phytosterol side chain biosynthesis. Lipids. 9: 582–595 [DOI] [PubMed] [Google Scholar]
- 23.McKean M. L., Nes W. R. 1977. Evidence for separate intermediates in the biosynthesis of 24α- and 24β-alkylsterols in tracheophytes. Phytochemistry. 16: 683–686 [Google Scholar]
- 24.Bard M., Wilson K. J., Thompson R. M. 1978. Isolation of sterol mutants in Chlamydomonas reinhardtii: chromatographic analysis. Lipids. 13: 533–539 [DOI] [PubMed] [Google Scholar]
- 25.Sanger R., Granick S. 1954. Nutritional control of sexuality in Chlamydomonas reinhardtii. J. Can. Microbiol. 18: 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q., Pan J., Snell W. J. 2006. Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell. 25: 549–562 [DOI] [PubMed] [Google Scholar]
- 27.Kanagasabai R., Zhou W., Liu J., Nguyen T. T., Veeramachaneni P., Nes W. D. 2004. Disruption of ergosterol biosynthesis, growth and the morphological transition in Candida albicans by sterol methyltransferase inhibitors containing sulfur at C-25 in the sterol side chain. Lipids. 39: 737–746 [DOI] [PubMed] [Google Scholar]
- 28.Xu S. H., Norton R. A., Crumley F. G., Nes W. D. 1988. Comparison of the chromatographic properties of sterols, select additional steroids and triterpenoids: gravity-flow liquid chromatography, thin-layer chromatography, gas-liquid chromatography, and high performance liquid chromatography. J. Chromatogr. A. 452: 377–398 [DOI] [PubMed] [Google Scholar]
- 29.Guo D. A., Venkatramesh M., Nes W. D. 1995. Developmental regulation of sterol biosynthesis in Zea mays. Lipids. 30: 203–219 [DOI] [PubMed] [Google Scholar]
- 30.Nes W. D., Nichols S. D. 2006. Phytosterol biosynthesis pathway in Mortierella alpina. Phytochemistry. 67: 1716–1721 [DOI] [PubMed] [Google Scholar]
- 31.Zhou W., Cross G. A. M., Nes W. D. 2007. Cholesterol import fails to prevent catalyst-based inhibition of ergosterol synthesis and cell proliferation in Trypanosoma brucei. J. Lipid Res. 48: 665–673 [DOI] [PubMed] [Google Scholar]
- 32.Goad L. J., Akihisa T., editors. 1997. Analysis of sterols Blackie Academic & Professional, New York [Google Scholar]
- 33.Gealt M. A., Adler J. A., Nes W. R. 1981. The sterols and fatty acids from purified flagella of Chlamydomonas reinhardtii. Lipids. 16: 133–135 [Google Scholar]
- 34.Matthew T., Zhou W., Rupprecht J., Lim L., Thomas-Hall S. R., Doebbe A., Kruse O., Hankamer B., Marx U. C., Smith S. M., et al. 2009. The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur depletion. J. Biol. Chem. 284: 23415–23425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwender J., Gemunden C., Licthentaler H. K. 2001. Chlorophyta exclusively use the 1-deoxyxylulose 5-phosphate/2-C-methylerthritol 4-phosphate pathway for the biosynthesis of isoprenoids. Planta. 212: 416–423 [DOI] [PubMed] [Google Scholar]
- 36.Giner J-L., Wunsche L., Andersen R. A., Djerassi C. 1991. Dinoflagellates cyclize squalene oxide to lanosterol. Biochem. Syst. Ecol. 19: 143–145 [Google Scholar]
- 37.Dennis A. L., Nes W. D. 2002. Sterol methyltransferase. Evidence for successive C-methyl transfer reactions generating Δ24(28)- and Δ25(27)-olefins by a single plant enzyme. Tetrahedron Lett. 43: 7017–7021 [Google Scholar]
- 38.Zhou W., Lepesheva G. I., Waterman M. R., Nes W. D. 2006. Mechanistic analysis of a multiple product sterol methyltransferase implicated in ergosterol biosynthesis in Trypanosoma brucei. J. Biol. Chem. 281: 6290–6296 [DOI] [PubMed] [Google Scholar]
- 39.Nes W. D., Zhou W., Ganapathy K., Liu J., Vatsyayan R., Chamala S., Herandez K., Miranda M. 2009. Sterol C24-methyltransferase: an enzymatic target for the disruption of ergosterol biosynthesis and homeostasis in Cryptococcus neoformans. Arch. Biochem. Biophys. 481: 210–218 [DOI] [PubMed] [Google Scholar]
- 40.Nes W. R. 1977. The biochemistry of plant sterols. Adv. Lipid Res. 15: 233–324 [Google Scholar]
- 41.Zhou W-X., Nes W. D. 2000. Stereochemistry of hydrogen introduction at C-25 in ergosterol synthesized by the mevalonate-independent pathway. Tetrahedron Lett. 41: 2791–2795 [Google Scholar]
- 42.Nes W. D., Le P. H. 1990. Evidence for separate intermediates in the biosynthesis of 24β-methyl sterol end products by Gibberella fujikuroi. Biochim. Biophys. Acta. 1042: 119–125 [Google Scholar]
- 43.Bouvier F., Rahier A., Camara B. 2005. Biogenesis, molecular recognition and function of plant isoprenoids. Prog. Lipid Res. 44: 357–429 [DOI] [PubMed] [Google Scholar]
- 44.Lombard J., Moreira D. 2011. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol. Biol. Evol. 28: 87–99 [DOI] [PubMed] [Google Scholar]
- 45.Lichtenthaler H. K. 1999. Evolution of carotenoid and isoprenoid biosynthesis in photosynthetic and non-photosynthetic organsisms. Annu. Rev. Plant Biol. 50: 47–65 [DOI] [PubMed] [Google Scholar]
- 46.Cvejic J. H., Rohmer M. 2000. CO2 as main carbon source for isoprenoid biosynthesis via the mevalonate-independent methylerthritol 4- phosphate route in the marine diatoms Phaeodactylum tricornutum and Nitschia ocalis. Phytochemistry. 53: 21–28 [DOI] [PubMed] [Google Scholar]
- 47.Rubinstein I., Goad L. G. 1974. Occurrence of (24S)-24-methylcholesta-5–22E-dien-3β-ol in the diatom Phaeodactylum tricornutum. Phytochemistry. 13: 485–487 [Google Scholar]
- 48.Kodner R. B., Summons R. E., Pearson A., King N., Knoll A. H. 2008. Sterols in a unicellular relative of the metazoans. Proc. Natl. Acad. Sci. USA. 105: 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pereira M., Song Z., Santos-Silva L. K., Richards M. H., Nguyen T. T. M., Liu J., de Almeida Soares C. M., da Silva Cruz A. H., Ganapathy K., Nes W. D. 2010. Cloning, mechanistic and functional analysis of a fungal sterol C24-methyltransferase implicated in brassicasterol biosynthesis. Biochim. Biophys. Acta. 1801: 1163–1174 [DOI] [PubMed] [Google Scholar]
- 50.Nes W. D., Koike K., Jia Z., Sakamoto Y., Satou T., Nikaido T., Griffin J. F. 1998. 9β,19-Cyclosterol analysis by 1H- and 13C-NMR, crystallographic observations and molecular mechanics calculations. J. Am. Chem. Soc. 120: 5970–5980 [Google Scholar]
- 51.Xue Z., Duan L. X., Liu D., Guo J., Dicks J., Omaille P., Osbourn A. 2012. Divergentevolution of oxidosqualene cyclases in plants. New Phytol. 193: 1022–1038 [DOI] [PubMed] [Google Scholar]
- 52.Kolesnikova M. D., Xiong Q., Lodeiro S., Hua L., Matsuda S. P. T. 2006. Lanosterol biosynthesis in plants. Arch. Biochem. Biophys. 447: 87–95 [DOI] [PubMed] [Google Scholar]
- 53.Ohyama K., Masahi S., Kikuchi J., Saito K., Muranaka T. 2009. Dual biosynthetic pathways to phytosterol via cycloartenol and lanosterol in Arabidopsis. Proc. Natl. Acad. Sci. USA. 106: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neelakanda A. K., Song Z., Wang J., Richards M. H., Wu X., Valliyodan B., Nguyen H. T., Nes W. D. 2009. Cloning, functional expression and phylogenetic analysis of plant sterol C24-methyltransferase involved in sitosterol biosynthesis. Phytochemistry. 70: 1982–1998 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.