Abstract
Scavenger receptor BI (SR-BI), an HDL receptor, plays a key role in reverse cholesterol transport. In mice, disruption of SR-BI results in hypersensitivity to lipopolysaccharide (LPS) and bacteria-induced septic shock due to adrenal insufficiency and abnormal hepatic pathogen clearance. In this study, we identify an anti-inflammatory role of macrophage SR-BI. Using bone marrow transplantation, we report an enhanced pro-inflammatory response to LPS in wild-type (WT) mice receiving SR-BI-null compared with WT bone marrow cells and a reduced response in SR-BI-null mice receiving WT compared with SR-BI-null cells. Although significant, SR-BI deficiency limited to bone marrow-derived cells promoted a relatively modest enhancement of the inflammatory response to LPS in mice compared with the effect of whole-body SR-BI deletion. Consistent with earlier findings, SR-BI-null primary macrophages exhibited a greater inflammatory cytokine response to LPS than control macro phages. In addition, we showed that overexpression of SR-BI in J774 macrophages attenuated the inflammatory response to LPS. The LPS-induced cytokine expression in both WT and SR-BI-null macrophages was dependent not only on NFκB as previously reported but also on JNK and P38 cell signaling pathways. The increased inflammatory signaling in SR-BI-null cells was not related to alterations in cellular cholesterol content. We conclude that SR-BI plays an important function in regulating the macrophage inflammatory response to LPS.
Keywords: scavenger receptor, inflammation, lipopolysaccharide
Sepsis is associated with substantial morbidity and mortality despite the use of various therapeutic approaches (1). Lipopolysaccharide (LPS), a well-characterized toxin present in gram-negative bacteria, is an important trigger of inflammation in sepsis, primarily through its interaction with the Toll-like receptor TLR4 (1, 2). LPS-induced, TLR4-mediated inflammatory signaling stimulates pro-inflammatory cytokine gene expression and cytokine production as part of the host defense against pathogens (3, 4). However, uncontrolled systemic inflammation causes serious systemic complications, such as tissue damage and endotoxic shock (1, 5).
Class B scavenger receptor BI (SR-BI), an HDL receptor, has been shown to mediate the cellular uptake of certain bacteria and may therefore play an important role in innate immunity (6). SR-BI mediates the binding and uptake of both LPS and lipoteichoic acid (LTA), cell wall components of gram-negative and gram-positive bacteria, respectively (6, 7). However, the physiological significance of this is unclear. Evidence has suggested that SR-BI facilitates bacterial invasion and proliferation in macrophages in a manner by which bacteria evade lysosomal processing (8). As a result, SR-BI might facilitate bacterial infection and sepsis. However, we and others reported that compared with wild-type (WT) mice, SR-BI-null mice are more susceptible to endotoxin-induced inflammation and death (9, 10). This was attributed to the protective effects of SR-BI in maintaining adrenal corticosteroid production, promoting LPS clearance by the liver and inhibiting nitric oxide (NO)-induced cell toxicity (9, 10). In line with these findings, pro-inflammatory cytokine production was markedly enhanced in SR-BI-null mice (9) Li et al. showed that SR-BI protected against septic death in mice and inhibited the inflammatory response to LPS in macrophages (11).
SR-BI is expressed in a variety of cell types, most abundantly in steroidogenic cells and in the liver (12). SR-BI is also expressed in macrophages (13); however, its function in macrophages and the impact of macrophage SR-BI on the integrated inflammatory response is not yet clear. SR-BI has been recognized as a mediator of bidirectional cholesterol flux between macrophages and extracellular acceptors (12, 14). When fed a Western diet, mice lacking both SR-BI and apoE showed enhanced macrophage cholesterol accumulation (15). The dysregulation of cholesterol homeostasis in these cells was attributed to a disruption of macrophage cholesterol efflux and lysosomal cholesterol trafficking (15). Recent studies from our laboratory revealed that SR-BI can promote macrophage cholesterol efflux to HDL (16). Deletion of SR-BI in bone marrow cells was shown to promote atherosclerosis in mice, at least during later-stage lesion development (17–19).
Macrophage inflammatory responses have been shown to be modulated by cellular membrane cholesterol levels. In ABCA1- and ABCG1-deficient macrophages, the increased cell membrane cholesterol content resulting from reduced cellular cholesterol efflux contributes to the hyperinflammatory responses in these cells in response to stimuli (20, 21). SR-BI is known to affect the content and distribution of cholesterol in plasma membranes, but the possibility that SR-BI modulates macrophage inflammatory responses by such an alteration remains to be explored.
In this study, we investigated the role of macrophage SR-BI in the LPS-stimulated inflammatory response. We present our findings that SR-BI expression in macrophages decreases the pro-inflammatory cytokine response to LPS in both mice and isolated macrophages. The increased cytokine expression in SR-BI-null cells involved both NFκB as well as JNK and P38 cell signaling pathways. The hyperinflammatory response in SR-BI-null macrophages is not associated with alterations in cellular cholesterol content.
MATERIALS AND METHODS
Animals and reagents
Mice were housed in the Veterans Affairs Medical Center (Lexington, KY), and all animal protocols received appropriate institutional approval (Animal Welfare Assurance Number, Veterans Affairs Medical Center, A3506-01; VMU IACUC protocols 2009-0005V, 2009-0006V). Animals were maintained in a pathogen-free facility with 12 h light/dark cycle and free access to food and water. SR-BI-null mice were obtained from Jackson Laboratory (Monty Krieger, MIT) (22) and backcrossed for 10 generations into a C57BL/6 background (MacRae F. Linton, Vanderbilt University) (15). ELISA sets for cytokine determination were purchased from BD Biosciences (San Jose, CA). Antibodies were purchased from Cell Signaling (Danvers, MA) unless otherwise indicated. The MAPK inhibitors PD98059, SB203580, and SP600125 as well as LPS and LTA were obtained from Sigma-Aldrich (St. Louis, MO). The NFκB inhibitor 1-pyrrolidinecarbodithioic acid (PDTC) was obtained from Calbiochem (San Diego, CA). J774 cells stably transfected with mouse SR-BI and control J774 cells were kindly provided by Dr. Theodore Mazzone (NorthShore University, Chicago, IL). The J774-SR-BI cells were transfected to stably express full-length murine SR-BI in pcDNA 3.1 zeo as previously described in detail (23). Clones were selected with zeocin, and cell viability was not altered by the expression of SR-BI.
Bone marrow transplantation
The procedure was performed as previously reported (24). Six- to 8-week-old SR-BI+/+ (WT) and SR-BI−/− (SR-BI-null) recipient mice were maintained on water containing an antibiotic (sulfratrim, 4 μg/ml) for 1 week prior to irradiation. Recipient mice were irradiated with 450 rad from a cesium (γ radiation) source twice with 3 h between exposures. Irradiated recipient mice were repopulated by a tail vein injection of 1 × 106 bone marrow cells harvested from tibia and femurs of age-matched SR-BI−/− and SR-BI+/+ mice. Following bone marrow transplantation, mice were maintained on water containing sulfratrim (4 μg/ml) for 4 weeks and then switched to regular water for 1 week. Mice were then injected intraperitoneally with LPS (0.25 μg/g body weight for SR-BI+/+ recipients, 0.1 μg/g body weight for SR-BI−/− recipients). Mice were euthanized 2 h after LPS injection, and blood and tissues were harvested. To assess engraftment of donor hematopoietic cells, DNA was isolated from bone marrow cells of recipient mice using a DNeasy Tissue Kit (Qiagen, Valencia, CA) and analyzed by PCR for SR-BI expression.
Bone marrow macrophage isolation
Bone marrow cells were harvested from tibia and femurs of age-matched SR-BI−/− and SR-BI+/+ mice 5 days after bio-gel elicitation [injected intraperitoneally with 1 ml/mouse of polyacrylamide gel P-100 (Bio-Rad, Hercules, CA) in a 2% w/v suspension in endotoxin-free water]. Bone marrow cells were then cultured and differentiated in 12-well dishes containing RPMI-1640 supplemented with 50 IU/ml penicillin G, 50 μg/ml streptomycin, 2 mM glutamine, 10% fetal bovine serum, and 15% (v/v) L929-cell conditioned medium for 7 days.
Inflammatory response in macrophages
Primary bone marrow macrophages (BMM) were treated with LPS in the presence or absence of specific inhibitors of cell signaling pathways in RPMI medium supplemented with 5% fetal bovine serum. These inhibitors included MAPK inhibitors SP600125 (JNK, 20 μM), SB203580 (P38, 20 μM), and PD98059 (ERK, 25 μM), and NFκB inhibitor PDTC (50 μM). BMMs were incubated overnight in serum-free RPMI, pretreated with the indicated inhibitor for 1 h, and then treated with the same conditions plus LPS for 4 h at 37°C. Media were collected for cytokine analysis by ELISA (Invitrogen, Carlsbad, CA), and cellular protein was harvested in lysis buffer containing protease inhibitor (Roche Diagnostics, Indianapolis, IN) and phosphatase inhibitor (Thermo Scientific, Rockford, IL) cocktails.
Western blotting of solubilized macrophage protein
Total cellular protein was harvested from BMMs using Ripa buffer (Sigma, St. Louis, MO) supplemented with protease inhibitors (Roche Diagnostics, Indianapolis, IN) and a phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL). Cell protein (15 μg/well) was separated in 4–16% SDS polyacrylamide gels and transferred to PVDF membranes. The blots were incubated overnight at 4°C with specific antibodies: rabbit anti-mouse SR-BI (Novus, Littleton, CO); rabbit anti-mouse P42/44 ERK or ERK; rabbit anti-mouse P-JNK or JNK; and rabbit anti-mouse P-P38 or P38. Following 1 hr incubation with the corresponding second antibody (horseradish peroxidase-linked anti-rabbit IgG), immunoblots were briefly treated with a chemiluminescent reagent (Thermo Scientific, Rockford, IL) and exposed to Kodak film. Gel bands were quantified using KODAK ID Image Analysis software.
Macrophage cholesterol depletion and repletion
Cholesterol depletion and repletion of macrophages was performed as previously described (20, 25). Briefly, BMMs were incubated with or without 10 mM methyl-β-cyclodextrin (MβCD) in prewarmed Hanks buffered salt solution (HBSS) supplemented with calcium and magnesium at 37°C for 30 min. To reload BMMs with cholesterol, cholesterol-depleted cells were washed with Hanks buffer and incubated in the presence of cholesterol (80 μg/ml) complexed to MβCD (1.5 mM) at 37°C for 1 h. Following treatment with 100 ng/ml LPS for 4 h, BMMs were treated with hexane/isopropanol to extract lipids, and cellular cholesterol levels were determined by commercially available kits (Wako Chemicals, Richmond, VA). BMM membrane lipid distribution was analyzed using CtxB staining (20). CtxB staining was performed using commercially available kits (Invitrogen, Carlsbad, CA) (20). Fluorescent signals were visualized on an Olympus BX51 microscope, and images were captured with equal exposure time. The intensity of fluorescent signal in BMMs was quantified by ImageJ public domain software (Wayne Rasband, National Institutes of Health, Bethesda, MD) (26). Individual cells were analyzed from five random fields.
Quantitative real-time PCR
Total RNA was isolated from mouse adipose tissue and liver using the standard TRIzol method (Invitrogen, Calsbad, CA) and was purified with DNase I (Roche) and RNeasy Mini Kits (Qiagen, Valencia, CA). Two micrograms of RNA was reverse-transcribed into cDNA using a reverse-transcription system (Promega, Madison, WI). After a 4-fold dilution, 5 μl was used as a template for Q-PCR. Amplification was done for 40 cycles using a Power SYBR Green PCR Master Mix Kit (Applied Biosystems, Carlsbad, CA) and DNA Engine Optical 2 System (MJ Research Inc., Ramsey, MN). Both an internal control (GAPDH) and a negative control (minus reverse transcriptase) were included in each analysis. Values of each RNA sample were the average of duplicate assays normalized to GAPDH RNA levels.
125I radionuclide labeling of LPS and BMM LPS uptake
LPS was iodinated by the chloramine T method as described previously (9). In brief, 1 mg of LPS was dissolved in 0.05 M borate buffer (pH = 8) and incubated with 50 mM p-OH-methylbenzimidate at 37°C for 18 h. After extensive dialysis, the M-LPS was radiolabeled with Na125I by the chloramine T method. After iodination, unincorporated Na125I was removed by extensive dialysis in sterile saline at 4°C. The bioactivity of labeled LPS was determined using a Limulus assay (LAL) kit. Nearly identical bioactivity was observed between unlabeled LPS and 125I-LPS. LPS uptake into BMMs was performed as described previously (9). In brief, BMMs were washed with phosphate-buffered saline (PBS) and incubated at 37°C for 30 min or 4°C for 2 h with 125I-LPS in RPMI medium supplemented with 5% fetal bovine serum. Following incubation, medium was removed and the cells were washed four times with PBS. The cells were then solubilized in 0.1 N NaOH for 1 h at room temperature, and the protein content and radioactivity of the lysates were determined.
Statistical analysis
Statistical significance in experiments comparing only two groups was determined by 2-tailed Student t-test. The significance of the difference in mean values between more than two groups was evaluated by one-way ANOVA, followed by post hoc analysis using Tukey's test. All significant differences (P < 0.05 or P < 0.01) are given in the figures. Statistical analyses were carried out using GraphPad Prism 4 (GraphPad Software, CA). Values are expressed as mean ± SD. A P value of less than 0.05 was considered significant.
RESULTS
Effect of SR-BI expression in transplanted bone marrow cells on the inflammatory response to LPS in mice
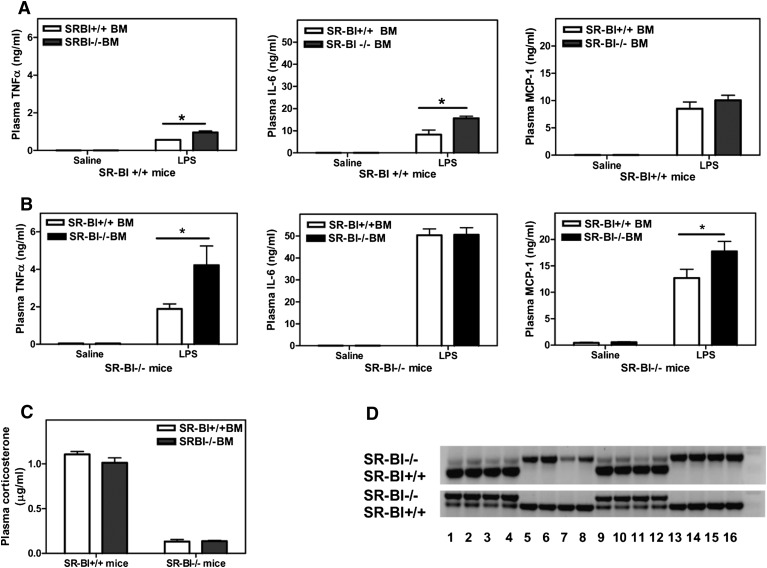
Our recent studies showed that mice lacking SR-BI are highly sensitive to LPS-induced toxic shock. The protective effect of SR-BI was largely due to its role in maintaining adrenal corticosteroid production and facilitating hepatic LPS clearance. To determine the possible role of macrophage-specific SR-BI in the inflammatory response, we investigated the response to LPS in mice transplanted with either SR-BI-null or WT bone marrow cells (Fig. 1).Given that SR-BI-null mice are hypersensitive to LPS (9), a lower dose of LPS was given to SR-BI-null mice (0.1 μg/g body weight) than to WT mice (0.25 μg/g body weight). Even at the relatively lower LPS dose, SR-BI-null mice (Fig. 1B) demonstrated significantly elevated pro-inflammatory cytokine secretion compared with the WT mice (Fig. 1A). However, the inflammatory response was also affected by SR-BI expression in transplanted bone marrow cells. As shown in Fig. 1A, WT mice that received SR-BI-null cells had significantly elevated tumor necrosis factor (TNF)α and interleukin (IL)-6 levels following LPS treatment compared with mice that received WT cells. This indicated that SR-BI expressed in macrophages reduced the pro-inflammatory response to LPS. In line with these findings, SR-BI-null mice that received WT cells showed decreased levels of pro-inflammatory cytokines and monocyte chemotactic protein-1 (MCP-1) in response to LPS compared with mice that received SR-BI-null cells (Fig. 1B).
Fig. 1.
Inflammatory response to LPS following bone marrow transplantation. Bone marrow cells from male SR-BI+/+ or SR-BI−/− mice were transfused into irradiated male (A) SR-BI+/+ and (B) SR-BI−/− mice through tail-vein injection. After 4 weeks of recovery on antibiotics, recipient SR-BI−/− and SR-BI+/+ mice were injected intraperitoneally with 0.25 μg/g body weight (A) or 0.1 μg/g body weight (B) of LPS. Mice were euthanized after 2 h. Plasma cytokine levels were determined by ELISA. (C) Plasma corticosterone levels were determined by RIA kits. Values shown are the mean ± SD, n = 5 mice/group. (D) Bone marrow cell DNA was analyzed for the presence of SR-BI+/+ and SR-BI−/− alleles by PCR. (Top panel) Lanes 1–4 and 9–12 SR-BI −/− mice receiving SR-BI +/+ bone marrow cells; lanes 5–8 and 13–16 SR-BI −/− mice receiving SR-BI−/− cells. (Bottom panel) Lanes 1,2,3,4, 9,10,11,12 SR-BI+/+ mice receiving SR-BI −/− bone marrow cells; lanes 5–8 and 13–16 SR-BI +/+ mice receiving SR-BI +/+ cells. (Upper band) Gene product from SR-BI −/− genotype. (Lower band) Gene product in SR-BI +/+ genotype. *P < 0.05, WT versus SR-BI KO.
Although the differences observed between mice transplanted with WT and SR-BI-null bone marrow are smaller than the differences observed between LPS-treated WT and SR-BI-null mice (about 3- to 4-fold) (9), these results confirm that SR-BI expressed in bone marrow-derived cells makes a significant contribution to the general inflammatory response. The differences between mice are independent of adrenal function because mice receiving bone marrow cells of different genotypes showed similar plasma corticosterone levels during inflammation (Fig. 1C). Successful bone marrow cell repopulation was achieved in mice receiving either WT or SR-BI-null bone marrow cells, such that expression of SR-BI in the bone marrow cells of the transplanted mice predominantly reflected the genotype of the donor bone marrow cells (Fig. 1D). As shown in Table 1, bone marrow transplantation did not alter plasma lipid levels. Therefore, the different inflammatory responses seen in the mice receiving different genotypes of bone marrow cells was likely not related to alterations in plasma lipids.
TABLE 1.
Plasma lipids in SR-BI+/+ and SR-BI−/− mice transplanted with SR-BI+/+ or SR-BI−/− bone marrow cells
| Plasma lipids (mg/dl) in SR-BI+/+ mice (recipients) | ||||
| Donor | SR-BI+/+ C | SR-BI−/− C | SR-BI+/+ LPS | SR-BI−/− LPS |
| TC | 82.9 ± 7.6 | 83.5 ± 5.5 | 79.7 ± 12.9 | 79.6 ± 8.7 |
| FC | 12.5 ± 1.7 | 12.7 ± 1.1 | 13.1 ± 3.7 | 14.4 ± 4.1 |
| PL | 268.2 ± 30.2 | 271.8 ± 21.7 | 255.3 ± 32.7 | 250.7 ± 31.2 |
| TG | 47.8 ± 8.6 | 49.6 ± 10.4 | 90.7 ± 29.7 | 88.2 ± 15.8 |
| Plasma lipids (mg/dl) in SR-BI−/− mice (recipients) | ||||
| TC | 269.9 ± 19.0 | 252.4 ± 25.3 | 176.0 ± 8.0 | 197.8 ± 15.4 |
| FC | 131.6 ± 5.6 | 114.8 ± 12.7 | 85.7 ± 5.1 | 73.2 ± 8.7 |
| PL | 609 ± 19.9 | 619.3 ± 32.9 | 376.0 ± 13.1 | 423.7 ± 32.6 |
| TG | 159.7 ± 8.6 | 111.5 ± 13.3 | 78.0 ± 13.5 | 86.5 ± 15.8 |
TC, total cholesterol; FC, free cholesterol; PL, phospholipid; TG, triglyceride.
Hyperinflammatory response in macrophages isolated from SR-BI-null mice and reduced response in macrophages overexpressing SR-BI
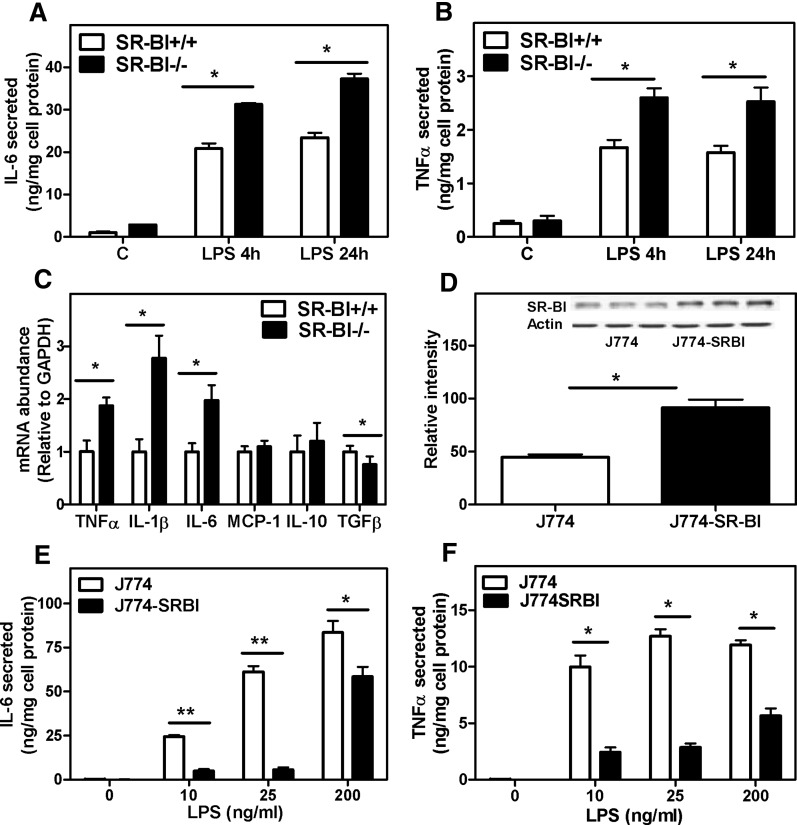
To further examine how the presence of SR-BI in macrophages influences the macrophage inflammatory response, primary BMMs isolated from SR-BI-null and WT mice were challenged with LPS. BMMs lacking SR-BI produced elevated levels of the pro-inflammatory cytokines TNFα and IL-6 compared with SR-BI-expressing BMMs (Fig. 2A, B). Consistent with cytokine protein expression, inflammatory cytokine gene expression was also markedly increased in SR-BI-null BMMs in response to LPS, confirming an earlier report (11) (Fig. 2C). Interestingly, the anti-inflammatory gene TGFβ, typically associated with the M2 response, was slightly but significantly downregulated in SR-BI-null BMMs.
Fig. 2.
Enhanced inflammatory responses in SR-BI−/− BMMs. BMMs were harvested and differentiated as described in Materials and Methods. (A, B) BMMs were incubated with LPS (20 ng/ml) in 5% FBS-containing medium for 24 h. Medium was harvested at 4 h and 24 h. Cytokines in the medium were determined by ELISA kits. (C) BMMs were treated with LPS (20 ng/ml) for 1 h in 5% FBS-containing medium, and gene expression was determined by Q-PCR. Relative values to GAPDH are shown as the mean ± SD of triplicate determinations. (D) SR-BI expression in J774 cells and stably transfected J774-SR-BI overexpressing cells. Cellular protein was harvested and SR-BI expression was determined by Western blotting. SR-BI-specific bands were quantified by densitometry and normalized to the actin value. (E, F) Cytokine secretion. Cells were treated with LPS at the doses indicated for 4 h. Medium was harvested, and cytokine was secretion determined by ELISA. Values shown are mean ± SD of triplicate determinations. *P < 0.05, **P < 0.01, WT versus SR-BI KO.
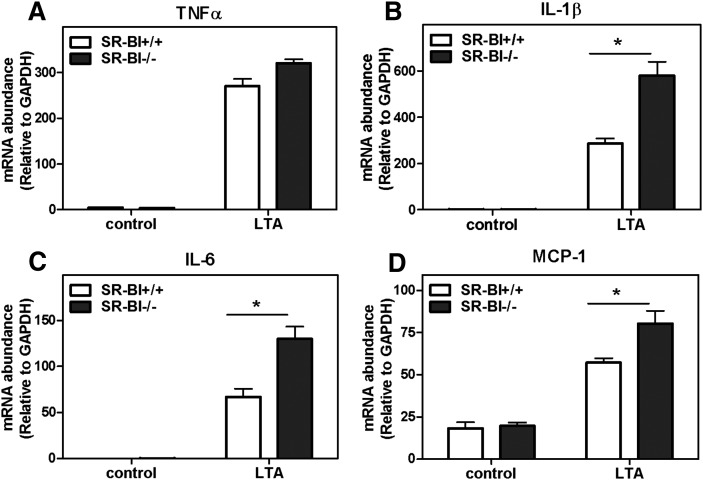
To substantiate the anti-inflammatory function of SR-BI observed in BMMs, the inflammatory response to LPS was determined in J774 macrophages stably overexpressing SR-BI. Stable transfection with an SR-BI-expressing vector resulted in approximately 2-fold increase in SR-BI expression in J774 cells (Fig. 2D). In comparison to the parent J774 cells, the SR-BI-overexpressing cells (J774-SR-BI) showed a markedly reduced secretion of the pro-inflammatory cytokines IL-6 and TNFα in response to LPS (Fig. 2E, F). Together, these results provide strong evidence that SR-BI expression modulates the inflammatory response in macrophages. To examine whether the effect of SR-BI on inflammatory signaling is restricted to LPS-induced signaling, we examined the inflammatory response to LTA, a ligand that induces cell signaling through its interaction with TLR2. As shown in Fig. 3, LTA induced TNFα, IL-6, IL-1β, and MCP-1 gene expression in WT BMMs and to an even greater extent in SR-BI-null BMMs, indicating that the anti-inflammatory role of SR-BI is not restricted to LPS-induced inflammation.
Fig. 3.
Enhanced inflammatory responses in SR-BI−/− BMM in response to LTA. BMMs were harvested and differentiated as described in Materials and Methods. BMMs were incubated with LTA (1 μg/ml) in 5% FBS-containing medium for 4 h. Cells were harvested, and RNA was extracted with Trizol-RT. Target gene expression was determined by Q-PCR. Relative values to GAPDH are shown as the mean ± SD of quadruplicate determinations. *P < 0.05, **P < 0.01. (A) TNFα. (B) IL-1β. (C) IL-6. (D) MCP-1.
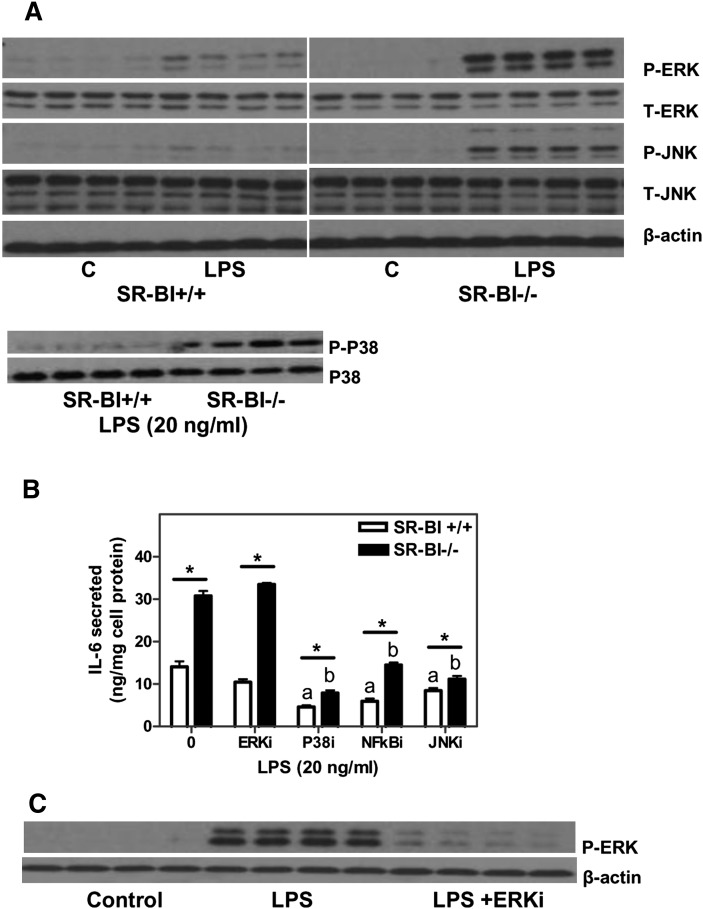
The release of pro-inflammatory cytokines such as TNFα and IL-1β in response to LPS is mediated by activation of the NFκB and MAP kinase pathways, and the hyperinflammatory response in SR-BI-null macrophages was shown to involve increased NFκB activation (11). We examined the ERK, P-38, and cJun-JNK signaling pathways. As shown in Fig. 4A, ERK and cJun-JNK were activated in BMMs exposed to LPS, as shown by increased phosphorylation. SR-BI-null BMMs demonstrated increased activation of ERK, P-38, and JNK signaling compared with WT control BMMs. The baseline levels of ERK and JNK activation in the absence of LPS were not significantly different between the two genotypes. Baseline levels of P-38 activation were also similar in the two genotypes as observed in similar experiments (data not shown). To investigate the contribution of these pathways to enhanced cytokine production in SR-BI-null cells, specific inhibitors were employed. As demonstrated in Fig. 4B, inhibition of ERK activity did not alter the pro-inflammatory response in either WT or SR-BI-null BMMs as measured by IL-6 secretion. Successful inhibition of ERK activation in response to LPS by the specific inhibitor was confirmed by Western blotting (Fig. 4C). In contrast, inhibition of JNK, P-38, or NFκB activity significantly reduced IL-6 production in both WT and SR-BI-null cells. Inhibition of P-38 or JNK signaling also partially reduced the difference between the two cell genotypes. These results indicated that both JNK and P-38 signaling pathways contribute to the hyperinflammatory response observed in SR-BI null BMMs but do not fully account for the differences in inflammatory response between the two genotypes.
Fig. 4.
Pro-inflammatory signaling in SR-BI+/+ and SR-BI −/− BMMs. (A) Pro-inflammatory signaling in differentiated BMMs. BMMs were incubated with LPS (20 ng/ml) for 1 h. Cellular protein was harvested and proteins [total (T) or phosphorylated (P) protein] were analyzed by Western blotting using commercially available antibodies. (B) Inhibition of inflammatory signaling pathways using specific inhibitors. BMMs were pretreated with inhibitors as indicated for 1 h at 37°C. Cells were then treated with LPS (20 ng/ml) for 1 h at 37°C. (C) Pro-inflammatory cytokine production in macrophages. Macrophages were pretreated with pathway inhibitors for 1 h at 37°C followed by LPS treatment (20 ng/ml) for 1 h. Medium was collected for cytokine determination by ELISA. Values shown are the mean ± SD of triplicate determinations. *P < 0.05. aSignificantly different from SR-BI+/+ LPS, P < 0.05. bSignificantly different from SR-BI−/− LPS, P < 0.01.
Cellular cholesterol content and LPS uptake in SR-BI-null macrophages
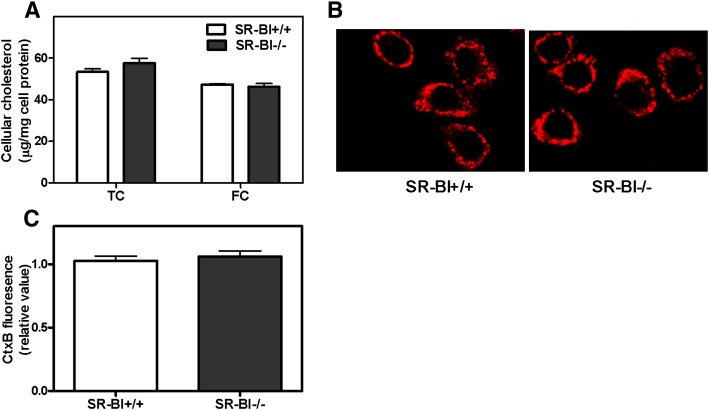
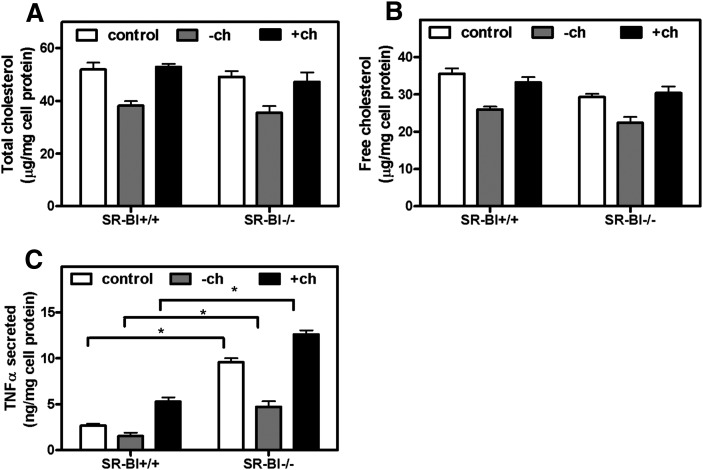
Recent studies have shown that altered membrane cholesterol content affects the inflammatory response to LPS in ABCA1-null macrophages (20, 21). To determine whether the enhanced inflammatory response in SR-BI-null BMMs is associated with altered cellular cholesterol content, we examined cellular cholesterol levels in WT control and SR-BI-null cells. As shown in Fig. 5, no difference was found in either cellular free cholesterol content or the level of cholesterol-rich lipid rafts in the plasma membrane between control and SR-BI-null BMMs (Fig. 5A–C). Plasma membrane cholesterol-rich lipid raft content was assessed by the cell association of Alexa-568-labeled cholera toxin B, which binds to lipid raft-associated lipid gangliosides (20). Neither depletion nor subsequent replenishment of cellular cholesterol with methyl-β-cyclodextrin normalized the hyperinflammatory response to LPS in SR-BI-null BMMs, as assessed by TNFα secretion (Fig. 6A–C). As expected from earlier studies (27), cholesterol depletion reduced TNFα expression, and cholesterol replenishment increased TNFα expression. However, the difference between control and SR-BI-null cells was maintained in each condition (Fig. 6A–C). These results suggest that the hyperinflammatory response observed in SR-BI-null BMMs was not due to altered cellular or membrane cholesterol content and point to a difference in the mechanism by which ABCA1 and SR-BI modulate LPS-induced inflammatory signaling.
Fig. 5.
Cholesterol content in SR-BI−/− and SR-BI +/+ BMMs. (A) Cellular cholesterol content in BMMs. Primary BMMs were harvested and differentiated in RPMI media supplemented with L929 cell-conditioned medium for seven days. Lipids were extracted from these fully differentiated cells, and cellular cholesterol levels were determined by Wako kits. Values shown are the mean ± SD of quadruplicate determinations. (B) BMMs were grown on cover slips and were allowed to differentiate for seven days. Cells were fixed and stained with Alexa-568-CtxB. Membrane lipid rafts are shown by a red fluorescent signal. Pictures were taken with equal exposure time, and representative pictures are shown. (C) Quantification of CtxB fluorescence signal in BMMs. Values shown are the mean ± SD of five fields. Similar results were found in two additional experiments.
Fig. 6.
Effect of cellular cholesterol levels on the inflammatory response in BMMs. SR-BI+/+ and SR-BI−/− BMMs were incubated with HBSS (control) or HBSS containing MβCD (−ch) at 37°C for 30 min. One group of cells was then incubated for 1 h in HBSS with cholesterol-loaded MβCD to replenish cellular cholesterol (+ch). Cells were then incubated with LPS (100 ng/ml) for 1 h. Medium was collected for TNFα determination. Cellular lipid and protein were also extracted and determined. (A) Cellular total cholesterol. (B) Cellular free cholesterol. (C) TNFα secreted. Values shown are the mean ± SD of quadruplicate determinations. *P < 0.05.
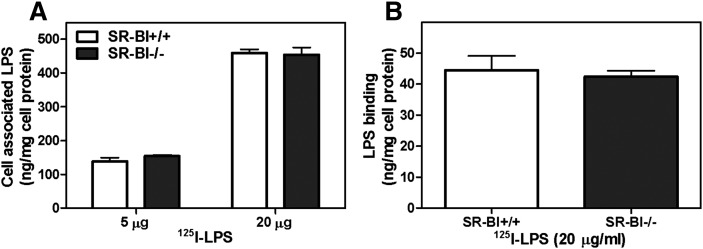
One possible mechanism by which SR-BI might exert its inhibitory effect on the response to LPS in macrophages is to bind and internalize LPS. Uptake and clearance of LPS could reduce the level of LPS available to activate TLR4 at the cell surface and therefore indirectly inhibit the inflammatory response. SR-BI is known to mediate LPS uptake into cells (7–9). However, SR-BI deficiency in BMMs did not alter LPS surface binding at 4°C or cellular uptake at 37°C (Fig. 7A, B). This indicates that the anti-inflammatory effect of macrophage SR-BI is likely independent of LPS binding and clearance.
Fig. 7.
LPS uptake in BMMs from WT and SR-BI-null mice. BMMs were isolated and differentiated as described in Materials and Methods. (A) 125I-LPS uptake in BMMs. BMMs were incubated with the indicated amount of 125I-LPS (2 μg/ml and 20 μg/ml) at 37°C for 30 min. Following washing of cells to remove the unbound LPS, cells were lysed in 0.1N NaOH, and 125I radioactivity was determined. (B) 125I -LPS bound to BMMs. Cells were incubated with 20 μg/ml of LPS at 4°C for 2 h after which cells were washed, lysed, and 125I radioactivity was determined. Values shown are the mean ± SD of triplicate determinations.
DISCUSSION
Novel findings are reported in this study examining the function of macrophage SR-BI. A bone marrow transplantation experiment showed that SR-BI expression in bone marrow-derived cells reduces the plasma inflammatory cytokine response to LPS in mice. However, the effect of SR-BI deletion in bone marrow-derived cells was substantially less than the effect of a whole-body knockout of SR-BI. We confirmed that SR-BI in cultured macrophages regulates the inflammatory cytokine response to LPS by showing that the response is affected by both SR-BI ablation as well as overexpression in cells. The LPS-induced cytokine expression in both WT and SR-BI-null cells involved JNK and P38 cell signaling pathways in addition to NFκB. Although the underlying mechanism of SR-BI modulation of the inflammatory response in macro phages is not yet fully understood, modulation was not associated with altered LPS uptake or cellular cholesterol levels.
In a previous study, we showed that both adrenal insufficiency and reduced hepatic LPS clearance contribute to the hypersensitivity to LPS in SR-BI-null mice (9). To assess the contribution of macrophage SR-BI to the general inflammatory response to LPS in mice, we performed bone marrow transplantation experiments in the current study. One advantage of this approach was that altered SR-BI expression in bone marrow-derived cells did not affect plasma lipoproteins or plasma corticosterone levels, alterations of which would exert significant effects on the general inflammatory response. Significantly elevated inflammatory cytokine levels were observed in WT mice receiving SR-BI-null bone marrow cells compared with WT cells in response to LPS, whereas reduced cytokine expression was found in SR-BI-null mice receiving WT bone marrow cells compared with SR-BI-null cells. These results demonstrate that SR-BI expression in bone marrow-derived cells attenuates inflammatory cytokine production in mice. Note that compared with the dramatic difference between SR-BI-null and WT mice in their response to LPS (9), relatively smaller differences in circulating TNFα and IL-6 levels were observed in the transplanted mice receiving SR-BI-null versus WT bone marrow.
The role of SR-BI in the LPS-induced inflammatory response was examined in primary BMMs isolated from WT and SR-BI-null mice. In agreement with a recent study (11), SR-BI-null BMMs displayed markedly enhanced pro-inflammatory cytokine production. In addition, the anti-inflammatory cytokine TGFβ was reduced in SR-BI-null BMMs. The regulatory role of SR-BI in the pro-inflammatory response was also confirmed by a gain-of-function approach in which SR-BI overexpression in J774 cells markedly reduced the pro-inflammatory response. These results provide strong evidence that SR-BI is an important modulator of the macrophage inflammatory response.
A previous study had shown that the hyperinflammatory response in SR-BI-null macrophages involved increased NFκB activation (11). In the present study, we also examined the potential involvement of the MAP kinase-signaling cascade in the SR-BI-regulated macrophage inflammatory response. We demonstrated enhanced phosphorylation of ERK, P38, and JNK signaling molecules in SR-BI-null BMMs, which implicated altered signaling through these alternative pathways in the regulation of cytokine production by SR-BI. The use of specific pathway inhibitors targeting ERK, P38, JNK, and NFκB identified JNK- and P38-mediated signaling, but not ERK, as additional key mediators responsible for enhanced LPS-induced cytokine production in macrophages that also may partially contribute to the difference in inflammatory responses between WT and SR-BI-null cells.
The mechanism(s) responsible for the enhanced inflammatory signaling in SR-BI−/− macrophages is not known. One possibility is that an interaction between SR-BI and the TLR4 complex might modulate TLR4 activation and subsequent intracellular signaling pathways. Such an interaction between TLRs and CD36, a class B scavenger receptor that is closely related to SR-BI, has been shown to regulate TLR signaling (28–30). As a coreceptor for TLR2, CD36 facilitates the TLR2-mediated NFκB activation through binding and clustering of TLR2 ligands, such as LTA (28, 29). CD36 was also shown to impact TLR4 signaling. CD36-dependent recognition of endogenous ligands is essential for stimulating the dimerization and activation of TLR4/TLR6, a complex required to initiate the innate immune activation of macrophages and microglia resulting in the activation of NFκB signaling (30). Possible interaction of TLR4 with SR-BI, shown in the current study to attenuate the inflammatory response to LPS, remains to be determined. In contrast to CD36, scavenger receptor A can attenuate TLR4-induced pro-inflammatory cytokine production by mediating rapid internalization and clearance of extracellular LPS, thus reducing LPS-induced TLR4 activation and inflammatory signaling (31). In the current study, LPS binding and cell association was not altered in SR-BI-null macrophages, suggesting that SR-BI does not modify TLR4 activation by affecting LPS clearance.
Alternatively, SR-BI may function as an independent signaling receptor capable of modulating intracellular signaling pathways upon ligand association as previously demonstrated for SR-BI expressed in nonmacrophage cell types (32, 33). For example, in endothelial cells, HDL stimulates the activation of e-NOS and Src family kinase(s) via SR-BI (34, 35). Src activation leads to the activation of phosphatidylinositol 3-kinase (PI3K), Akt kinase, and MAP kinase pathways (32, 36). In addition, interaction of HDL and SR-BI in endothelial cells inhibits the activation of NFκB and subsequent expression of the adhesion molecules vascular cell adhesion molecule 1 and intercellular adhesion molecule 1 to inflammatory stimuli (37). However, TLR4-independent SR-BI signaling in macrophages and the implication for its role in inflammation has yet to be examined.
In the current study, we addressed a third possible mechanism in which SR-BI impacts inflammatory signaling through its ability to facilitate free cholesterol flux between cells and lipoproteins and to influence the levels and distribution of plasma membrane cholesterol. Macrophage lipid content, especially the membrane cholesterol distribution, has been shown to regulate inflammatory responses in macrophages (20, 21). Free cholesterol accumulation in membranes, as observed in ABCA1 or ABCG1-null macrophages as a result of defective cholesterol efflux, stimulates the inflammatory response to LPS in a MyD88-dependent pathway (20, 21). Free cholesterol loading, especially in the endoplasmic reticulum, induces NFκB and MAPK stress signaling in response to LPS, promoting IL-6 and TNFα production (38). We therefore investigated whether macrophage SR-BI functions to regulate the inflammatory response by modulating cellular cholesterol content. However, the unaltered cellular cholesterol and membrane cholesterol-rich raft content observed in SR-BI-null macrophages suggests that SR-BI, unlike ABCA1, may modulate inflammatory responses independently of changes in macrophage cholesterol content.
Our findings may shed light on understanding the mechanism of influence of SR-BI in atherosclerotic lesion formation. The protective effect of SR-BI has been mainly attributed to its role in the reverse cholesterol transport pathway (39). However, liver-specific SR-BI-null mice exhibit less atherosclerotic lesion formation than SR-BI whole-body-deficient mice (40), suggesting an extra-hepatic protective effect(s) of SR-BI. Studies of SR-BI-null mice overexpressing cholesteryl ester transfer protein have also indicated that SR-BI plays an atheroprotective role that is unrelated to its role in mediating plasma HDL cholesterol clearance (41). Despite having normalized plasma HDL levels, SR-BI-null × CETP tg mice were not protected (41) or only partially protected (42) against atherosclerotic lesion formation compared with SR-BI-null mice. Given the fact that macrophage inflammation is a key feature in the development of atherosclerosis, our findings provide evidence that SR-BI may exert a protective effect against atherosclerotic lesion formation through modulating macrophage inflammatory responses.
This study establishes that SR-BI in macrophages serves to regulate the inflammatory response to LPS, an effect that is distinct from its known protective role in the adrenals and liver. The anti-inflammatory function of SR-BI appears to be mediated through a dampening of LPS-induced signaling through the MAP kinase and NFκB pathways, resulting in reduced inflammatory cytokine production. Our results therefore strongly suggest that SR-BI may play a key role in limiting pathological tissue inflammation and the macrophage response to pro-inflammatory stimuli, such as LPS. A more defined understanding of macrophage SR-BI may contribute to improved strategies for the treatment of acute and chronic inflammatory disease, such as sepsis and atherosclerosis.
Acknowledgments
The authors thank Xuebing Wang, Yingxia Li, Xin Shi, and Susan Bridges for excellent technical support. The authors thank Dr. MacRae F. Linton and Dr. Theodore Mazzone for kindly providing SR-BI-null mice and J774-SRBI cells, respectively. We acknowledge Dr. Nancy R. Webb and Dr. Erik E. Eckhardt for helpful discussion and comments.
Footnotes
Abbreviations:
- BMM
- bone marrow macrophage
- ERK
- extracellular-signal-regulated kinase
- IL
- interleukin
- JNK
- c-Jun N-terminal kinase
- KO
- knockout
- LPS
- lipopolysaccharide
- LTA
- lipoteichoic acid
- MAP
- mitogen-activated protein
- MCP-1
- monocyte chemotactic protein-1
- NF-κB
- nuclear factor-kappaB
- SR-BI
- scavenger receptor BI
- TGFβ
- transforming growth factor beta
- TNF
- tumor necrosis factor
- WT
- wild-type
This work was supported National Institutes of Health Grant P01-HL-086670-04 (to D. R. van der Westhuyzen) and VA Merit Award IO1-BX-000672 (to D. R. van der Westhuyzen). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Russell J. A. 2006. Management of sepsis. N. Engl. J. Med. 355: 1699–1713 [DOI] [PubMed] [Google Scholar]
- 2.Cohen J. 2002. The immunopathogenesis of sepsis. Nature. 420: 885–891 [DOI] [PubMed] [Google Scholar]
- 3.Aderem A., Ulevitch R. J. 2000. Toll-like receptors in the induction of the innate immune response. Nature. 406: 782–787 [DOI] [PubMed] [Google Scholar]
- 4.Trinchieri G., Sher A. 2007. Cooperation of Toll-like receptor signals in innate immune defence. Nat. Rev. Immunol. 7: 179–190 [DOI] [PubMed] [Google Scholar]
- 5.Rittirsch D., Flierl M. A., Ward P. A. 2008. Harmful molecular mechanisms in sepsis. Nat. Rev. Immunol. 8: 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philips J. A., Rubin E. J., Perrimon N. 2005. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 309: 1251–1253 [DOI] [PubMed] [Google Scholar]
- 7.Vishnyakova T. G., Bocharov A. V., Baranova I. N., Chen Z., Remaley A. T., Csako G., Eggerman T. L., Patterson A. P. 2003. Binding and internalization of lipopolysaccharide by Cla-1, a human orthologue of rodent scavenger receptor B1. J. Biol. Chem. 278: 22771–22780 [DOI] [PubMed] [Google Scholar]
- 8.Vishnyakova T. G., Kurlander R., Bocharov A. V., Baranova I. N., Chen Z., Abu-Asab M. S., Tsokos M., Malide D., Basso F., Remaley A., et al. 2006. CLA-1 and its splicing variant CLA-2 mediate bacterial adhesion and cytosolic bacterial invasion in mammalian cells. Proc. Natl. Acad. Sci. USA. 103: 16888–16893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai L., Ji A., de Beer F. C., Tannock L. R., van der Westhuyzen D. R. 2008. SR-BI protects against endotoxemia in mice through its roles in glucocorticoid production and hepatic clearance. J. Clin. Invest. 118: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X. A., Guo L., Asmis R., Nikolova-Karakashian M., Smart E. J. 2006. Scavenger receptor BI prevents nitric oxide-induced cytotoxicity and endotoxin-induced death. Circ. Res. 98: e60–e65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo L., Song Z., Li M., Wu Q., Wang D., Feng H., Bernard P., Daugherty A., Huang B., Li X. A. 2009. Scavenger receptor BI protects against septic death through its role in modulating inflammatory response. J. Biol. Chem. 284: 19826–19834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigotti A., Miettinen H. E., Krieger M. 2003. The role of the high-density lipoprotein receptor SR-BI in the lipid metabolism of endocrine and other tissues. Endocr. Rev. 24: 357–387 [DOI] [PubMed] [Google Scholar]
- 13.Chinetti G., Gbaguidi F. G., Griglio S., Mallat Z., Antonucci M., Poulain P., Chapman J., Fruchart J. C., Tedgui A., Najib-Fruchart J., et al. 2000. CLA-1/SR-BI is expressed in atherosclerotic lesion macrophages and regulated by activators of peroxisome proliferator-activated receptors. Circulation. 101: 2411–2417 [DOI] [PubMed] [Google Scholar]
- 14.Ji Y., Jian B., Wang N., Sun Y., Moya M. L., Phillips M. C., Rothblat G. H., Swaney J. B., Tall A. R. 1997. Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J. Biol. Chem. 272: 20982–20985 [DOI] [PubMed] [Google Scholar]
- 15.Yancey P. G., Jerome W. G., Yu H., Griffin E. E., Cox B. E., Babaev V. R., Fazio S., Linton M. F. 2007. Severely altered cholesterol homeostasis in macrophages lacking apoE and SR-BI. J. Lipid Res. 48: 1140–1149 [DOI] [PubMed] [Google Scholar]
- 16.Ji A., Meyer J. M., Cai L., Akinmusire A., de Beer M. C., Webb N. R., van der Westhuyzen D. R. 2011. Scavenger receptor SR-BI in macrophage lipid metabolism. Atherosclerosis. 217: 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Covey S. D., Krieger M., Wang W., Penman M., Trigatti B. L. 2003. Scavenger receptor class B type I-mediated protection against atherosclerosis in LDL receptor-negative mice involves its expression in bone marrow-derived cells. Arterioscler. Thromb. Vasc. Biol. 23: 1589–1594 [DOI] [PubMed] [Google Scholar]
- 18.Van Eck M., Bos I. S., Hildebrand R. B., Van Rij B. T., Van Berkel T. J. 2004. Dual role for scavenger receptor class B, type I on bone marrow-derived cells in atherosclerotic lesion development. Am. J. Pathol. 165: 785–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W., Yancey P. G., Su Y. R., Babaev V. R., Zhang Y., Fazio S., Linton M. F. 2003. Inactivation of macrophage scavenger receptor class B type I promotes atherosclerotic lesion development in apolipoprotein E-deficient mice. Circulation. 108: 2258–2263 [DOI] [PubMed] [Google Scholar]
- 20.Zhu X., Lee J. Y., Timmins J. M., Brown J. M., Boudyguina E., Mulya A., Gebre A. K., Willingham M. C., Hiltbold E. M., Mishra N., et al. 2008. Increased cellular free cholesterol in macrophage-specific Abca1 knock-out mice enhances pro-inflammatory response of macrophages. J. Biol. Chem. 283: 22930–22941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yvan-Charvet L., Welch C., Pagler T. A., Ranalletta M., Lamkanfi M., Han S., Ishibashi M., Li R., Wang N., Tall A. R. 2008. Increased inflammatory gene expression in ABC transporter-deficient macrophages: free cholesterol accumulation, increased signaling via toll-like receptors, and neutrophil infiltration of atherosclerotic lesions. Circulation. 118: 1837–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigotti A., Trigatti B. L., Penman M., Rayburn H., Herz J., Krieger M. 1997. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc. Natl. Acad. Sci. USA. 94: 12610–12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Z. H., Mazzone T. 2002. ApoE-dependent sterol efflux from macrophages is modulated by scavenger receptor class B type I expression. J. Lipid Res. 43: 375–382 [PubMed] [Google Scholar]
- 24.King V. L., Szilvassy S. J., Daugherty A. 2002. Interleukin-4 deficiency decreases atherosclerotic lesion formation in a site-specific manner in female LDL receptor−/− mice. Arterioscler. Thromb. Vasc. Biol. 22: 456–461 [DOI] [PubMed] [Google Scholar]
- 25.Powers K. A., Szaszi K., Khadaroo R. G., Tawadros P. S., Marshall J. C., Kapus A., Rotstein O. D. 2006. Oxidative stress generated by hemorrhagic shock recruits Toll-like receptor 4 to the plasma membrane in macrophages. J. Exp. Med. 203: 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins T. J. 2007. ImageJ for microscopy. Biotechniques. 43: 25–30 [DOI] [PubMed] [Google Scholar]
- 27.Koseki M., Hirano K., Masuda D., Ikegami C., Tanaka M., Ota A., Sandoval J. C., Nakagawa-Toyama Y., Sato S. B., Kobayashi T., et al. 2007. Increased lipid rafts and accelerated lipopolysaccharide-induced tumor necrosis factor-alpha secretion in Abca1-deficient macrophages. J. Lipid Res. 48: 299–306 [DOI] [PubMed] [Google Scholar]
- 28.Hoebe K., Georgel P., Rutschmann S., Du X., Mudd S., Crozat K., Sovath S., Shamel L., Hartung T., Zahringer U., et al. 2005. CD36 is a sensor of diacylglycerides. Nature. 433: 523–527 [DOI] [PubMed] [Google Scholar]
- 29.Stuart L. M., Deng J., Silver J. M., Takahashi K., Tseng A. A., Hennessy E. J., Ezekowitz R. A., Moore K. J. 2005. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J. Cell Biol. 170: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart C. R., Stuart L. M., Wilkinson K., van Gils J. M., Deng J., Halle A., Rayner K. J., Boyer L., Zhong R., Frazier W. A., et al. 2010. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 11: 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay S., Varin A., Chen Y., Liu B., Tryggvason K., Gordon S. 2011. SR-A/MARCO-mediated ligand delivery enhances intracellular TLR and NLR function, but ligand scavenging from cell surface limits TLR4 response to pathogens. Blood. 117: 1319–1328 [DOI] [PubMed] [Google Scholar]
- 32.Al-Jarallah A., Trigatti B. L. 2010. A role for the scavenger receptor, class B type I in high density lipoprotein dependent activation of cellular signaling pathways. Biochim. Biophys. Acta. 1801: 1239–1248 [DOI] [PubMed] [Google Scholar]
- 33.Saddar S., Mineo C., Shaul P. W. 2010. Signaling by the high-affinity HDL receptor scavenger receptor B type I. Arterioscler. Thromb. Vasc. Biol. 30: 144–150 [DOI] [PubMed] [Google Scholar]
- 34.Yuhanna I. S., Zhu Y., Cox B. E., Hahner L. D., Osborne-Lawrence S., Lu P., Marcel Y. L., Anderson R. G., Mendelsohn M. E., Hobbs H. H., et al. 2001. High-density lipoprotein binding to scavenger receptor-BI activates endothelial nitric oxide synthase. Nat. Med. 7: 853–857 [DOI] [PubMed] [Google Scholar]
- 35.Zhu W., Saddar S., Seetharam D., Chambliss K. L., Longoria C., Silver D. L., Yuhanna I. S., Shaul P. W., Mineo C. 2008. The scavenger receptor class B type I adaptor protein PDZK1 maintains endothelial monolayer integrity. Circ. Res. 102: 480–487 [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Ahmed A. M., McFarlane N., Capone C., Boreham D. R., Truant R., Igdoura S. A., Trigatti B. L. 2007. Regulation of SR-BI-mediated selective lipid uptake in Chinese hamster ovary-derived cells by protein kinase signaling pathways. J. Lipid Res. 48: 405–416 [DOI] [PubMed] [Google Scholar]
- 37.Kimura T., Tomura H., Mogi C., Kuwabara A., Damirin A., Ishizuka T., Sekiguchi A., Ishiwara M., Im D. S., Sato K., et al. 2006. Role of scavenger receptor class B type I and sphingosine 1-phosphate receptors in high density lipoprotein-induced inhibition of adhesion molecule expression in endothelial cells. J. Biol. Chem. 281: 37457–37467 [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Schwabe R. F., DeVries-Seimon T., Yao P. M., Gerbod-Giannone M. C., Tall A. R., Davis R. J., Flavell R., Brenner D. A., Tabas I. 2005. Free cholesterol-loaded macrophages are an abundant source of tumor necrosis factor-alpha and interleukin-6: model of NF-kappaB- and map kinase-dependent inflammation in advanced atherosclerosis. J. Biol. Chem. 280: 21763–21772 [DOI] [PubMed] [Google Scholar]
- 39.Van Eck M., Pennings M., Hoekstra M., Out R., Van Berkel T. J. 2005. Scavenger receptor BI and ATP-binding cassette transporter A1 in reverse cholesterol transport and atherosclerosis. Curr. Opin. Lipidol. 16: 307–315 [DOI] [PubMed] [Google Scholar]
- 40.Huby T., Doucet C., Dachet C., Ouzilleau B., Ueda Y., Afzal V., Rubin E., Chapman M. J., Lesnik P. 2006. Knockdown expression and hepatic deficiency reveal an atheroprotective role for SR-BI in liver and peripheral tissues. J. Clin. Invest. 116: 2767–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hildebrand R. B., Lammers B., Meurs I., Korporaal S. J., De Haan W., Zhao Y., Kruijt J. K., Pratico D., Schimmel A. W., Holleboom A. G., et al. 2010. Restoration of high-density lipoprotein levels by cholesteryl ester transfer protein expression in scavenger receptor class B type I (SR-BI) knockout mice does not normalize pathologies associated with SR-BI deficiency. Arterioscler. Thromb. Vasc. Biol. 30: 1439–1445 [DOI] [PubMed] [Google Scholar]
- 42.El Bouhassani M., Gilibert S., Moreau M., Saint-Charles F., Treguier M., Poti F., Chapman M. J., Le Goff W., Lesnik P., Huby T. 2011. Cholesteryl ester transfer protein expression partially attenuates the adverse effects of SR-BI receptor deficiency on cholesterol metabolism and atherosclerosis. J. Biol. Chem. 286: 17227–17238 [DOI] [PMC free article] [PubMed] [Google Scholar]