Abstract
Omega-3-PUFAs, eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA), are associated with prevention of various aspects of metabolic syndrome. In the present studies, the effects of oil rich in EPA on gene expression and activation of nuclear receptors was examined and compared with other ω3-PUFAs. The EPA-rich oil (EO) altered the expression of FA metabolism genes in THP-1 cells, including stearoyl CoA desaturase (SCD) and FA desaturase-1 and -2 (FASDS1 and -2). Other ω3-PUFAs resulted in a similar gene expression response for a subset of genes involved in lipid metabolism and inflammation. In reporter assays, EO activated human peroxisome proliferator-activated receptor α (PPARα) and PPARβ/γ with minimal effects on PPARγ, liver X receptor, retinoid X receptor, farnesoid X receptor, and retinoid acid receptor γ (RARγ); these effects were similar to that observed for purified EPA. When serum from a 6 week clinical intervention with dietary supplements containing olive oil (control), DHA, or two levels of EPA were applied to THP-1 cells, the expression of SCD and FADS2 decreased in the cells treated with serum from the ω3-PUFA-supplemented individuals. Taken together, these studies indicate regulation of gene expression by EO that is consistent with treating aspects of dyslipidemia and inflammation.
Keywords: omega-3 polyunsaturated fatty acid, steroyl coenzyme A desaturase, fatty acid desaturase, peroxisome proliferator-activated receptor, nuclear receptor
Cardiovascular disease (CVD) is the leading cause of death in industrialized countries and is of rising concern worldwide. The relationship between CVD and diet has been studied for nearly 100 years, essentially since the first observation of high-fat and high-cholesterol diets producing atherosclerosis in rabbits (1, 2). Diets rich in ω3-PUFAs such as α linolenic acid (ALA), eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA) are associated with decreased incidence and severity of several chronic diseases, including CVD and cancer. At least some of the beneficial effects of these dietary FAs are via metabolites such as prostaglandins, leukotrienes, thromboxanes, and resolvins. The effects of ω3-PUFAs are in contrast with those of FAs that differ mainly in position of the double bonds in the acyl chain, such as linoleic acid and arachidonic acid (AA) (ω6-PUFAs) and their corresponding metabolites. PUFAs in general are important for maintaining membrane integrity and as precursors to bioactive prostaglandins that regulate inflammation, blood clotting, and lipid metabolism. Thus, it is necessary to have diets sufficient in PUFAs (ω3 and ω6) to maintain a variety of biological processes. Positive effects of diets high in ω-3 FAs include reducing abdominal fat, preventing cardiac arrhythmia, lowering serum triacylglycerol levels, decreasing thrombosis, and improving endothelial function. As noted in Hu and Willett (3), several studies have shown association of fish intake and/or flaxseed oil (high in ALA) with decreased fatalities from CVD. Importantly, blood levels of EPA and DHA are strongly associated with decreased risk of death, myocardial infarction, and stroke (3).
In addition to playing a major role in CVD, chronic inflammation is a contributor to many human diseases. Omega-3 PUFAs play an important role in the regulation of inflammation by decreasing the production of inflammatory eicosanoids, cytokines, and reactive oxygen species and the expression of adhesion molecules (4, 5). EPA and DHA supplementation has proven effective in decreasing intestinal damage and improving gut histology in inflammatory bowel disease (6, 7). Fish oil supplementation decreases joint pain, number of tender and swollen joints, duration of morning stiffness and, as a result, use of nonsteroidal anti-inflammatory drugs (8).
Proteins of the nuclear receptor (NR) superfamily act as intracellular transcription factors that directly regulate gene expression in response to lipophilic molecules (9–12). They affect a wide variety of functions, including FA metabolism, inflammatory responses, cancer, reproductive development, and detoxification of foreign substances. Several NRs respond to dietary lipids and include the FA receptors peroxisome proliferator-activated receptors (PPARs), liver X receptors (LXRs), retinoid X receptors (RXRs), and farnesoid X receptors (FXRs). This particular subset of proteins may be considered constituents of a large group of NRs, the “metabolic nuclear receptors” that act as overall sensors of metabolic intermediates, xenobiotics, and compounds in the diet and allow cells to respond to environmental changes by inducing the appropriate metabolic genes and pathways (9, 12). The goals of the present study were to use multiple approaches to examine the effects of ω-3 PUFAs on gene expression in macrophages and to extrapolate these findings to a clinical study examining a dietary supplement enriched in EPA. Altered gene expression by several ω3-PUFAs was examined in a commonly employed human macrophage cell line (differentiated THP-1 cells), followed by an inflammatory challenge [lipopolysaccharide (LPS) stimulation]. In addition, nuclear receptor activity profiling was employed to compare potential molecular targets of n3-PUFAs. Together, these studies show differences in gene expression patterns and NR specificity among the ω3-PUFAs. Also, genes particularly sensitive to ω3-PUFA were identified that could aid in examining the clinical effectiveness of dietary supplements.
METHODS
Chemicals
ALA, steariodonic acid (SDA), EPA, docosapentaenoic acid (DPA), and DHA were purchased from Sigma-Aldrich (St. Louis, MO). FBS was purchased from HyClone (Logan, UT). Geneticin was purchased from Invitrogen (Grand Island, NY). The EPA-rich oil (EO, triglyceride form) used in this study is a proprietary product manufactured under contract and was provided by the study sponsor, DuPont Applied Biosciences, Wilmington, DE. DuPont has developed an oleaginous yeast that produces an oil rich in EPA at 35% of FA content and linoleic acid at 20% of content and which is low in all other FAs (<7%).
Preparation of BSA-conjugated FAs
Due to the aqueous insolubility, all free FAs were conjugated to FA-free BSA (BSA) for treatments (molar ratio of 4:1 FA:BSA). The EO was saponified, and FAs were extracted as described previously (13). Molecular weight of EO was determined by average of the components and estimated at 290 g/mol. FAs were weighed and dissolved in ethanol as a stock concentration of 0.5 M. A total of 32 μl stock solution was transferred to a brown glass vial and dried under argon, whereas an equal volume of ethanol was dried in another vial as a vehicle control. A total of 132 μl of 0.15 M KOH was added to both vials, vortexed, and incubated for 1 h at 70°C under argon. Following the incubation, 2 ml of filter-sterilized BSA (2 mM) in PBS was added to the FAs and the vehicle control to achieve a final concentration of 8 mM. The pH was adjusted to 7.2 to 7.4. The BSA-conjugated FA and its BSA control were stored at −20°C under argon until use.
Cell culture and treatments
THP-1 (Homo sapiens monocyte) cells were obtained from the American Type Culture Collection (ATCC; Rockville, MD) and cultured in RPMI 1640 with 10% heat-inactivated FBS, 50 μM 2-mercaptoethanol, 1 mM sodium pyruvate, and antibiotics. These cells were seeded in 24-well plates at a density of 3 × 105/well and differentiated into macrophages with 100 nmol/l phorbol 12-myristate 13-acetate (Sigma) for 48 h. For in vitro treatment experiments, THP-1 cells were grown to 75% confluency and treated with 0, 1, 10, and 100 µM of the EPA-enriched oil or FAs as BSA conjugates. Eighteen hours after treatment, the cells were stimulated with 10 ng/ml LPS for 6 h. For ex vivo experiments, THP-1 cells were cultured as described above and treated with 10% (v/v) human serum from individual subjects for 18 h, after which the cells were stimulated with LPS for 6 h.
RNA extraction, reverse transcription, and real-time PCR
Total RNA was isolated using a Qiagen RNeasy Mini Kit (Qiagen; Valencia, CA) according to the manufacturer's instructions. The total RNA was reverse transcribed using the ABI high-capacity cDNA archive kit (Applied Biosystems; Foster City, CA). Standard curves were made using serial dilutions from pooled cDNA samples. Real-time PCR was performed with the use of the SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol and amplified on the ABI Prism 7000 sequence detection system. Detailed information on primer sequences is provided in Supplementary Materials and Methods.
Microarray experiments and statistical analysis
THP-1 cells were treated with EO at three concentrations (1, 10, and 100 μM) or control (BSA) for 16 h, followed by stimulation with LPS for 6 h. RNA was extracted using Qiagen RNeasy, and quality was assessed by RNA Nano Chips on the Agilent Bioanalyzer. Each sample was labeled using the Affymetrix IVT Express Kit according to the manufacturer's protocol. The GeneChip Human Genome U133A 2.0 (Affymterix), representing 14,500 well-characterized genes, was hybridized with the labeled RNA using GeneChip Hybridization Wash and Stain Kit (#702232) in the Affymetrix GeneChip Hybridization Oven 640, according to the manufacturer's instructions. Following hybridization the arrays were washed and stained using the Affymetrix GeneChip Fluidics Station 450 according to the manufacturer's protocol and scanned using the GeneChip Scanner 3000 7G. The scanned image file (DAT) and the intensity data (CEL) were imported into GeneSpring 10.0 (Agilent Technologies). The Robust Multi-array Average was used to normalize the data (22,277 entities) and was filtered on expression (>20% percentile in at least 1 of 12 samples, 18,497 entities). The 12 slides were grouped based on dose (four doses; 0, 1, 10, 100 μM, n = 3 per dose) and one-way ANOVA with asymptotic P value, and Benjamini-Hochberg multiple corrections was performed. At a P value of 0.01, a total of 798 entities were significantly regulated, of which 58 exhibited expression differences of 2-fold when compared with the control group. The group of genes was examined by hierarchical clustering using complete linkage analysis of the normalized data (JMP 7.0, SAS Institute; Cary, NC).
Nuclear receptor reporter assays
Human PPARα, PPARβ/δ, PPARγ, LXRβ, RXRα, and FXR reporter assay systems were purchased from INDIGO Biosciences, Inc. (State College, PA). Assays were performed according to the manufacturer's instructions.
Effects of omega-3 FA supplementation, ex vivo
Serum samples for ex vivo studies were collected from subjects enrolled in a double-blinded, placebo-controlled, parallel-design study of 121 subjects randomized into four treatment groups. There were 26, 27, 29, and 28 completers in the placebo olive oil group, low-dose EPA group (600 mg/day), high-dose EPA group (1,800 mg/day), and DHA group (600 mg/day), respectively. Of the completing subjects, the mean age was 52 years, 67% were male, 33% were female (all postmenopausal), 70% were white, 26% were black, 3% were Asian, and 1% were Hispanic. The mean body mass index of the group was 27.4 kg/m2.The human protocol was approved by Schulman Associates Institutional Review Board, Cincinnati, OH. This study was registered at ClinicalTrials.gov as NCT00988585, and was given the identification number DuPont-0609. Study capsules were manufactured under contract and were provided by the study sponsor, DuPont Applied Biosciences, Wilmington, DE. DuPont has developed an oleaginous yeast that produces an oil rich in EPA at 38% of FA content and linoleic acid at 20% of content and which is low in all other FAs (<7%). At the time of the enrollment visit, all qualifying and consenting subjects were randomly allocated into a protocol, where they were required to take two 1 g capsules three times daily, which contained a total of either: 1) 6 g/day of olive oil placebo, 2) 600 mg/day of EPA/day and 4.42 g/day of olive oil, 3) 1,800 mg of EPA/day and 1.26 g/day of olive oil, and 4) 600 mg of DHA/day. The olive oil placebo, the low-dose EPA oil, the high-dose EPA oil, and the DHA oil supplements, in total milligrams, contained: oleic acid 3,954, 2,906, 852, and 145 mg; palmitic acid 696, 542, 218, and 187 mg; stearic acid 26.1, 26.4, 25.7, and 0.0 mg; myristic acid 0, 1.8, 3.0, and 223 mg; linoleic acid 92.3, 120.9, 176.0, and 0.0 mg; ALA 5.6, 14.6, 31.8, and 0.0 mg; EPA 0.0, 627, 1,869, and 1.73 mg; and DHA 0.0, 0.0, 4.8, and 600 mg, respectively (see supplementary Table II for additional details). Over the entire 42 days, the study subjects were expected to have consumed a total of 252 capsules. Compliance was calculated as a percentage of consumed capsule count/expected capsule count based on the number of days the subject was in the study.
At the time of the enrollment visit, all qualifying and consenting subjects were randomly allocated into the four treatment groups described above. Subjects were asked to fast for 12 h prior to blood draws that occurred prior to intervention (baseline) and after 6 weeks of consuming the capsules. Serum was prepared and frozen until examined. THP-1 cells were cultured and treated with serum obtained from the clinical study diluted in media (10% v/v) (13) for 16 h, followed by treatment with LPS for 6 h. RNA was extracted, and quantitative PCR was performed as described above.
Specialized laboratory measures
Using plasma aliquots, frozen at −80°C and never thawed, obtained at the baseline and at 6 week visits, we carried out a wide variety of biochemical assays. Total cholesterol, triglyceride, and HDL cholesterol were measured as previously described (15). Direct LDL cholesterol and small, dense LDL cholesterol levels were measured using kits obtained from Denka-Seiken Corporation, Tokyo, Japan (16). Triglyceride-rich lipoprotein cholesterol was calculated by subtraction of direct LDL and HDL cholesterol values from total cholesterol. Analysis of the FA profile of total serum phospholipids was determined at Nutrasource Diagnostics, Guelph, Canada in a blinded fashion as described (17, 18) Serum was subjected to Folch extraction, and phospholipids were separated by thin-layer chromatography. FA methyl esters were prepared from the phospholipid fraction and measured on a Varian 3400 gas-liquid chromatograph using a 60 meter DB-23 capillary column (0.32 mm internal diameter).
Statistical analysis
One-way ANOVA, followed by Dunnett's posthoc test, was used to test the difference between treatments (P < .05). The values were expressed as mean ± SEM. All data analyses were performed by JMP 7.0 (SAS Institute; Cary, NC) and data were plotted by Prism 5.01 (GraphPad Software, Inc.; San Diego, CA).
RESULTS
Comprehensive analysis of gene expression
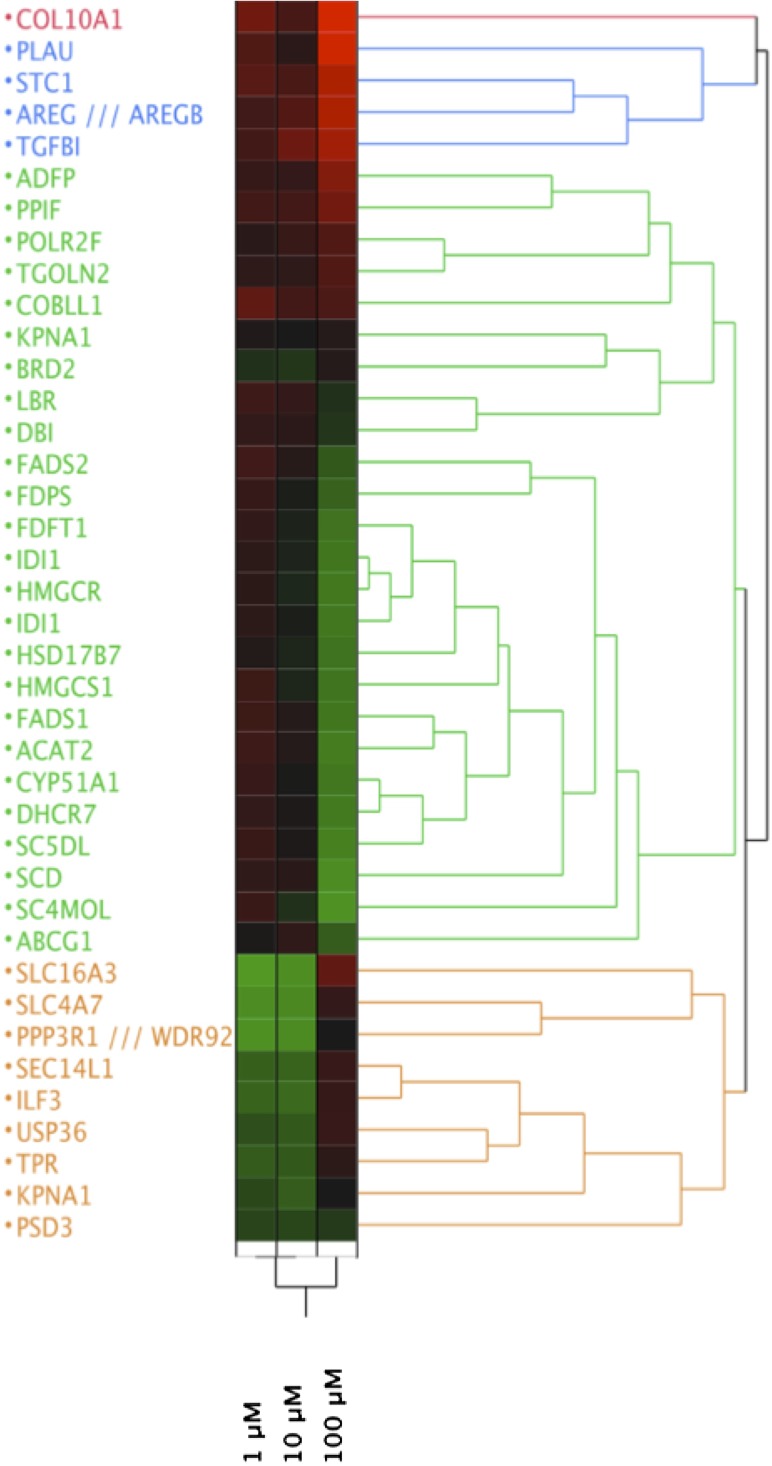
The design of the present experiments was aimed to understand the anti-inflammatory effects of ω3-PUFA oil and to identify sensitive biomarkers for subsequent studies. The THP-1 monocytes were treated with EO for 16 h, followed by stimulation with LPS, and the RNA was used to examine gene expression via high-density microarray. A total of 798 genes were significantly regulated (corrected P value 0.01) by EO in a dose-dependent manner, as assessed by one-way ANOVA, of which 58 were 2-fold different than control at a minimum of one dose of EO (Table 1, Fig. 1) . The dose-dependent regulation of gene expression followed three predominant trends: genes that are increased at the highest dose of EO (i.e., PLAU, TGFβ1, COL10A); genes that are decreased at the highest dose of EO [i.e., stearoyl CoA desaturase 1 (SCD-1), CYP51A, IDI1, ACAT2]; and genes that are decreased at low doses but increased at high doses of EO (i.e., ILF3, SLC16A3) (Fig. 1). The predominant function of EO-regulated genes was steroid, sterol, and lipid isoprenoid biosynthesis and metabolism (see supplementary Table II). This subset of genes was examined for common transcriptional regulators using Ingenuity Pathway Analysis (IPA 9.0, Ingenuity Systems, Inc.; Redwood City, CA), as shown in supplementary Fig. I. Several nuclear receptors, including the PPARs and the LXRs, as well as the nuclear factor κB complex, were implicated in the gene expression patterns observed.
TABLE 1.
Genes significantly affected by EO in THP-1 cells
| Fold Change (relative to control) | ||||
| Gene Symbol | Gene Title | 1 μM | 10 μM | 100 μM |
| COL10A1 | Collagen, type X, α 1 | 1.3278 | 1.1086 | 2.5391 |
| PLAU | Plasminogen activator, urokinase | 1.1471 | 0.9655 | 1.9077 |
| STC1 | Stanniocalcin 1 | 1.1964 | 1.1177 | 1.6985 |
| AREG /// AREGB | Amphiregulin /// amphiregulin B | 1.0694 | 1.1683 | 1.6936 |
| TGFBI | Transforming growth factor, β-induced, 68 kDa | 1.0750 | 1.3081 | 1.6264 |
| ADFP/ADRP | Adipose differentiation-related protein | 1.0160 | 1.0060 | 1.4352 |
| PPIF | Peptidylprolyl isomerase F | 1.0781 | 1.0807 | 1.3483 |
| SLC16A3 | Solute carrier family 16, member 3 (monocarboxylic acid transporter 4) | 0.2643 | 0.3233 | 1.2493 |
| POLR2F | Polymerase (RNA) II (DNA directed) polypeptide F | 0.9596 | 1.0271 | 1.1591 |
| TGOLN2 | Trans-Golgi network protein 2 | 0.9787 | 0.9838 | 1.1552 |
| COBLL1 | COBL-like 1 | 1.2353 | 1.0838 | 1.1328 |
| SEC14L1 | SEC14-like 1 (S. cerevisiae) | 0.5690 | 0.5472 | 1.0216 |
| USP36 | Ubiquitin-specific peptidase 36 | 0.6582 | 0.5994 | 1.0188 |
| ILF3 | Interleukin enhancer binding factor 3, 90 kDa | 0.5487 | 0.5171 | 1.0136 |
| SLC4A7 | Solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 0.3262 | 0.3462 | 1.0044 |
| TPR | Translocated promoter region (to activated MET oncogene) | 0.5943 | 0.6034 | 0.9741 |
| KPNA1 | Karyopherin α 1 (importin α 5) | 0.9225 | 0.8926 | 0.9395 |
| BRD2 | Bromodomain-containing 2 | 0.8018 | 0.7723 | 0.9390 |
| KPNA1 | Karyopherin α 1 (importin α 5) | 0.6928 | 0.5825 | 0.8933 |
| PPP3R1 /// WDR92 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, α isoform /// WD repeat domain 92 | 0.3005 | 0.3412 | 0.8904 |
| LBR | Lamin B receptor | 1.0578 | 1.0050 | 0.8025 |
| DBI | Diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) | 0.9965 | 0.9613 | 0.7820 |
| PSD3 | Pleckstrin and Sec7 domain-containing 3 | 0.7057 | 0.7027 | 0.7524 |
| FADS2 | FA desaturase 2 | 1.0707 | 0.9498 | 0.6029 |
| ABCG1 | ATP-binding cassette, sub-family G (WHITE), member 1 | 0.9029 | 0.9907 | 0.5864 |
| FDPS | Farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) | 1.0144 | 0.8790 | 0.5572 |
| FDFT1 | Farnesyl-diphosphate farnesyltransferase 1 | 1.0011 | 0.8560 | 0.4714 |
| HSD17B7 | Hydroxysteroid (17-β) dehydrogenase 7 | 0.9309 | 0.8483 | 0.4649 |
| HMGCS1 | 3-Hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) | 1.0462 | 0.8465 | 0.4643 |
| FADS1 | FA desaturase 1 | 1.0469 | 0.9361 | 0.4585 |
| IDI1 | Isopentenyl-diphosphate delta isomerase 1 | 0.9725 | 0.8530 | 0.4530 |
| IDI1 | Isopentenyl-diphosphate delta isomerase 1 | 0.9725 | 0.8795 | 0.4468 |
| CYP51A1 | Cytochrome P450, family 51, subfamily A, polypeptide 1 | 1.0189 | 0.8881 | 0.4406 |
| HMGCR | 3-Hydroxy-3-methylglutaryl-CoA reductase | 0.9683 | 0.8389 | 0.4321 |
| DHCR7 | 7-Dehydrocholesterol reductase | 0.9987 | 0.9105 | 0.4267 |
| ACAT2 | Acetyl-CoA acetyltransferase 2 | 1.0593 | 0.9382 | 0.4142 |
| SC5DL | Sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like | 1.0299 | 0.9073 | 0.3937 |
| SCD | Stearoyl-CoA desaturase (delta-9-desaturase) | 0.9913 | 0.9573 | 0.3298 |
| SC4MOL | Sterol-C4-methyl oxidase-like | 1.0343 | 0.8020 | 0.3001 |
Fig. 1.
Altered gene expression by EO in THP-1 cells stimulated with LPS. Comprehensive analysis of gene expression was performed by microarrays as described in Methods. Average expression values (n = 3 arrays per group) were exported to JMP (SAS Institute, Cary, NC), and hierarchical clustering was performed. Data are expressed relative to the DMSO-treated group, with green representing a decrease and red an increase relative to control (depicted as black).
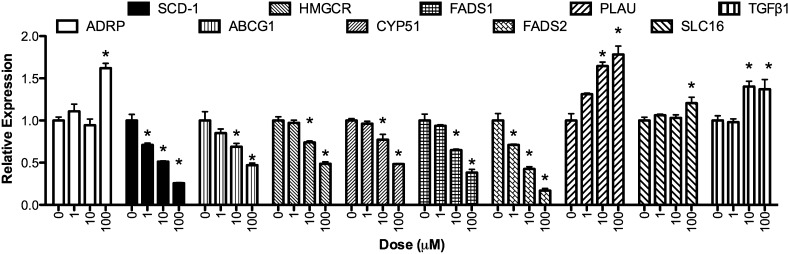
Quantitative real-time PCR was used to confirm a subset of transcripts identified by the microarray experiments. Care was taken to choose genes that were both increased and decreased by EO treatment, as well as those with known biological functions. In addition, because the desire was to find sensitive biomarkers of EO response genes that were significantly affected at the 1 μM EO in the microarray experiment were of particular interest. As shown in Fig. 2, all genes studied were significantly regulated by EPA oil, albeit to a different extent and with varying potency. Some mRNAs, such as ADRP and SLC16A, were induced only at the highest concentration of EO (100 µM), whereas SCD-1 and FADS2 were significantly repressed at the lowest dose (1 µM). Transcripts for ABCG1, HMCGR, CYP51, FADS1, PLAU, and TGFβ2 showed intermediate sensitivity, with significant alterations in quantity starting at 10 µM EO.
Fig. 2.
Expression of selected genes identified as responsive to EO in THP-1 cells. Genes were selected from Table 1 and Fig. 1 for validation by real-time PCR. Cells were treated, and RNA was extracted as described in Methods. Gene expression following treatment with EO (0, 1, 10, 100 μM) is expressed relative to a housekeeping gene (GAPDH) and normalized to vehicle control (DMSO). Asterisks denote significantly different from control (P < 0.05, n = 3). Bars represent mean and SEM.
Structure-activity relationships
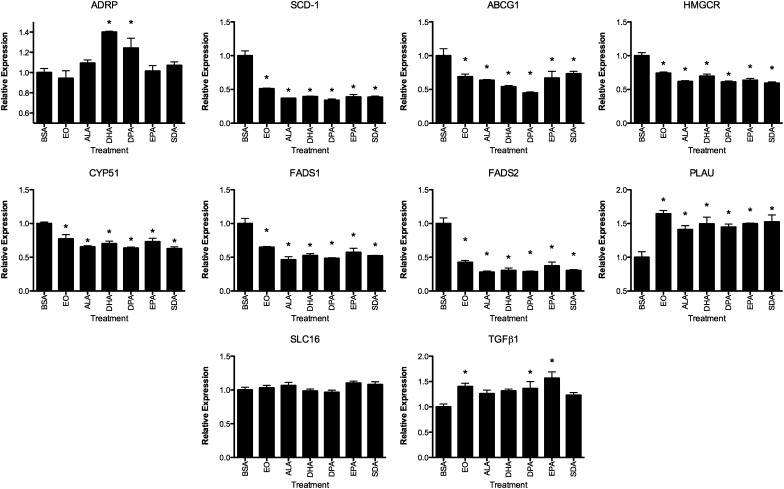
The genes regulated by EO were compared with that of an equal dose (100 µM) of the ω-3 PUFAs ALA, DHA, DPA, EPA, and SDA. THP-1 cells were treated with the FAs and subsequently stimulated with LPS or control. Figure 3 depicts the effects of ω-3 PUFA treatment on the LPS-stimulated cells. The repression of gene expression for SCD-1, ABCG1, HMGCR, CYP51, and FADS1 and -2 was similar among the FAs and EO in that all treatments significantly decreased mRNA amounts. Similarly, PLAU mRNA was induced by EO, and the FAs were examined. ADRP mRNA was increased by DHA, and DPA and was not affected by other ω-3 PUFAs, whereas TGFβ1 mRNA was increased by EO, DPA, and EPA treatment. The effects of EO (1, 10, and 100 µM) and ω-3 PUFAs at 100 µM, with or without LPS stimulation of gene expression, are shown in supplementary Fig. I. In general, the alterations in gene expression were more evident in the LPS-treated THP-1 cells, due in part to higher variability in the unstimulated cells.
Fig. 3.
Structure-activity relationships of altered gene expression in THP-1 cells. THP-1 cells were treated with the indicated FAs or EO at 100 μM and stimulated with LPS. See legend to Fig. 2 for details. Asterisks denote significantly different from control (P < 0.05, n = 3). Bars represent mean and SEM.
Transcription factor profiling
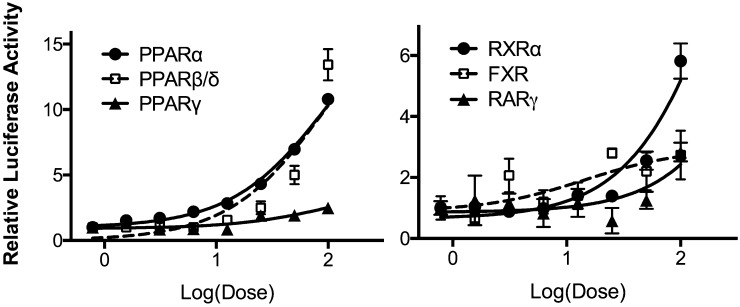
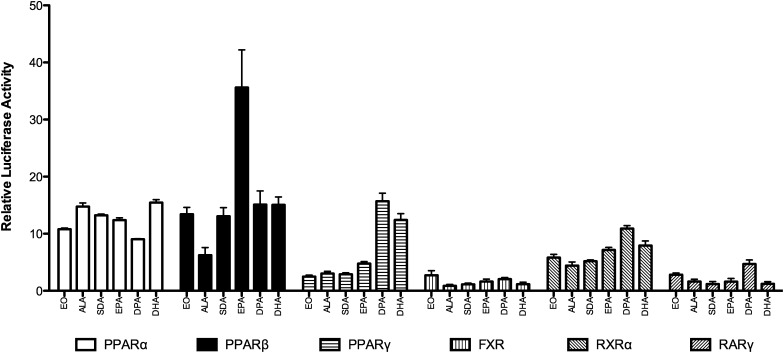
The subset of genes regulated by EO in the microarray experiments was examined for common transcriptional regulators using Ingenuity Pathway Analysis (see supplementary Fig. II). Several NRs were identified as being transcriptional regulators of EO-responsive genes, including LXRα and -β (NR1H3 and 2), glucocorticoid receptor, NR3C1, thyroid hormone receptor β (TRβ), PPARα and -γ (NR1C1 and -3), and estrogen receptor (ER) α (NR3B1). These ligand-activated transcription factors and several others identified or speculated to be FA receptors were examined in whole-cell receptor assays for regulation by EO. Significant dose-response regulation by EO of the PPARs (α, β/δ, and γ), RXRβ, farnesoid X receptor (FXR), and retinoic acid receptor γ (RARγ) was seen (Fig. 4). In contrast, ERα, ERβ, LXRα, LXRβ, TRα, vitamin D receptor (VDR), and constitutive androstane receptor, variant 3 (CAR3) activity was not affected by EPA oil treatment (data not shown). PPARα (10-fold) and PPARβ/δ (13-fold) were the most activated by EO, followed by RXRα (6-fold), RARγ (3-fold), and FXR (2-fold). The dose-response activation of these six nuclear receptors was examined for ω-3 PUFAs (see supplementary Fig. III), and the activation in comparison to EO at the 100 µM concentration is shown in Fig. 5. PPARα was responsive to all FAs examined with activation of approximately 10-fold. The largest response observed was for EPA activation of PPARβ/δ (35-fold). PPARγ showed a distinct preference for DHA and DPA relative to the other FAs. EO was the most efficacious activator of FXR, albeit only a 2-fold activation.
Fig. 4.
Activation of nuclear receptors by EO. Whole-cell reporter assays were used to examine the activation of various nuclear receptors by EO. See Methods for details. Data are expressed relative to vehicle control. Shown is mean and SEM, n = 4.
Fig. 5.
Structure-activity relationships for activation of nuclear receptors by ω-3 PUFAs. FAs were examined at 100 μM, as described in Methods, and luciferase data are expressed relative to vehicle control. Bars represent mean and SEM (n = 4).
Effects of omega-3 PUFAs on gene expression ex vivo
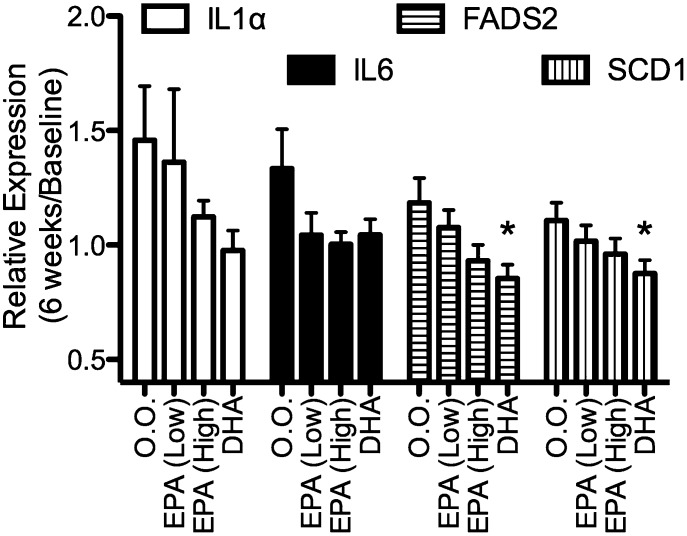
The hypothesis tested herein was that bioactive molecules are present in human serum following supplementation with EPA that affects gene expression. Studies performed in vitro indicated that FADS2 and SCD-1 are sensitive to EO and individual ω3-PUFAs, and those were chosen for these studies along with two inflammatory genes [interleukin 1α (IL1α) and IL6] that are affected in vitro and in vivo by FAs. Serum from baseline and following 6 weeks of dietary supplementation was used as a treatment medium (10% v/v) for LPS-stimulated THP-1 cells. As shown in Fig. 6, the general trend is for a decreased expression of IL1α, IL6, FADS2, and SCD-1 mRNA in THP-1 cells treated with serum obtained from the ω3-PUFA-supplemented individuals compared with the olive oil-administered group; the differences were significant for serum from the DHA-treated individuals.
Fig. 6.
Serum from individuals given ω3-PUFA supplements affected gene expression in THP-1 cells. Serum was diluted in treatment media (10% v/v) and applied to THP-1 cells, followed by LPS stimulation. Data are expressed as gene expression, with serum obtained at 6 weeks of treatment relative to the effects seen with serum obtained at baseline. (Mean and SEM, n = 20–25 per group). Asterisks denote significantly different from the olive oil (O.O.)-administered group (P < 0.05, Dunnett's t-test).
Alteration in serum FA profile and correlation to ex vivo gene expression
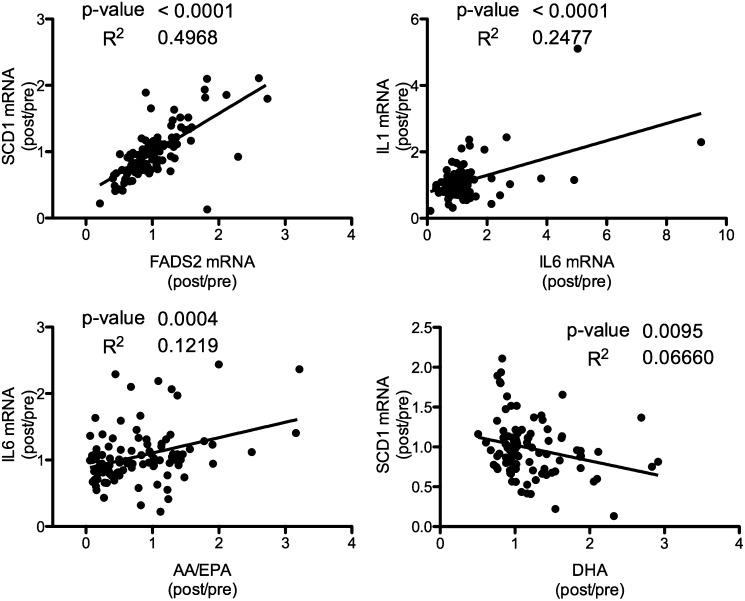
The effects of supplementation with EPA and DHA on serum FA profile is shown in Table 2. The placebo control had no effect on absolute or relative ω3-PUFA levels. The lower dose of EPA resulted in significant increases in total ω-3 and ω3/ω6 and EPA and decreased AA/EPA ratio. When the dose of EPA was increased, in addition to the previously observed effects, the total AA and the SCD-18 index [ratio of 18:1(n-9)/18:0] decreased. DHA administration increased total ω3, ω3/ω6, and DHA while decreasing the AA/DHA, AA, EPA, and SCD-18 index, as well as SCD-16 [ratio of 16:1(n-7)/16:0]. To examine potential mediators for the ex vivo observations, the gene expression values were correlated with each other and with the serum FA profile (see supplementary Table IV and supplementary for more details). As shown in Fig. 7, SCD-1 and FADS2 mRNA alterations as well as IL6 and IL1 mRNA changes were significantly correlated. In addition, the ratio of AA to EPA correlated with IL6 mRNA, and serum DHA affected SCD-1 mRNA expression ex vivo.
TABLE 2.
Serum FA profile from individuals given FA supplements for 6 weeks
| 0.0. |
EPA (Low) |
EPA (High) |
DHA |
|||||||||
| Lipid | Pre | Post | Pa | Pre | Post | P | Pre | Post | P | Pre | Post | P |
| Omega3 | 5.678 ± 0.474 | 5.635 ± 0.526 | 0.847 | 5.376 ± 0.340 | 6.657 ± 0.354 | <0.001 | 5.554 ± 0.438 | 9.255 ± 0.607 | <0.001 | 5.386 ± 0.258 | 7.628 ± 0.323 | <0.001 |
| AA/EPA | 17.06 ± 2.211 | 17.13 ± 1.747 | 0.971 | 17.51 ± 2.066 | 9.029 ± 1.321 | <0.001 | 18.85 ± 2.144 | 5.833 ± 1.530 | <0.001 | 16.18 ± 1.627 | 15.97 ± 1.323 | 0.822 |
| AA/DHA | 4.084 ± 0.420 | 4.158 ± 0.444 | 0.576 | 4.347 ± 0.294 | 4.267 ± 0.339 | 0.600 | 4.161 ± 0.343 | 3.867 ± 0.327 | 0.114 | 3.854 ± 0.271 | 2.076 ± 0.215 | <0.001 |
| N3/N6 | 0.151 ± 0.014 | 0.152 ± 0.017 | 0.827 | 0.139 ± 0.010 | 0.18 ± 0.012 | <0.001 | 0.146 ± 0.013 | 0.266 ± 0.021 | <0.001 | 0.141 ± 0.007 | 0.209 ± 0.010 | <0.001 |
| AA | 11.60 ± 0.495 | 11.52 ± 0.559 | 0.731 | 12.86 ± 0.455 | 12.42 ± 0.548 | 0.082 | 12.63 ± 0.475 | 11.40 ± 0.462 | <0.001 | 11.74 ± 0.354 | 10.90 ± 0.361 | 0.022 |
| EPA | 0.959 ± 0.111 | 0.937 ± 0.130 | 0.851 | 0.95 ± 0.100 | 1.901 ± 0.177 | <0.001 | 0.895 ± 0.123 | 3.871 ± 0.371 | <0.001 | 0.891 ± 0.082 | 0.770 ± 0.050 | 0.030 |
| DHA | 3.586 ± 0.353 | 3.579 ± 0.379 | 0.937 | 3.287 ± 0.251 | 3.258 ± 0.231 | 0.865 | 3.566 ± 0.315 | 3.316 ± 0.219 | 0.220 | 3.332 ± 0.182 | 5.973 ± 0.331 | <0.001 |
| SCD-18 | 0.86 ± 0.041 | 0.874 ± 0.036 | 0.447 | 0.818 ± 0.028 | 0.808 ± 0.024 | 0.551 | 0.839 ± 0.024 | 0.768 ± 0.023 | <0.001 | 0.853 ± 0.022 | 0.794 ± 0.022 | 0.004 |
| SCD-16 | 0.010 ± 0.001 | 0.010 ± 0.001 | 0.774 | 0.009 ± 0.000 | 0.009 ± 0.000 | 0.638 | 0.010 ± 0.001 | 0.008 ± 0.000 | 0.057 | 0.011 ± 0.001 | 0.009 ± 0.001 | 0.003 |
Data shown as means ± standard error of 26 individuals. All concentrations are expressed as mg/dl. Omega 3, total DHA plus EPA; AA, arachidonic acid; SCD-18, 18:l(n-9)/18:0; SCD-16,16:l(n-7)/16:0.
Paired t-test (two-tailed).
Fig. 7.
Correlation of ex vivo gene expression with serum FA concentrations. Individual data for gene expression and clinical FA concentrations were examined by pair-wise comparison and linear regression. The ratio of the post-to-pretreatment was examined for all treatment groups (n = 101–110).
DISCUSSION
Animal experiments and human studies have shown that ω3-PUFAs regulate genes in various tissues, including adipose tissue and peripheral blood mononuclear cells (PBMCs). There have been several comprehensive analyses of transcription responses to ω3-PUFAs, including in PBMC following fish oil supplementation (19), in adipose tissue following a high-PUFA diet in humans (20) and mice (21), in breast cancer cell lines treated with EPA and DHA (22), and in colon cancer cells treated with DHA (23), to name a few. In the present studies, the model system was biased to examine the anti-inflammatory responses of ω3-PUFAs by using a human monocytic cell line challenged with LPS. Despite this choice of model, many of the genes regulated by EO were involved in cholesterol, sterol, and lipid metabolism. This is consistent with clinical observations whereby the predominant effect of ω3-PUFA dietary supplementation is the lowering of circulating triglyceride levels (24–27). In fact, the lipid-lowering effect is often seen in the absence of an anti-inflammatory response (28). In general, the favorable effects of 3-PUFAs mainly result from the combined effects of decreased expression of lipogenesis-related genes and stimulation of FA oxidation transcripts. Fish oil decreases expression of sterol response element binding protein-1c (29), a key enzyme in controlling lipogenesis. Other lipogenic genes are decreased by ω3-PUFAs, including FA synthase, malic enzyme, and glucose-6-phosphate dehydrogenase (11). In the liver, several genes involved in FA metabolism are increased by PUFAs, including apolipoproteins A-I and A-II A, acyl-CoA synthetase, acyl-CoA oxidase, liver FA-binding protein, carnitine palmitoyl transferase, and cytochrome P450 4A1 (11). Although the genomic response to EO in THP-1 cells showed a preponderance of lipid metabolism genes, the pathways affected were generally lipogenic genes and were decreased upon treatment. This pattern is more reminiscent of the response of adipocytes (30) than of hepatocytes to ω3-PUFA treatment.
FA elongation and desaturation are two key metabolic routes for the synthesis of saturated, monounsaturated, and polyunsaturated FAs. Of these, FA desaturases have received considerable attention for their regulation by hormones and nutrients and their capacity to generate specific unsaturated FAs. One of these enzymes, SCD-1, or delta 9 desaturase, have emerged as key enzymes in the control of whole-body lipid composition (31). Although oleate is found ubiquitously throughout the body, endogenously derived oleate from SCD is special in terms of its preferential trafficking through acyl-CoA:diacylglycerol acyltransferase 2 and driving triglyceride (TG) synthesis. Omega-3 FAs decrease SCD-1 mRNA expression in liver, and this effect is correlated with decreased circulating TG (32). In addition, decreased SCD-1 expression in macrophages by ALA is associated with increased cholesterol efflux (33), a beneficial response in terms of reverse cholesterol transport. Delta-5 desaturase [D5D, FA desaturase 2 (FADS1)] and delta-6 desaturase (D5D, FADS2) are the key enzymes for the synthesis of PUFAs such as AA and DHA. Elovl-1 (Ssc1) and Elovl-6 (LCE, FACE, rElo2) elongate saturated and monounsaturated FAs. Omega-3 PUFAs decease the expression of D6D and D5D (decreased levels of PUFAs increases their expression) as well as Elovl-6; however, the conversion of ALA to longer chain PUFAs is regulated by substrate levels to a greater extent than to expression of the synthetic enzymes (34). Nonetheless, the repression of FADS1 and FADS2 as well as SCD-1 in macrophages is among the most-sensitive biomarkers for EO response.
The sensitivity of FADS2 and SCD-1 to EO treatment in vitro made them likely choices for examination in the ex vivo experiments. In addition, the inflammatory markers IL1 and IL6 are typically decreased by ω3-PUFA treatment in vitro and in vivo (28, 34). In these studies, serum from individuals given various supplements were used as a treatment to THP-1 cells; this approach has been used previously by our laboratory and others as a means to assess alterations in circulating bioactive molecules following a treatment or diet (14, 35, –39). The expression of these genes would be expected to be lower following administration of serum to THP-1 cells from the 6-week treatment with EO compared with serum taken at baseline. In fact, there is a dose-dependent trend for decreased ex vivo expression of FADS2 and SCD-1, although it only reached statistical significance in the DHA group. Individuals’ ex vivo SCD-1 and FADS2 responses are significantly correlated, as are the IL1 and IL6 responses. In contrast, the FADS2 and SCD-1 responses were not indicative of changes in interleukin expression (see supplementary Table IV). This may be reflective of similar mechanisms of gene expression for SCD-1 and FADS2 that differ from those of IL1 and 6. Of note is the fact the one's serum DHA change (post treatment/pre-treatment) is inversely associated with the ex vivo SCD-1 mRNA (post/pre). Similarly, the change in ratio of AA to EPA in the serum is correlated with the ex vivo expression of IL1 and IL6. Principal component analysis of the individual post/pre-treatment values for the FA concentration as well as the ex vivo gene expression showed a similar observation with AA/EPA being more associated with IL1 and IL6 mRNA, whereas AA/DHA clustered with SCD-1 and FADS2, as well as the SCD-16 and -18 ratios (see supplementary ). Together, these data show that altering DHA and EPA concentrations may impact different endpoints preferentially.
Often the biological responses of the predominant ω3-PUFAs, ALA, EPA, and DHA, are considered to be equivalent or interchangeable, although it is generally held that the marine-based FAs are more beneficial than their plant-based counterparts (5, 38). In the present studies, the altered gene expression of a small subset of genes (Fig. 3) suggests that they do, in fact, lead to similar genomics responses in vitro. However, more-comprehensive analysis of gene expression following treatment with ω3-PUFAs shows that there are subsets of genes that are affected by particular FA structures, in particular at higher doses (Vanden Heuvel, unpublished observations). This may be the result of differential metabolism of ALA, EPA, and DHA to various bioactive molecules and/or interaction with specific transcription factors, enzymes, and receptors. In addition, as mentioned above, the ex vivo studies suggest that serum from individuals on the EO supplements differed from that from DHA-supplemented cohorts, further illustrating that these two ω3-PUFAs are similar but are not fully interchangeable.
Of the several identified FA receptors, perhaps the family that can best explain the effects of ω3-PUFAs are the PPARs. The PPARs exist as three subtypes (α, β, and γ) that vary in expression, ligand recognition, and biological function. PPARα was the first transcription factor identified as a prospective FA receptor (as reviewed in Refs. 39–41) and is involved in FA transport, FA binding proteins, fatty acyl CoA synthesis, microsomal, peroxisomal and mitochondrial FA oxidation, ketogenesis, and FA desaturation. PPARβ/δ is ubiquitously expressed and is often found in higher abundance than PPARα or γ. Examination of PPARβ/δ-null mice has shown a role for this NR in development, myelination of the corpus callosum, lipid metabolism, and epidermal cell proliferation (42). PPARγ is expressed in many tissues, including adipose, muscle, vascular cells, macrophages, and epithelial cells of the mammary gland, prostate, and colon (as reviewed in Ref. 43). Activated PPARγ induces LPL and FA transporters (CD36), and enhances adipocyte differentiation, as well as inhibits cytokine and cylcooxygenase 2 (COX-2) expression. The EPA-enriched oil and individual ω3-PUFAs activated PPARα to a similar extent, with little distinction. In fact, this NR is activated by ω6-PUFAs (44), and metabolites of FAs, including epoxyeicosatrienoic acids, hydroxyepoxyeicosatrienoic acids (HETEs) (45), leukotriene B4 (46), and prostaglandin D2 (PGD2) (47). The least-studied member of the PPAR family, PPARβ/δ, was preferentially activated by EPA and showed the highest level of activation of any NR by EO. ALA was the weakest activator of PPARβ/δ among the ω3-PUFAs, with induction similar to the ω6-PUFA linoleic acid (not shown). Prostaglandin A1 (PGA1), PGD2, and PGD1 can activate PPARβ/δ in reporter assays (47). 15-HETE and the toxic lipid 4-hydroxynonenol 4-HNE are also PPARβ/δ activators (48). PPARγ has received considerable attention as a target of anti-diabetic and -inflammatory drugs. This NR was only marginally affected by EO and ω3-PUFAs, with the noted exception of DPA and DHA. Interestingly, the 5-lipoxygenase metabolite of DHA (4-hydroxy DHA) (49) and the COX-2 metabolites electrophilic oxo-derivatives (50), are more-potent PPARγ activators than the parent FA and may be responsible for effects of this ω3-PUFA on angiogenesis and inflammation. Less-distinctive structure-activity relationships were noted for RXRα, a previously noted DHA receptor (51), and RARγ, a heretofore-unidentified FA receptor. The differences in gene expression and transcription factor activation among the members of the ω3-PUFA family can explain some of the differences in potency of biological and therapeutic responses and can point to specific recommendations for the individual FAs. For example, EPA may be of more benefit for diseases with a PPARβ/δ etiology, whereas DHA is more-amenable for PPARγ-related therapies. This is especially important because genetic modification has been used to develop a new generation of plants (e.g, corn and soy-beans) that produce seeds with a modified FA profile for use as dietary supplements with specific health benefits in mind.
Taken together, these studies have shown that EO alters gene expression in a human monocytic cell line, consistent with altered lipid metabolism and inflammation. This particular dietary supplement contrasts with others on the market that are rich in other ω3-PUFAs, such as DHA, SDA, and ALA. Most of the responses observed for each ω3-PUFA supplement are similar and have beneficial properties against many aspects of metabolic syndrome. However, due in part to specificity of nuclear receptor activation as well as differential metabolism, each ω3-PUFA and corresponding supplement or diet must be considered a unique entity. Evidence is provided that EOs may preferentially affect PPARβ/δ-associated therapies, whereas DHA and other ω3-PUFA supplementation have their own subset of benefits. Hence, additional research is warranted to provide recommendations of ω3-PUFA supplements for specific outcomes.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- ALA
- α linolenic acid
- COX-2
- cylcooxygenase 2
- CVD
- cardiovascular disease
- DHA
- docosahexaenoic acid
- DPA
- docosapentaenoic acid
- EO
- EPA-rich oil
- EPA
- eicosapentaenoic acid
- ER
- estrogen receptor
- FXR
- farnesoid X receptor
- HETE
- hydroxyepoxyeicosatrienoic acid
- IL1
- interleukin 1
- LPS
- lipopolysaccharide
- LXR
- liver X receptor
- NR
- nuclear receptor
- PBMC
- peripheral blood mononuclear cell
- PGD2
- prostaglandin D2
- PPAR
- peroxisome proliferator-activated receptor
- RAR
- retinoic acid receptor
- RXR
- retinoid X receptor
- SCD
- stearoyl CoA desaturase
- SDA
- steariodonic acid
- TG
- triglyceride
This work was conducted under a contract between DuPont, Inc. and INDIGO Biosciences, Inc. J.V.H. is an employee of Penn State University and has a financial stake with INDIGO Biosciences, Inc., which may constitute a conflict of interest.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures and four tables.
REFERENCES
- 1.Renaud S., Lanzmann-Petithory D. 2001. Coronary heart disease: dietary links and pathogenesis. Public Health Nutr. 4: 459–474 [DOI] [PubMed] [Google Scholar]
- 2.Willett W. C. 2007. The role of dietary n-6 fatty acids in the prevention of cardiovascular disease. J. Cardiovasc. Med. (Hagerstown). 8 (Suppl.): 42–45 [DOI] [PubMed] [Google Scholar]
- 3.Hu F. B., Willett W. C. 2002. Optimal diets for prevention of coronary heart disease. J. Am. Med. Assoc. 288: 2569–2578 [DOI] [PubMed] [Google Scholar]
- 4.Calder P. C. 2003. N-3 polyunsaturated fatty acids and inflammation: from molecular biology to the clinic. Lipids. 38: 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simopoulos A. P. 2002. Omega-3 fatty acids in inflammation and autoimmune diseases. J. Am. Coll. Nutr. 21: 495–505 [DOI] [PubMed] [Google Scholar]
- 6.Calder P. C. 2008. Polyunsaturated fatty acids, inflammatory processes and inflammatory bowel diseases. Mol. Nutr. Food Res. 52: 885–897 [DOI] [PubMed] [Google Scholar]
- 7.Wild G. E., Drozdowski L., Tartaglia C., Clandinin M. T., Thomson A. B. R. 2007. Nutritional modulation of the inflammatory response in inflammatory bowel disease–from the molecular to the integrative to the clinical. World J. Gastroenterol. 13: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James M., Proudman S., Cleland L. 2010. Fish oil and rheumatoid arthritis: past, present and future. Proc. Nutr. Soc. 69: 316–323 [DOI] [PubMed] [Google Scholar]
- 9.Schulman I. G. 2010. Nuclear receptors as drug targets for metabolic disease. Adv. Drug Deliv. Rev. 62: 1307–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaven S. W., Tontonoz P. 2006. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu. Rev. Med. 57: 313–329 [DOI] [PubMed] [Google Scholar]
- 11.Sampath H., Ntambi J. M. 2005. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu. Rev. Nutr. 25: 317–340 [DOI] [PubMed] [Google Scholar]
- 12.Francis G. A., Fayard E., Picard F., Auwerx J. 2003. Nuclear receptors and the control of metabolism. Annu. Rev. Physiol. 65: 261–311 [DOI] [PubMed] [Google Scholar]
- 13.Torres C. F., Garcia H. S., Ries J. J., Charles J., Hill G. 2001. Esterification of glycerol with conjugated linoleic acid and long-chain fatty acids from fish oil. J. Am. Oil Chem. Soc. 78: 1093–1098 [Google Scholar]
- 14.Zhang J., Grieger J. A., Kris-Etherton P. M., Thompson J. T., Gillies P. J., Fleming J. A., Vanden Heuvel J. P. 2011. Walnut oil increases cholesterol efflux through inhibition of stearoyl CoA desaturase 1 in THP-1 macrophage-derived foam cells. Nutr. Metab. (Lond). 8: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otokozawa S., Ai M., Asztalos B. F., White C. C., Demissie-Banjaw S., Cupples L. A., Nakajima K., Wilson P. W. F., Schaefer E. J. 2010. Direct assessment of plasma low density lipoprotein and high density lipoprotein cholesterol levels and coronary heart disease: results from the Framingham Offspring Study. Atherosclerosis. 213: 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ai M., Otokozawa S., Asztalos B. F., Ito Y., Nakajima K., White C. C., Cupples L. A., Wilson P. W., Schaefer E. J. 2010. Small dense LDL cholesterol and coronary heart disease: results from the Framingham Offspring Study. Clin. Chem. 56: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dewailly E., Blanchet C., Gingras S., Lemieux S., Holub B. J. 2002. Cardiovascular disease risk factors and n-3 fatty acid status in the adult population of James Bay Cree. Am. J. Clin. Nutr. 76: 85–92 [DOI] [PubMed] [Google Scholar]
- 18.Asztalos B. F., Cupples L. A., Demissie S., Horvath K. V., Cox C. E., Batista M. C., Schaefer E. J. 2004. High-density lipoprotein subpopulation profile and coronary heart disease prevalence in male participants of the Framingham Offspring Study. Arterioscler. Thromb. Vasc. Biol. 24: 2181–2187 [DOI] [PubMed] [Google Scholar]
- 19.Bouwens M., van de Rest O., Dellschaft N., Bromhaar M. G., de Groot L. C. P. G. M., Geleijnse J. M., Müller M., Afman L. A. 2009. Fish-oil supplementation induces antiinflammatory gene expression profiles in human blood mononuclear cells. Am. J. Clin. Nutr. 90: 415–424 [DOI] [PubMed] [Google Scholar]
- 20.Radonjic M., van Erk M. J., Pasman W. J., Wortelboer H. M., Hendriks H. F. J., van Ommen B. 2009. Effect of body fat distribution on the transcription response to dietary fat interventions. Genes Nutr. 4: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flachs P., Horakova O., Brauner P., Rossmeisl M., Pecina P., Franssen-van Hal N., Ruzickova J., Sponarova J., Drahota Z., Vlcek C., et al. 2005. Polyunsaturated fatty acids of marine origin upregulate mitochondrial biogenesis and induce beta-oxidation in white fat. Diabetologia. 48: 2365–2375 [DOI] [PubMed] [Google Scholar]
- 22.Hammamieh R., Chakraborty N., Miller S-A., Waddy E., Barmada M., Das R., Peel S. A., Day A. A., Jett M. 2007. Differential effects of omega-3 and omega-6 fatty acids on gene expression in breast cancer cells. Breast Cancer Res. Treat. 101: 7–16 [DOI] [PubMed] [Google Scholar]
- 23.Narayanan B. A., Narayanan N. K., Reddy B. S. 2001. Docosahexaenoic acid regulated genes and transcription factors inducing apoptosis in human colon cancer cells. Int. J. Oncol. 19: 1255–1262 [DOI] [PubMed] [Google Scholar]
- 24.Duda M. K., O'Shea K. M., Stanley W. C. 2009. Omega-3 polyunsaturated fatty acid supplementation for the treatment of heart failure: mechanisms and clinical potential. Cardiovasc. Res. 84: 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka N., Zhang X., Sugiyama E., Kono H., Horiuchi A., Nakajima T., Kanbe H., Tanaka E., Gonzalez F. J., Aoyama T. 2010. Eicosapentaenoic acid improves hepatic steatosis independent of PPARα activation through inhibition of SREBP-1 maturation in mice. Biochem. Pharmacol. 80: 1601–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kjaer M. A., Vegusdal A., Gjøen T., Rustan A. C., Todorcević M., Ruyter B. 2008. Effect of rapeseed oil and dietary n-3 fatty acids on triacylglycerol synthesis and secretion in Atlantic salmon hepatocytes. Biochim. Biophys. Acta. 1781: 112–122 [DOI] [PubMed] [Google Scholar]
- 27.Skulas-Ray A. C., West S. G., Davidson M. H., Kris-Etherton P. M. 2008. Omega-3 fatty acid concentrates in the treatment of moderate hypertriglyceridemia. Expert Opin. Pharmacother. 9: 1237–1248 [DOI] [PubMed] [Google Scholar]
- 28.Skulas-Ray A. C., Kris-Etherton P. M., Harris W. S., Vanden Heuvel J. P., Wagner P. R., West S. G. 2011. Dose-response effects of omega-3 fatty acids on triglycerides, inflammation, and endothelial function in healthy persons with moderate hypertriglyceridemia. Am. J. Clin. Nutr. 93: 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ide T. 2005. Interaction of fish oil and conjugated linoleic acid in affecting hepatic activity of lipogenic enzymes and gene expression in liver and adipose tissue. Diabetes. 54: 412–423 [DOI] [PubMed] [Google Scholar]
- 30.Muhlhausler B. S., Cook-Johnson R., James M., Miljkovic D., Duthoit E., Gibson R. Opposing effects of omega-3 and omega-6 long chain polyunsaturated fatty acids on the expression of lipogenic genes in omental and retroperitoneal adipose depots in the rat. J. Nutr. Metab. Epub ahead of print. August 5, 2010; doi:10.1155/2010/927836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen P., Friedman J. M. 2004. Leptin and the control of metabolism: role for stearoyl-CoA desaturase-1 (SCD-1). J. Nutr. 134: 2455S–2463S [DOI] [PubMed] [Google Scholar]
- 32.Velliquette R. A., Gillies P. J., Kris-Etherton P. M., Green J. W., Zhao G., Vanden Heuvel J. P. 2009. Regulation of human stearoyl-CoA desaturase by omega-3 and omega-6 fatty acids: implications for the dietary management of elevated serum triglycerides. J. Clin. Lipidol. 3: 281–288 [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., Kris-Etherton P. M., Thompson J. T., Hannon D. B., Gillies P. J., Vanden Heuvel J. P. 2012. Alpha-linolenic acid increases cholesterol efflux in macrophage-derived foam cells by decreasing stearoyl CoA desaturase 1 expression: evidence for a farnesoid-X-receptor mechanism of action. J. Nutr. Biochem. 23: 400–409 PubMed [DOI] [PubMed] [Google Scholar]
- 34.Tu W. C., Cook-Johnson R. J., James M. J., Mühlhäusler B. S., Gibson R. A. 2010. Omega-3 long chain fatty acid synthesis is regulated more by substrate levels than gene expression. Prostaglandins Leukot. Essent. Fatty Acids. 83: 61–68 [DOI] [PubMed] [Google Scholar]
- 35.Jenkins D. J., Kendall C. W., Vidgen E., Agarwal S., Rao A. V., Rosenberg R. S., Diamandis E. P., Novokmet R., Mehling C. C., Perera T., et al. 1999. Health aspects of partially defatted flaxseed, including effects on serum lipids, oxidative measures, and ex vivo androgen and progestin activity: a controlled crossover trial. Am. J. Clin. Nutr. 69: 395–402 [DOI] [PubMed] [Google Scholar]
- 36.Henning S. M., Aronson W., Niu Y., Conde F., Lee N. H., Seeram N. P., Lee R. P., Lu J., Harris D. M., Moro A., et al. 2006. Tea polyphenols and theaflavins are present in prostate tissue of humans and mice after green and black tea consumption. J. Nutr. 136: 1839–1843 [DOI] [PubMed] [Google Scholar]
- 37.Singhal R., Badger T. M., Ronis M. J. 2007. Reduction in 7,12-dimethylbenz[a]anthracene-induced hepatic cytochrome-P450 1A1 expression following soy consumption in female rats is mediated by degradation of the aryl hydrocarbon receptor. J. Nutr. 137: 19–24 [DOI] [PubMed] [Google Scholar]
- 38.Gebauer S. K., Psota T. L., Harris W. S., Kris-Etherton P. M. 2006. n-3 fatty acid dietary recommendations and food sources to achieve essentiality and cardiovascular benefits. Am. J. Clin. Nutr. 83 (Suppl.): 1526–1535 [DOI] [PubMed] [Google Scholar]
- 39.Hihi A. K., Michalik L., Wahli W. 2002. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell. Mol. Life Sci. 59: 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montagner A., Rando G., Degueurce G., Leuenberger N., Michalik L., Wahli W. 2011. New insights into the role of PPARs. Prostaglandins Leukot. Essent. Fatty Acids. 85: [DOI] [PubMed] [Google Scholar]
- 41.Wahli W. 2002. Peroxisome proliferator-activated receptors (PPARs): from metabolic control to epidermal wound healing. Swiss Med. Wkly. 132: 83–91 [DOI] [PubMed] [Google Scholar]
- 42.Peters J. M., Lee S. S., Li W., Ward J. M., Gavrilova O., Everett C., Reitman M. L., Hudson L. D., Gonzalez F. J. 2000. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta). Mol. Cell. Biol. 20: 5119–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spiegelman B. M. 1997. Peroxisome proliferator-activated receptor gamma: a key regulator of adipogenesis and systemic insulin sensitivity. Eur. J. Med. Res. 2: 457–464 [PubMed] [Google Scholar]
- 44.Latruffe N., Vamecq J. 1997. Peroxisome proliferators and peroxisome proliferator activated receptors (PPARs) as regulators of lipid metabolism. Biochimie. 79: 81–94 [DOI] [PubMed] [Google Scholar]
- 45.Cowart L. A., Wei S., Hsu M-H., Johnson E. F., Krishna M. U., Falck J. R., Capdevila J. H. 2002. The CYP4A isoforms hydroxylate epoxyeicosatrienoic acids to form high affinity peroxisome proliferator-activated receptor ligands. J. Biol. Chem. 277: 35105–35112 [DOI] [PubMed] [Google Scholar]
- 46.Krey G., Braissant O., L'Horset F., Kalkhoven E., Perroud M., Parker M. G., Wahli W. 1997. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 11: 779–791 [DOI] [PubMed] [Google Scholar]
- 47.Yu K., Bayona W., Kallen C. B., Harding H. P., Ravera C. P., McMahon G., Brown M., Lazar M. A. 1995. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J. Biol. Chem. 270: 23975–23983 [DOI] [PubMed] [Google Scholar]
- 48.Coleman J. D., Prabhu K. S., Thompson J. T., Reddy P. S., Peters J. M., Peterson B. R., Reddy C. C., Vanden Heuvel J. P. 2007. The oxidative stress mediator 4-hydroxynonenal is an intracellular agonist of the nuclear receptor peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta). Free Radic. Biol. Med. 42: 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sapieha P., Stahl A., Chen J., Seaward M. R., Willett K. L., Krah N. M., Dennison R. J., Connor K. M., Aderman C. M., Liclican E., et al. 2011. 5-Lipoxygenase metabolite 4-HDHA is a mediator of the antiangiogenic effect of ω-3 polyunsaturated fatty acids. Sci. Transl. Med. 3: 69ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Groeger A. L., Cipollina C., Cole M. P., Woodcock S. R., Bonacci G., Rudolph T. K., Rudolph V., Freeman B. A., Schopfer F. J. 2010. Cyclooxygenase-2 generates anti-inflammatory mediators from omega-3 fatty acids. Nat. Chem. Biol. 6: 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Urquiza A. M., Liu S., Sjoberg M., Zetterstrom R. H., Griffiths W., Sjovall J., Perlmann T. 2000. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science. 290: 2140–2244 [DOI] [PubMed] [Google Scholar]