Abstract
Peroxisome proliferator-activated receptor α (PPARα) is a nuclear receptor that regulates fatty acid transport and metabolism. Previous studies revealed that PPARα can affect bile acid metabolism; however, the mechanism by which PPARα regulates bile acid homeostasis is not understood. In this study, an ultraperformance liquid chromatography coupled with electrospray ionization qua dru pole time-of-flight mass spectrometry (UPLC-ESI-QTOFMS)-based metabolomics approach was used to profile metabolites in urine, serum, and bile of wild-type and Ppara-null mice following cholic acid (CA) dietary challenge. Metabolomic analysis showed that the levels of several serum bile acids, such as CA (25-fold) and taurocholic acid (16-fold), were significantly increased in CA-treated Ppara-null mice compared with CA-treated wild-type mice. Phospholipid homeostasis, as revealed by decreased serum lysophos phati dylcholine (LPC) 16:0 (1.6-fold) and LPC 18:0 (1.6-fold), and corticosterone metabolism noted by increased urinary excretion of 11β-hydroxy-3,20-dioxopregn-4-en-21-oic acid (20-fold) and 11β,20α-dihydroxy-3-oxo-pregn-4-en-21-oic acid (3.6-fold), were disrupted in CA-treated Ppara-null mice. The hepatic levels of mRNA encoding transporters Abcb11, Abcb4, Abca1, Abcg5, and Abcg8 were diminished in Ppara-null mice, leading to the accumulation of bile acids in the liver during the CA challenge. These observations revealed that PPARα is an essential regulator of bile acid biosynthesis, transport, and secretion.
Keywords: PPARα, bile acid metabolism, phospholipid metabolism, cholesterol and corticosterone metabolism
Peroxisome proliferator-activated receptor α (PPARα) is a nuclear hormone receptor regulating genes involved in lipid homeostasis, including fatty acid transport and catabolism, lipoprotein metabolism, glucose homeostasis, and inflammation (1, 2). Recent studies have suggested a role for PPARα in bile acid biosynthesis. A previous study reported that the expression level of cholesterol 7α-hydroxylase (Cyp7a1) and 8β-hydroxylase (Cyp8b1) was upregulated in wild-type mice during [4-chloro-6-(2,3-xylidino)-2-pyrimidinylthio] acetic acid (Wy-14,643) treatment and fasting conditions; this effect was diminished in Ppara-null mice, thus revealing the involvement of PPARα (3). Another study found that both human CYP7A1 and mouse Cyp7a1 promoters were stimulated by fatty acids and Wy-14,643 treatment (4). These studies provided compelling evidence that PPARα can positively regulate bile acid biosynthesis. In contrast to the above observations, it was reported that 0.5% bezafibrate treatment leads to a decrease in liver bile acid and Cyp7a1 expression in wild-type mice and Ppara-null mice (5). Others found that Cyp7a1 and cholesterol 27α-hydroxylase (Cyp27a1) were decreased in wild-type mice by ciprofibrate treatment but not in Ppara-null mice (6). These results indicate that the activation of PPARα by fibrates can inhibit bile acid biosynthesis; however, bezafibrate and ciprofibrate show different effects on bile acid biosynthesis enzymes in Ppara-null mice. Thus, the mechanism by which PPARα controls bile acid homeostasis remains uncertain. Additionally, limited studies revealed that PPARα exert its effect on the bile acid conjugation and transport in the liver and intestine (7, 8).
Recent studies have demonstrated the power of mass spectrometry-based metabolomics to profile metabolic pathways to reveal the mechanism of action of nuclear receptors and the metabolic fate of drugs (9–12). Metabolomics has revealed the downregulated tryptophan-nicotinamide pathways following Wy-14,643 treatment (9) and the decreased excretion of carnitine-conjugated metabolites by fenofibrate treatment in humans (13). Furthermore, an increase in the excretion of glycine conjugated metabolites was observed in Ppara-null mice (9), and the long-chain fatty acid carnitines, including palmitoylcaritine, myristolcarnitine, oleoylcarnitine, and palmitoleoylcarnitine, were elevated in the serum after suppression of PPARα signal transduction by acetaminophen (14).
In the present study, cholic acid (CA)-treated wild-type and Ppara-null mice were used to evaluate the functional role of PPARα in bile acid homeostasis. Metabolomics analysis showed that severe liver dysfunction in Ppara-null mice was induced during CA challenge, including the disruption of bile acids, phospholipids, and cholesterol homeostasis. Additionally, the excretion of several endogenous cationic and anionic metabolites was significantly affected. Further results demonstrated that PPARα exerted a role in bile acid homeostasis via regulation of bile acid biosynthesis, transport, and secretion.
MATERIALS AND METHODS
Chemicals and reagents
Cholic acid (CA), taurocholic acid (TCA), chenodeoxycholic acid (CDCA), tauro-α/β-muricholic acid (T-α/β-MCA), taurochenodeoxycholic acid (TCDCA), taurodeoxycholic acid (TDCA), tauroursodeoxycholic acid (TUDCA), tauromurideoxycholic acid (TMDCA), chlorpropamide, creatinine, creatine, nicotimamide 1-oxide, xanthurenic acid, and phenylacetylglycine were purchased from Sigma (St. Louis, MO). 11β-Hydroxy-3,20-dioxopregn-4-en-21-oic acid (HDOPA) and 11β,20α-dihydroxy-3-oxo-pregn-4-en-21-oic acid (DHOPA) were ordered from Anbilaunch Consulting (San Meteo, CA). 1-Palmitoyl-sn-glycero-3-phosphocholine (LPC 16:0) and 1-stearoyl-sn-glycero-3-phosphocholine (LPC 18:0) were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL). All solvents and organic reagents were of the highest obtainable grade.
Animal study
Ppara-null mice and wild-type mice on a C57BL/6N genetic background were maintained under a standard 12 h light/12 h dark cycle with water and a normal diet (NIH-31) provided ad libitum. Handling was in accordance with an animal study protocol approved by the National Cancer Institute Animal Care and Use Committee. Groups of 8- to 12-week-old male mice were put on a synthetic purified diet (AIN-93G, Bio-Serv, NJ) for 5 days before experiment. Ppara-null mice and wild-type mice were fed with CA diet (AIN-93G supplemented with 1% CA) and control diet (AIN-93G) for 12 days based on previous studies (15–17). Urine samples were collected and food intake was measured from mice placed individually in metabowls (Jencons, Leighton Buzzard, UK) for 24 h, before treatment, and on days 1, 3, 6, 9, and 12 following the treatment. The mice had ad libitum access to diet and water in these metabolic chambers. All urine samples were stored at −80°C until analyzed. The serum, bile, liver, and kidney tissue were harvested and frozen at −80°C for further analysis at the end of the study.
UPLC-ESI-QTOFMS analysis
Samples for injection were prepared by diluting 20 µl urine with 180 µl 50% aqueous acetonitrile. Serum samples were prepared by 10 µl serum mixed with 190 µl 67% aqueous acetonitrile, and bile samples were prepared by 1 µl bile mixed with 1,000 µl 67% aqueous acetonitrile. The samples were vortexed for 5 min in shaker and centrifuged at 14,000 rpm for 20 min at 4°C to remove particulates and precipitate protein. The supernatant was transferred to an autosampler vial for analysis. A 5 µl aliquot of supernatant samples was injected into the system of ultraperformance liquid chromatography coupled quadrupole time-of-flight mass spectroscopy (UPLC-ESI-QTOFMS). The liquid chromatography system was ACQUITY UPLC® equipment (Waters) consisting of a reverse-phase 2.1 × 50 mm ACQUITY UPLC® BEH C18 1.7 µm column (Waters Corp., Milford, MA) with a gradient mobile phase comprising 0.1% formic acid solution (A) and acetonitrile containing 0.1% formic acid solution (B). The gradient was maintained at 100% A for 0.5 min, increased to 100% B over the next 7.5 min, and returned to 100% A in last 2 min. Data were collected in positive mode and negative mode on a Waters Q-TOF, which was operated in full-scan mode at m/z 100 to 1,000. Nitrogen was used as both cone gas (50 l/h) and desolvation gas (600 l/h). Source temperature and desolvation temperature were set at 120°C and 350°C, respectively. The capillary voltage and cone voltage were 3,000 and 20 V, respectively. Chlorpropamide (5 µM) was added in the sample as the internal standard.
Data processing and multivariate data analysis
Raw data from UPLC-ESI-QTOFMS system were processed using MarkerLynx software (Waters) to generate a data matrix consisting of peak areas corresponding to a unique m/z and retention time (RT) without normalization. After the generation of a multivariated data matrix, this data set was exported into SIMCA-P+12.0 (Umetrics, Kinnelon, NJ) for principal component analysis (PCA). The loadings scatter plots and the contribution lists were used to determine the candidate biomarkers in CA-fed Ppara-null mice compared with other group mice.
Identification and quantitation of urinary and serum metabolites
To identify the structure of high-contribution score metabolites, metabolomics databases (Madison Metabolomics Consortium Database and METLIN) were searched to find potential candidates. Seven Golden Rules (18) were used to calculate the mass error based on the elemental compositions of each metabolite. To confirm the identities of markers, authentic standards at 5 to 20 μM in 50% acetonitrile were compared with the urine and serum sample on the condition of MS/MS fragmentation with collision energy ramping from 15 to 35 V. Identities of the ions were further confirmed by comparison of fragmentation pattern and retention time with authentic compounds.
Quantitation of metabolites in urine and serum was performed using an ACQUITY UPLC system coupled with a XEVO triple-quadrupole tandem mass spectrometer (Waters). The detection and quantitation of biomarkers were accomplished by multiple reaction monitoring (MRM) mass spectrometry. The following MRM transitions were monitored for each metabolite: xanthurenic acid (206→156; ESI+), nicotinamide 1-oxide (139→106; ESI+), phenylacetylglycine (192→74; ESI−), HDOPA (361→121; ESI−), DHOPA (363→121; ESI−), creatinine (114→86; ESI+), creatine (132→90; ESI+), CA (407→343; ESI−), TCA (514→80; ESI−), LPC 16:0 (496→104; ESI+), and LPC 18:0 (524→104; ESI+). The concentrations of metabolites in urine and serum were determined by use of calibration curves constructed with authentic standards. To obtain the accurate concentration of metabolites, 0.5 μM chlorpropamide (277→111; ESI+) was used as the internal standard.
Gene expression analysis
Total RNA was extracted from approximately 100 mg portions of frozen liver and kidney using TRIzol reagent (Invitrogen, Carlsbad, CA). Quantitative real-time PCR (qPCR) was carried out using SYBR green PCR master mix (Superarray) in an ABI Prism 7900HT sequence detection system (Applied Biosystems). QPCR primer sequences are shown in the supplementary Table I. Measured mRNA levels were normalized to those of β-actin (liver) or 18S (kidney) rRNA and expressed as fold change relative to those of wild-type mice fed a control diet.
Serum chemistry
Serum alanine aminotransferase transaminase (ALT), alkaline phosphatase (ALP), and other physiological serum parameters were measured using the VetScan VS2 Comprehensive Diagnostic Profile (Abaxis, Inc., Union City, CA).
Histological analysis
Fresh livers were fixed in 10% neutral-buffered formalin, and then subjected to dehydration in different concentrations of alcohol and xylene for paraffin embedding. Four-micrometer serial sections were made through the entire tissue. Histological findings were examined using a light microscope after hematoxylin and eosin staining.
Statistical analysis
Experimental values are presented as mean ± SD. Statistical analysis was performed using GraphPad Prism (San Diego, CA). The significance of metabolite concentration and mRNA levels was determined using two-tailed Student t-test or the one-way ANOVA with Bonferroni correction for multiple comparisons. P values of less than 0.05 were considered significant.
RESULTS
Phenotypes of Ppara-null mice fed with CA diet
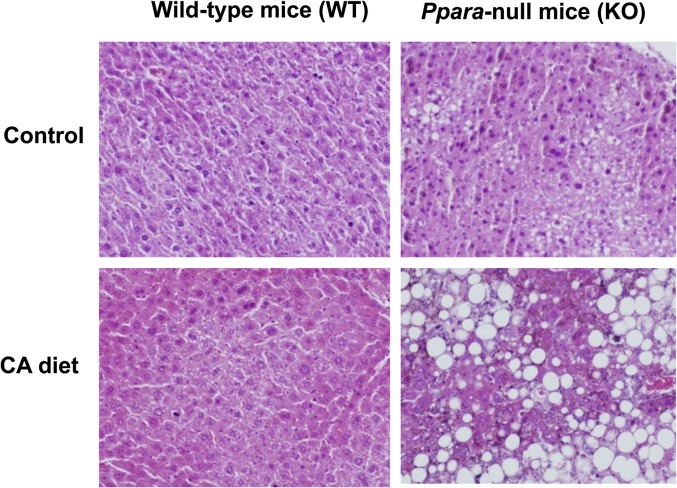
Male wild-type and Ppara-null mice fed a control diet showed no significant differences in body weight. However, Ppara-null mice exhibited significant body weight loss after feeding a CA diet for 12 days, whereas wild-type mice were slightly decreased (Table 1). There were no significant differences in food intake and urine volume between the four groups of mice (supplementary Fig. I-A, B). Histological analysis showed severe fatty liver and cholestasis in CA-treated Ppara-null mice (Fig. 1). In contrast, no fatty liver and cholestasis was induced in wild-type mice fed the CA diet. Further examination indicated that the major lipid composition of hepatic steatosis in CA-treated Ppara-null mice was cholesterol and triglyceride (supplementary Fig. II-A, B). In addition, serum chemistry analysis indicated that CA-treated Ppara-null mice showed higher ALP, ALT, and serum total bilirubin levels than CA-treated wild-type mice (Table 1). These results indicate that Ppara-null mice are susceptible to liver injury induced by dietary CA overload.
TABLE 1.
Physiological characteristics and serum chemistry
| Control |
CA-Fed |
|||
| Wild-type mice | Ppara-null mice | Wild-type mice | Ppara-null mice | |
| Body mass (%) | 100 ± 10.2 | 104 ± 1.7 | 91.1 ± 6.1 | 67 ± 0.5a |
| Liver/body mass (%) | 3.2 ± 0.1 | 3.6 ± 0.2 | 3.6 ± 0.2 | 6.1 ± 0.7a |
| ALT (U/l) | 28.5 ± 4.4 | 29 ± 8.7 | 335 ± 148 | 3660 ± 389a |
| ALP (U/l) | 104 ± 29.2 | 65 ± 12.4 | 67 ± 6.8 | 396 ± 63.3a |
| Glucose (mg/dl) | 198 ± 29 | 150 ± 46.4 | 157 ± 16.2 | 85 ± 8.4a |
| Total bilirubin (mg/dl) | 0.4 ± 0 | 0.4 ± 0 | 0.4 ± 0.1 | 0.7 ± 0.1b |
| BUN (mg/dl) | 18 ± 3.6 | 21 ± 3.8 | 20.5 ± 3.4 | 30 ± 3.3a |
| Potassium (mmol/l) | 7.1 ± 0.5 | 7.7 ± 0.6 | 7.2 ± 0.5 | 10.9 ± 0.6a |
| Total protein (g/dl) | 6 ± 0.7 | 5.6 ± 0.1 | 5.6 ± 0.3 | 4.9 ± 0.2b |
Comparison of diagnostic biomarkers between wild-type and Ppara-null mice fed with CA diet (n = 4). The percentage of body weight was calculated by comparing the body weight at day 12 to the starting weight at day 0. The liver/body mass was calculated by comparing the liver weight at day 12 to the body weight at day 12.
P< 0.01.
P< 0.05.
Fig. 1.
Histological examination of liver. Liver sections were stained by the hematoxylin and eosin method. CA treatment caused severe fatty liver and cholestasis in Pparα-null mice, but wild-type mice showed no liver abnormalities following CA treatment.
Metabolomic analysis of mouse bile, urine, and serum
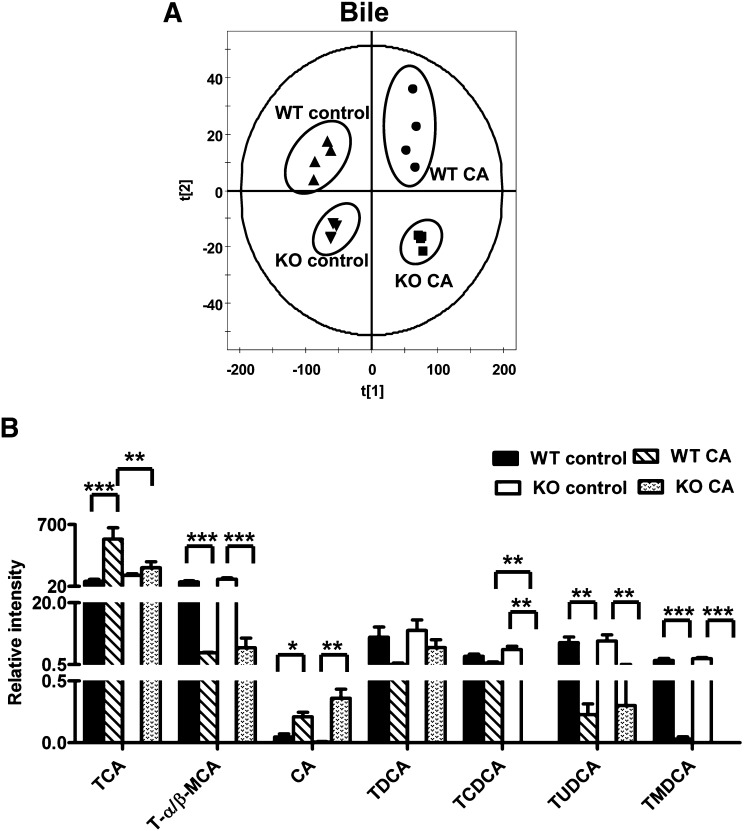
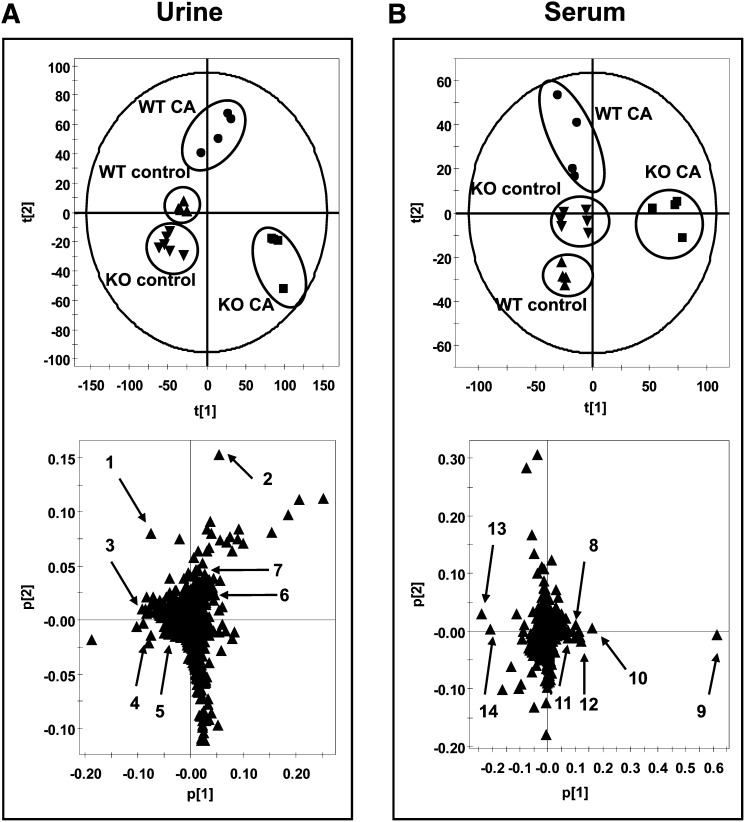
To gain an understanding for why Ppara-null mice were more sensitive to CA challenge than wild-type mice, metabolomic analysis was used to profile the metabolites in the serum, urine, and bile from the four groups. Unsupervised PCA was used to analyze the data sets from the control and CA-fed groups in both wild-type and Ppara-null mice. The PCA model shows that bile samples between wild-type and Ppara-null mice fed with control or CA diet were distributed in four different quadrants. CA Treatment can significantly increase bile volume in both wild-type and Ppara-null mice gallbladder; however, there were no significant differences in bile volume between wild-type and Ppara-null mice or between CA-treated wild-type and Ppara-null mice (supplementary Fig. I-C). These data suggested that different bile compositions were present in the four groups of mice (Fig. 2A). Significant separation of urine and serum samples were observed in the CA-treated group and control group on day 12 (Fig. 3A, B). Compared with the urine sample, a more significant difference was observed between CA-treated Ppara-null and wild-type mouse serum in component 1 (X-axis) of the PCA model, suggesting that serum metabolites were more directly affected by CA treatment in Ppara-null mice.
Fig. 2.
Metabolomics analysis of bile in control and CA-treated wild-type and Pparα-null mice. (A) Scores plot of an PCA model of bile ions from wild-type (WT control, triangle; CA-fed, dot) and Pparα-null (KO control, inverted triangle; CA-fed, box) mice fed with control diet or CA diet for 12 days. Each point represents an individual mice bile sample. The t[1] and t[2] correspond to principal components 1 and 2, respectively. Four groups are distributed in four different quadrants of the PCA model. (B) The level of bile acids in bile. *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. 3.
Identification of urine and serum metabolites significantly altered in Pparα-null mice with CA diet treatment using UPLC-ESI-QTOFMS-based metabolomics. (A) Scores plot of a PCA model and PCA loadings scatter plot of urinary ions from wild-type (WT control, triangle; CA-fed, dot) and Pparα-null (KO control, inverted triangle; CA-fed, box) mice fed with control diet or 1% CA diet for 12 days. Each point represents an individual mouse urine sample (top) and a urinary ion (bottom). Urine metabolites are labeled in the scatter plot (1–7). (B) Scores plot of an PCA model and PCA loadings scatter plot of serum ions from wild-type (control, closed triangle; CA-fed, closed circle) and Pparα-null (control, open triangle; CA-fed, open circle) mice fed with control diet or 1% CA diet for 12 days. Each point represents an individual mouse serum sample (top) and a serum ion (bottom). Serum metabolites are labeled in the scatter plot (8–14). The t[1] and t[2] correspond to principal components 1 and 2, respectively. The p[1] values represent the interclass difference, and p[2] values represent the relative abundance of the ions. Urinary data were obtained in positive mode (ESI+), while serum data were obtained in negative mode (ESI−).
After PCA modeling of wild-type and Ppara-null mice in the CA- and control-fed groups, a loadings scatter plot revealed ions with the greatest contribution to separation between CA-fed Ppara-null mice and other groups. The significant ions increased in Ppara-null mice after CA loading were in the first and fourth quadrants, and they decreased in the second and third quadrants (Fig. 3A, B). To identify the ions that contribute to the separation, accurate mass values of each ion were used for chemical formula calculations using element composition and mass-based searches in metabolomics databases (Madison Metabolomics Consortium Database and METLIN). The ions m/z 114.0662+, 132.0783+, 139.0500+, 361.2002+, 363.2171+, and 206.0453+ in urine correspond to creatine, creatinine, nicotinamide 1-oxide, HDOPA, DHOPA, and xanthurenic acid. The ions m/z 407.2795−, 391.2827−, 514.2827−, 514.2820−, 498.2885−, 540.3303−, and 568.3623− in serum correspond to CA, CDCA, TCA, T-α/β-MCA-MCA, TCDCA, LPC 16:0, and LPC 18:0 (Table 2).
TABLE 2.
Summary of ions altered in CA-treated Ppara-null mice compared with CA-treated wild-type mice
| No | RT (min) | Mass (m/z) | Empirical Formula | Mass Error (ppm) | Identity | Up/Down | Fold Change |
| Urine | |||||||
| 1 | 0.32 | 132.0783 | C4H9N3O2 [H+] | 7.6 | Creatine | ↑ | 8.4 |
| 2 | 0.31 | 114.0662 | C4H7N3O [H+] | −4.4 | Creatinine | ↓ | 2.6 |
| 3 | 1.80 | 206.0453 | C10H7NO4 [H+] | 0 | Xanthurenic acid | ↓ | 3.2 |
| 4 | 0.33 | 139.0500 | C6H6N2O2 [H+] | −5.7 | Nicotinamide 1-oxide | ↓ | 8.3 |
| 5 | 2.61 | 194.0812 | C10H11NO3 [H+] | −2.5 | Phenylacetylglycine | ↓ | 3.9 |
| 6 | 5.33 | 361.2002 | C21H28O5 [H+] | −3.6 | HDOPA | ↑ | 20 |
| 7 | 5.22 | 363.2171 | C21H30O5 [H+] | 0 | DHOPA | ↑ | 3.6 |
| Serum | |||||||
| 8 | 3.53 | 407.2795 | C24H40O5 [H–] | −0.5 | CA | ↑ | 25 |
| 9 | 2.92 | 514.2827 | C26H45NO7S [H–] | −2.1 | TCA | ↑ | 16 |
| 10 | 2.56 | 514.2820 | C26H45NO7S [H–] | −3.5 | T-α/β-MCA | ↑ | 12 |
| 11 | 4.13 | 391.2827 | C24H40O4 [H–] | −5.3 | CDCA | ↑ | 7 |
| 12 | 3.40 | 498.2885 | C26H45NO6S [H–] | −0.8 | TCDCA | ↑ | 10 |
| 13 | 5.38 | 568.3623 | C27H56NO9P [H–] | 1.6 | LPC 18:0 | ↓ | 1.6 |
| 14 | 4.76 | 540.3303 | C25H52NO9P [H–] | 0.4 | LPC 16:0 | ↓ | 1.6 |
↑, upregulation; ↓, downregulation.
Disrupted bile acid homeostasis in CA-treated Ppara-null mice
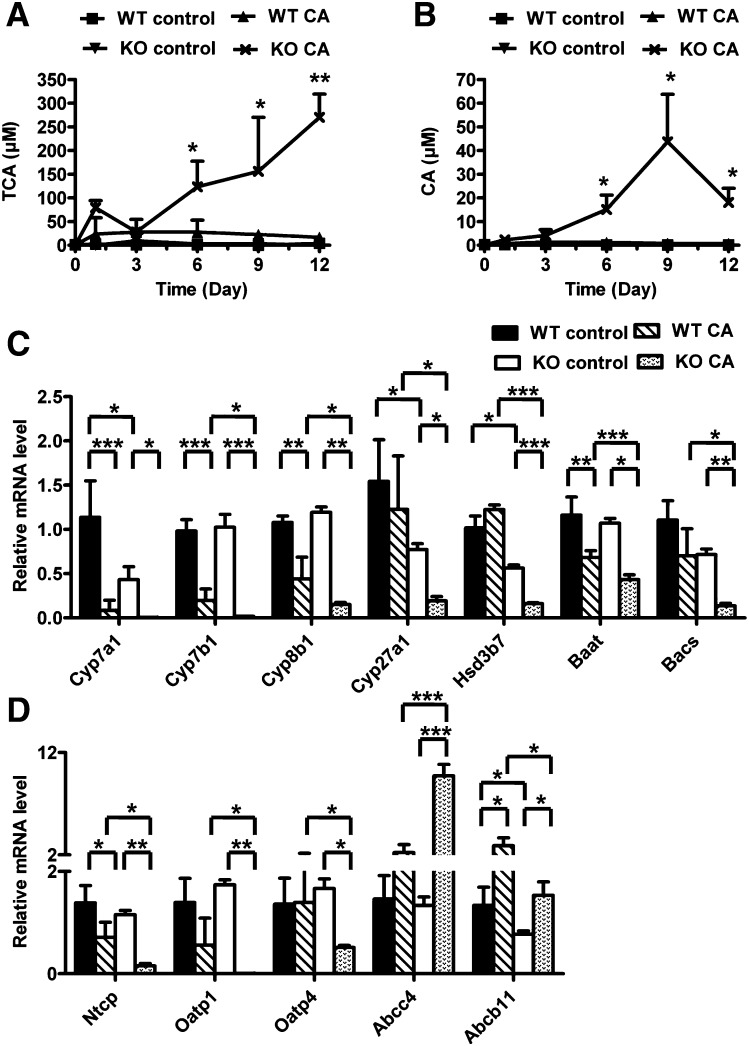
Metabolomic analysis indicated that TCA was the most important ion increased in the serum of CA-fed Ppara-null mice. The level of TCA was increased from day 6 of treatment, and the concentration was nearly 16-fold higher at day 12 of treatment in CA-fed Ppara-null mice compared with CA-fed wild-type mice (Fig. 4A). Along with TCA, CA was markedly elevated in CA-fed Ppara-null mice from day 6 (P < 0.05) to 12 (P < 0.01) (Fig. 4B). On day 12, there was a 25-fold increase in CA-treated Ppara-null mice compared with CA-treated wild-type mice. In addition, the levels of serum CDCA, T-α/β-MCA, and TCDCA were increased at day 12 of treatment in Ppara-null mice fed with CA diet (data no shown). Further gene expression analysis indicated that the major enzymes involved in the bile acid biosynthesis, including Cyp7a1, Cyp7b1, Cyp8b1, Cyp27a1, and hydroxy-delta-5-steroid dehydrogenase 3b7 (Hsd3b7) were decreased in the CA-fed Ppara-null mice at day 12 of treatment compared with CA-treated wild-type mice and untreated Ppara-null mice (Fig. 4C). Two enzymes involved in the bile acid conjugation, bile acid-CoA synthetase (Bacs, also called Slc27a5) and bile acid-CoA amino acid N-acetyltransferase (Baat), were significantly diminished in the liver of CA-fed Ppara-null mice. The bile acid transporters sodium-taurocholate cotransporting polypeptide (Ntcp), anion transporting polypeptide 1 (Oatp1), and anion transporting polypeptide 4 (Oatp4), which absorb bile acids from blood into liver, were decreased in Ppara-null mice, whereas the transporter Abcc4, which carries bile acids from liver to blood, was dramatically increased (Fig. 4D). In addition, the mRNA encoding bile acid transporter ATP-binding cassette subfamily B member 11 (Abcb11, also called Bsep) located in liver canaliculus that transports bile acids from liver to bile duct, was increased by CA treatment in wild-type and Ppara-null mice. However, lower levels of Abcb11 were observed in CA-treated Ppara-null mice compared with CA-treated wild-type mice. Metabolomic analysis of bile indicated that CA treatment significantly changes the bile acid composition of bile in both wild-type and Ppara-null mice, notably T-α/β-MCA, TUDCA, and TMDCA (Fig. 2B). Since CA treatment significantly altered the bile volume in both wild-type and Ppara-null mice, the relative concentrations of bile acids in bile were determined (Fig. 2B). However, compared with CA-treated wild-type mice, TCA and TCDCA were lower in CA-treated Ppara-null mice. These results suggested that the bile acid homeostasis was markedly disrupted in CA-treated Ppara-null mice compared with CA-treated wild-type mice and control mice.
Fig. 4.
Effect of CA diet on bile acid metabolism in Pparα-null mice. (A, B) Serum levels of CA (A) and TCA (B) between wild-type (WT) and Pparα-null (KO) mice fed with control diet and CA diet from day 0 to 12. (C) Hepatic mRNA levels of genes associated with bile acid biosynthesis and transport in mice with day 12 of treatment. *P < 0.05; **P < 0.01; ***P < 0.001.
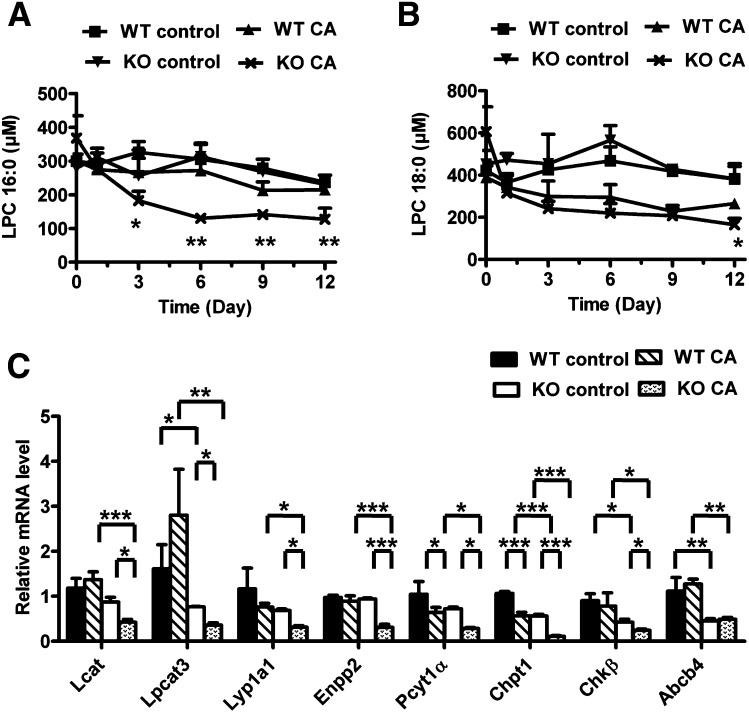
Alteration of phospholipid homeostasis in CA-treated Ppara-null mice
Along with the accumulation of bile acids, metabolomic analysis showed the decreased levels of two phospholipids, LPC 16:0 and LPC 18:0 in the serum of CA-treated Ppara-null mice. Triplequadrupole quantitation further confirmed that the level of LPC 16:0 was decreased in CA-treated Ppara-null mice from day 3 (P < 0.05) to 12 (P < 0.01) (Fig. 5A). Compared with LPC 16:0, the level of LPC 18:0 was significantly reduced in CA-treated Ppara-null mice on day 12 (P < 0.05) (Fig. 5B). On day 12, there was a 1.6-fold depletion of both phospholipids in CA-treated Ppara-null mice compared with CA-treated wild-type mice. Phospholipids have been reported to be biomarkers for LCA induced cholestasis (19). Liver gene expression indicated that the mRNAs encoding enzymes involved in the LPC biosynthesis [lecithin-cholesterol acyltransferase (Lcat), phosphate cytidylyltransferase 1 (Pcyt1α), choline phosphotransferase 1 (Chpt1), and choline kinase β (Chkβ)] were significantly decreased in CA-treated Ppara-null mice compared with CA-treated wild-type mice and untreated Ppara-null mice (Fig. 5C). Similarly, the expression of mRNAs encoding proteins associated with LPC metabolism [lysophosphatidylcholine acyltransferase 3 (Lpcat3), lysophospholipase A1 (Lyp1a1), and ectonucleotide pyrophosphatase/ phosphodiesterase 2 (Enpp2)] were reduced in CA-treated Ppara-null mice (Fig. 5C). In addition, the mRNA encoding the phospholipid transporter ATP-binding cassette subfamily B member 4 (Abcb4) located in the liver canaliculus, was diminished in CA-treated Ppara-null mice compared with CA-treated wild-type mice. These data indicate that phospholipids homeostasis was also disrupted in CA-treated Ppara-null mice.
Fig. 5.
Effect of CA diet on phospholipid metabolism in Pparα-null mice. (A, B) Serum levels of LPC 16:0 (A) and 18:0 (B) between wild-type (WT) and Pparα-null (KO) mice fed control diet and CA diet from day 0 to 12. (C) Hepatic mRNA levels of genes associated with LPC biosynthesis, metabolism, and transport in mice with day 12 of treatment. *P < 0.05; **P < 0.01; ***P < 0.001.
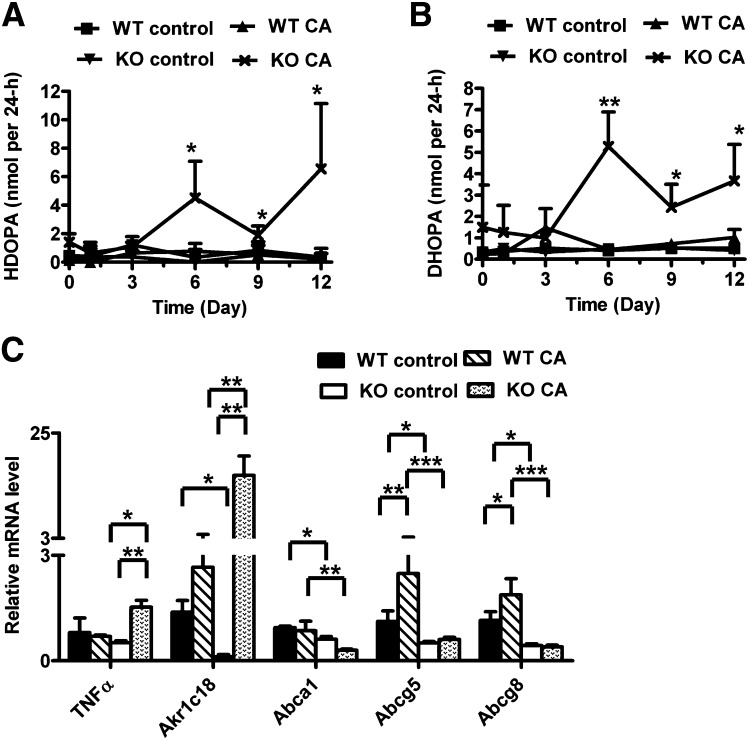
Change in cholesterol and corticosterone metabolism in CA-treated Ppara-null mice
The generation of bile acids from cholesterol is a metabolic pathway for cholesterol degradation in vivo. To determine whether other metabolic pathways involving cholesterol were also affected, the metabolites associated with cholesterol and steroid hormone metabolism were examined in serum and urine. Although there was no significant change in serum corticosterone and progesterone levels (supplementary Fig. III), two corticosterone derivatives, HDOPA and DHOPA, showed a significant increase in urine of CA-treated Ppara-null mice (Fig. 6A, B). Both metabolites were significantly increased from day 6 to 12. On day 12, HDOPA and DHOPA were increased by 20- and 3.6-fold, respectively, in CA-treated Ppara-null mice compared with CA-treated wild-type mice. In addition, hepatic expression levels of the PPARα target gene aldo-keto reductase family 1c18 (Akr1c18) was significantly diminished in the control diet-treated Ppara-null mice compared with wild-type mice (20). The mRNA encoding Akr1c18 involved in the generation of HDOPA and DHOPA was greatly increased (6-fold) in CA-treated Ppara-null mice (Fig. 6C), whereas genes associated with corticosterone metabolism, such as aldo-keto reductase family 1c14 (Akr1c14), steroid 5 α-reductase 1 (Srd5a1), hydroxysteroid (17-β) dehydrogenase 3 (Hsd17b3), and aldehyde dehydrogenase family 3, subfamily A2 (Aldh3a2), were decreased (supplementary Fig. IV). These data suggested that enhanced Akr1c18 expression was responsible for the increase of HDOPA and DHOPA in the CA-treated Ppara-null mice urine. In addition, three cholesterol transporters, ATP-binding cassette subfamily A member 1 (Abca1), ATP-binding cassette subfamily G member 5 (Abcg5), and ATP-binding cassette subfamily G member 8 (Abcg8), were significantly decreased in CA-treated Ppara-null mice compared with CA-treated wild-type mice. Although the mRNA levels of Abcg5 and Abcg8 are elevated by CA treatment in wild-type mice, there was no significant difference between CA-treated and untreated Ppara-null mice.
Fig. 6.
Effect of CA diet on cholesterol and corticosterone metabolism in Pparα-null mice. (A, B) Urinary excretion of HDOPA (A) and DHOPA (B) between wild-type (WT) and Pparα-null (KO) mice fed with control diet and CA diet from day 0 to 12. (C) Hepatic mRNA level of genes associated with corticosterone metabolism and cholesterol transport in mice with day 12 of treatment. *P < 0.05; **P < 0.01; ***P < 0.001.
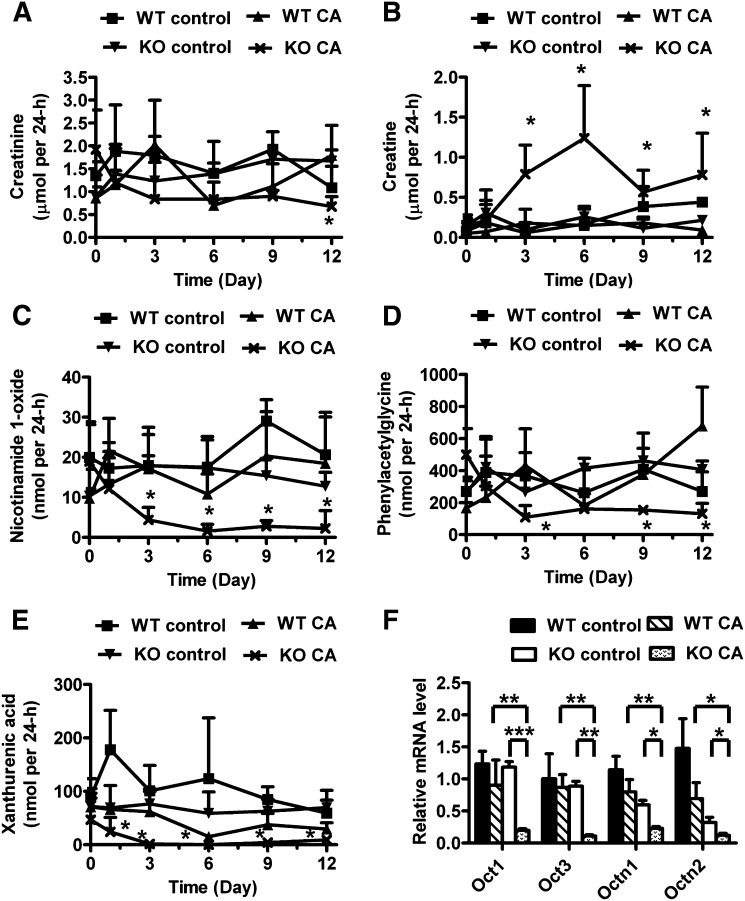
Disorder of cation and anion transport in CA-treated Ppara-null mice
Along with the increase in the HDOPA and DHOPA excretion, metabolomic analysis revealed altered levels of endogenous cationic and anionic metabolites, such as nicotinamide 1-oxide and xanthurenic acid, in urine of CA-treated Ppara-null mice. As shown in Fig. 7A, the excretion of creatinine was significantly decreased in CA-treated Ppara-null mice. Therefore, the level of urinary metabolites was presented as total mass in 24 h. The excretion of creatine, one endogenous cation, was significantly increased in CA-treated Ppara-null mice from day 3 (P < 0.05) to 12 (P < 0.05) (Fig. 7B), whereas creatinine excretion was decreased on day 12 (P < 0.05) (Fig. 7A). The excretion of another cation nicotinamide 1-oxide also was significantly reduced from day 3 (P < 0.05) to 12 (P < 0.05) (Fig. 7C). Similarly, the urinary excretion of two endogenous anion metabolites, phenylacetylglycine (3.9-fold) and xanthurenic acid (3.2-fold), was decreased in CA-treated Ppara-null mice on day 12 (Fig. 7D, E). qPCR analysis indicated that the expression of several organic cation transporters (Octs) and organic anion transporters (Oats) was dramatically diminished in the liver and kidney of CA-treated Ppara-null mice compared with untreated Ppara-null mice and CA-treated wild-type mice, including organic cation transporter 1 (Oct1/Slc22A1), organic cation transporter 2 (Oct2/Slc22A2), organic cation transporter 3 (Oct3/Slc22A3), organic cation/carnitine transporter 1 (Octn1/Slc22A4), organic cation/carnitine transporter 2 (Octn2/Slc22A5), Oatp1, and Oatp4 (Figs. 3D and 7F and supplementary Fig. V). These results demonstrated that CA challenge also leads to a disruption of endogenous cation and anion transport in Ppara-null mice.
Fig. 7.
Effect of CA diet on endogenous cations and anions transport in Pparα-null mice. (A–E) Urinary excretion of creatinine (A), creatine (B), nicotinamide 1-oxide (C), phenylacetylglycine (D), and xanthurenic acid (E) between wild-type (WT) and Pparα-null (KO) mice fed with control diet and CA diet in 24 h from day 0 to 12. (F) Hepatic mRNA levels of organic cation transporters (Octs) in mice with day 12 of treatment. *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
To clarify the functional role of PPARα in bile acid homeostasis, the present study investigated the change in endogenous metabolites and associated genes in Ppara-null mice during exposure to a CA diet. Metabolomics indicated disruption of bile acid homeostasis, phospholipid homeostasis, cholesterol metabolism, and endogenous cations and anions transport in CA-treated Ppara-null mice. Further gene expression analysis revealed that the bile acid disorder in CA-treated Ppara-null mice might be due to a deficiency in expression of the Abc transporters (Abcb11, Abcb4, Abca1, Abcg5, and Abcg8) in the liver canliculus. Additionally, the levels of bile acid synthesis enzymes (Cyp7a1, Cyp27a1, and Hsd3b7) were lower in untreated Ppara-null mice than in untreated wild-type mice. These results demonstrated that PPARα regulates bile acid homeostasis not only via its effect on bile acid biosynthesis and transport but also via its effect on biliary secretion.
When Ppara-null mice were treated with a CA diet, one significant observation was liver injury. The change in phenotype of the CA-treated Ppara-null mice included body weight loss, enhanced serum ALP and ALT levels, and cholestasis. These phenotypes are similar with those previously reported for the farnesoid X receptor (Fxr)-null mice after feeding CA (21). In addition, higher total bilirubin and lower glucose levels were also observed in CA-fed Ppara-null mice. Further metabolomic analysis revealed an alteration of three important metabolic pathways, bile acid metabolism, phospholipid metabolism, and cholesterol metabolism, in CA-treated Ppara-null mice. First, a high level of bile acids, including CA, CDCA, TCA, T-α/β-CA, and TCDCA, were noted in CA-treated Ppara-null mouse serum, and the genes involved in bile acid synthesis and transport were diminished in the liver, thus suggesting significant liver injury after feeding Ppara-null mice CA. Numerous studies have demonstrated that increased levels of serum bile acids were induced in cholestasis models (16, 22, 23). Second, two phospholipids, LPC 16:0 and LPC 18:0, were decreased in CA-treated Ppara-null mice serum. LPCs, which are derived from phosphatidylcholine by hydrolysis, are the major phospholipid species in serum. Phospholipids are important components in bile that are sensitive to liver bile acid levels. In the clinic, abnormal levels of serum LPCs are typically associated with liver injury (24, 25). Similarly, it was reported that a series of LPCs were significantly decreased in cholestasis induced by LCA treatment and nonalcoholic steatohepatitis induced by methionine- and choline-deficient diet treatment (19, 26). Third, the excretion of HDOPA and DHOPA was increased in the CA-treated Ppara-null mice urine. HDOPA and DHOPA were generated from corticosterone under the effect of cytochrome P450s (Cyps), which was confirmed by in vitro and in vivo experiments (9, 20). Recently, HDOPA and DHOPA were reported as biomarkers for PPARα activation (9). However, the expression of PPARα target genes suppressed in Ppara-null mice suggested that the increase in both metabolites in this study was not likely due to the activation of PPARα. It was reported that in cholestastic animals and humans, the hypothalamic-pituitary-adrenal axis can be activated by endotoxin and cytokines, such as tumor necrosis factor α (TNFα), resulting in the elevation of glucocorticoids (27). Two studies indicated that the excretion of HDOPA and DHOPA was increased in the hepatotoxicity model of hepatocyte nuclear factor 1α (Hnf1α)-null mice (28) and CA-treated Fxr-null mice (21). Here, the expression of TNFα was activated in CA-treated Ppara-null mice, suggesting that accelerated corticosterone metabolism in CA-treated Ppara-null mice also results from liver cholestasis. The change in the bile acids, phospholipids, and cholesterol homeostasis provides compelling evidence that hepatotoxicity was induced in Ppara-null mice in response to CA challenge.
Another observation in this study is the disrupted transport of endogenous cations and anions in the liver and kidney. The possibility exists that altered levels of creatine, creatinine, and nicotinamide 1-oxide observed in the CA-treated Ppara-null mice is due to inhibition of Octs function in vivo. In the liver and kidney, the Octs family mediates the reabsorption and excretion of various cationic compounds, including endogenous cations (choline and carnitine), cationic toxins (1-methyl-4-phenylpyridium), and cationic drugs (quinidine and procainamide) (29). Several studies have shown that Oct2 plays an important role in renal secretion of creatinine in humans and mice (30, 31). Additionally, methylnicotinamide, used as a model substrate for studying organic cation transport, can be regulated by Oct1 and Oct2 (32, 33). In vivo, nicotinamide 1-oxide and methylnicotinamide are derived from nicotinamide. These studies suggest that inhibition of Octs might directly affect the transport of creatinine, creatine, and nicotinamide 1-oxide in vivo. Additionally, two organic anionic metabolites, phenylacetylglycine and xanthurenic acid, were significantly decreased in CA-treated Ppara-null mice. A recent study using untargeted metabolomics revealed that both metabolites were identified as biomarkers associated with the deficiency of Oat1 (34). Another earlier study conducted by gas chromatography-mass spectrometry (GC-MS), also indicated that the excretion of endogenous organic anions was decreased in Oat1 knockout mouse urine (35). Here, CA treatment decreased Oatp1 and Oatp4 in CA-fed Ppara-null mouse liver and kidney. Oatp1 mRNA is found in various tissues (36) including the liver, kidney, and brain, whereas Oatp4 is exclusively in the liver (37). Along with the transport of bile acids, both transporters play a major role in the uptake and secretion of other organic anions in the kidney and liver, such as bromosulfophthalein and estrone sulfate (38, 39). Thus, it is likely that the diminished expression of Oatp1 and Oatp4 in the liver and kidney is responsible for the decrease of phenylacetylglycine and xanthurenic acid in urine.
Previous studies have demonstrated that the activation/repression of PPARα can affect bile acid biosynthesis. However, different activators, such as Wy-14,643 and fibrates, show completely different effects on the rate-limiting enzyme in bile acid synthesis enzyme Cyp7a1 (3-5). Compared with fenofibrate and other fibrates, Wy-14,643 is a more potent ligand of PPARα. One possible reason for different effects is that the level of Cyp7a1 might be regulated by other nuclear receptors, such as liver X receptor (LXR) (40) and hepatocyte nuclear factor 4α (HNF4α), during the activation of PPARα (41, 42). In this study, in comparison with control diet-treated wild-type mice, the enzymes involved in biosynthesis of bile acids from cholesterol were reduced in control diet-treated Ppara-null mice, including Cyp7a1, Cyp27a1, and Hsd3b7. One early study reported that the mRNA level of Cyp7a1 also could be reduced in Ppara-null mice compared with wild-type mice (3). Although earlier studies showed that ciprofibrate (43) and perfluorinated fatty acids (7) cause a decrease in hepatic transporters Ntcp and Oatp1, the level of Ntcp and Oatps were unchanged in control diet-treated Ppara-null mice compared with wild-type mice. In the liver, the expression of Abcb11 was decreased in control diet-treated Ppara-null mice, consistent with the observations that Ppara-null mice show lower hepatic Abcb11 level than wild-type mice (5). Thus, the deficiency of PPARα can directly affect the bile acid transporter Abcb11 in the liver. More importantly, the mRNA levels of the phospholipid transporter Abcb4 and cholesterol transporters Abca1, Abcg5, and Abcg8 also were diminished in the liver following knockout of PPARα. Various studies have indicated that treatment with Wy-14,643 and other PPARα activators can stimulate the expression of Abcb4, Abca1, Abcg5, and Abcg8 (43–45). Actually, it was reported that PPARα can regulate hepatic lipid homeostasis via its target effect on these Abc transporters (46, 47). During CA challenge, the reduced mRNA expression of these Abc transporters greatly affected the transport of bile acids in Ppara-null mice, finally causing severe liver cholestasis. Taken together, the deficiency of PPARα can cause decreased bile acid biosynthesis, transport, and secretion.
Although the expression of several genes involved in the bile acid metabolism was reduced in Ppara-null mice, metabolomics did not reveal abnormal bile acid levels in untreated urine and serum in these mice. To better evaluate the role of PPARα in the regulation of bile acid homeostasis, CA challenge was conducted in Ppara-null mice. The results showed severe hepatotoxicity in Ppara-null mice during CA challenge that was not observed in wild-type mice. Numerous studies have reported that CA challenge can lead to severe liver injury in transgenic mice in which specific genes involved in bile acid metabolism had been knocked out, such as Fxr-null mice (48), Abcb11-null mice (49), and Shp-null mice (50). The current studies demonstrated a key role of PPARα in the regulation of bile acid homeostasis. The hepatotoxicity in CA-treated Ppara-null mice might be due to the decrease in expression of the Abc transporters in the liver canaliculus. The reduced biliary secretion causes the accumulation of bile acids in Ppara-null mice liver during CA exposure. Indeed, a metabolic disorder was observed in CA-treated Ppara-null by metabolomics, including altered bile acid metabolism, phospholipid metabolism, cholesterol metabolism, and transport of endogenous cations and anions. The data presented herein demonstrate the power of mass spectrometry-based metabolomic analysis in combination with traditional serum chemistry analysis and histological examination using a transgenic mouse model to profile metabolic pathways and uncover the physiological function of nuclear receptor.
Supplementary Material
Footnotes
Abbreviations:
- Abst
- apical sodium-dependent bile salt transporter
- ALT
- alanine aminotransferase
- ALP
- alkaline phosphatase
- Bacs
- bile acid-CoA synthetase
- Baat
- bile acid-CoA amino acid N-acetyltransferase
- BUN
- blood urea nitrogen
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- Cyp
- cytochrome P450
- DHOPA
- 11β,20α-dihydroxy-3-oxo-pregn-4-en-21-oic acid
- Enpp2
- ectonucleotide pyrophosphatase/phosphodiesterase 2
- Fxr
- farnesoid X receptor
- HDOPA
- 11β
- -Hydroxy-3
- 20-dioxopregn-4-en-21-oic acid
- Hsd
- hydroxy-delta-5-steroid dehydrogenase
- LPC
- lysophosphatidylcholine
- Lpcat
- lysophosphatidylcholine acyltransferase
- Lypla1
- lysophospholipase A1
- Oat
- organic anion transporter
- Oatp1
- organic anion transporting polypeptide 1
- Ntcp
- sodium-taurocholate cotransporting polypeptide
- Oct
- organic cation transporter
- PPARα
- peroxisome proliferator-activated receptor α
- PCA
- principal component analysis
- RT
- retention time
- Slc
- solute carrier family
- TCA
- taurocholic acid
- TCDCA
- taurochenodeoxycholic acid
- T-α/β-MCA
- tauro-α/β-muricholic acid
- TDCA
- taurodeoxycholic acid
- TMDCA
- tauromurideoxycholic acid
- TNFα
- tumor necrosis factor α
- TUDCA
- tauroursodeoxycholic acid
- UPLC-ESI-QTOFMS
- ultraperformance liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry
- Wy-14
- 643, [4-chloro-6-(2,3-xylidino)-2-pyrimidinylthio] acetic acid
This work was supported in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures and one tables.
REFERENCES
- 1.Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V., Li Y., Goetz R., Mohammadi M., Esser V., et al. 2007. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 5: 415–425 [DOI] [PubMed] [Google Scholar]
- 2.Mandard S., Muller M., Kersten S. 2004. Peroxisome proliferator-activated receptor alpha target genes. Cell. Mol. Life Sci. 61: 393–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunt M. C., Yang Y. Z., Eggertsen G., Carneheim C. M., Gafvels M., Einarsson C., Alexson S. E. 2000. The peroxisome proliferator-activated receptor alpha (PPARalpha) regulates bile acid biosynthesis. J. Biol. Chem. 275: 28947–28953 [DOI] [PubMed] [Google Scholar]
- 4.Cheema S. K., Agellon L. B. 2000. The murine and human cholesterol 7alpha-hydroxylase gene promoters are differentially responsive to regulation by fatty acids mediated via peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 275: 12530–12536 [DOI] [PubMed] [Google Scholar]
- 5.Hays T., Rusyn I., Burns A. M., Kennett M. J., Ward J. M., Gonzalez F. J., Peters J. M. 2005. Role of peroxisome proliferator-activated receptor-alpha (PPARalpha) in bezafibrate-induced hepatocarcinogenesis and cholestasis. Carcinogenesis. 26: 219–227 [DOI] [PubMed] [Google Scholar]
- 6.Post S. M., Duez H., Gervois P. P., Staels B., Kuipers F., Princen H. M. 2001. Fibrates suppress bile acid synthesis via peroxisome proliferator-activated receptor-alpha-mediated downregulation of cholesterol 7alpha-hydroxylase and sterol 27-hydroxylase expression. Arterioscler. Thromb. Vasc. Biol. 21: 1840–1845 [DOI] [PubMed] [Google Scholar]
- 7.Cheng X., Klaassen C. D. 2008. Critical role of PPAR-alpha in perfluorooctanoic acid- and perfluorodecanoic acid-induced downregulation of Oatp uptake transporters in mouse livers. Toxicol. Sci. 106: 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung D., Fried M., Kullak-Ublick G. A. 2002. Human apical sodium-dependent bile salt transporter gene (SLC10A2) is regulated by the peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 277: 30559–30566 [DOI] [PubMed] [Google Scholar]
- 9.Zhen Y., Krausz K. W., Chen C., Idle J. R., Gonzalez F. J. 2007. Metabolomic and genetic analysis of biomarkers for peroxisome proliferator-activated receptor alpha expression and activation. Mol. Endocrinol. 21: 2136–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patterson A. D., Bonzo J. A., Li F., Krausz K. W., Eichler G. S., Aslam S., Tigno X., Weinstein J. N., Hansen B. C., Idle J. R., et al. 2011. Metabolomics reveals attenuation of the SLC6A20 kidney transporter in nonhuman primate and mouse models of type 2 diabetes mellitus. J. Biol. Chem. 286: 19511–19522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F., Patterson A. D., Hofer C. C., Krausz K. W., Gonzalez F. J., Idle J. R. 2011. A comprehensive understanding of thioTEPA metabolism in the mouse using UPLC-ESI-QTOFMS-based metabolomics. Biochem. Pharmacol. 81: 1043–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F., Patterson A. D., Hofer C. C., Krausz K. W., Gonzalez F. J., Idle J. R. 2010. Comparative metabolism of cyclophosphamide and ifosfamide in the mouse using UPLC-ESI-QTOFMS-based metabolomics. Biochem. Pharmacol. 80: 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson A. D., Slanar O., Krausz K. W., Li F., Hofer C. C., Perlik F., Gonzalez F. J., Idle J. R. 2009. Human urinary metabolomic profile of PPARalpha induced fatty acid beta-oxidation. J. Proteome Res. 8: 4293–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C., Krausz K. W., Shah Y. M., Idle J. R., Gonzalez F. J. 2009. Serum metabolomics reveals irreversible inhibition of fatty acid beta-oxidation through the suppression of PPARalpha activation as a contributing mechanism of acetaminophen-induced hepatotoxicity. Chem. Res. Toxicol. 22: 699–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J. 2000. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 102: 731–744 [DOI] [PubMed] [Google Scholar]
- 16.Teng S., Piquette-Miller M. 2007. Hepatoprotective role of PXR activation and MRP3 in cholic acid-induced cholestasis. Br. J. Pharmacol. 151: 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song P., Zhang Y., Klaassen C. D. 2011. Dose-response of five bile acids on serum and liver bile Acid concentrations and hepatotoxicty in mice. Toxicol. Sci. 123: 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kind T., Fiehn O. 2007. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 8: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsubara T., Tanaka N., Patterson A. D., Cho J. Y., Krausz K. W., Gonzalez F. J. 2011. Lithocholic acid disrupts phospholipid and sphingolipid homeostasis leading to cholestasis in mice. Hepatology. 53: 1282–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T., Shah Y. M., Matsubara T., Zhen Y., Tanabe T., Nagano T., Fotso S., Krausz K. W., Zabriskie T. M., Idle J. R., et al. 2010. Control of steroid 21-oic acid synthesis by peroxisome proliferator-activated receptor alpha and role of the hypothalamic-pituitary-adrenal axis. J. Biol. Chem. 285: 7670–7685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho J. Y., Matsubara T., Kang D. W., Ahn S. H., Krausz K. W., Idle J. R., Luecke H., Gonzalez F. J. 2010. Urinary metabolomics in Fxr-null mice reveals activated adaptive metabolic pathways upon bile acid challenge. J. Lipid Res. 51: 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiorucci S., Clerici C., Antonelli E., Orlandi S., Goodwin B., Sadeghpour B. M., Sabatino G., Russo G., Castellani D., Willson T. M., et al. 2005. Protective effects of 6-ethyl chenodeoxycholic acid, a farnesoid X receptor ligand, in estrogen-induced cholestasis. J. Pharmacol. Exp. Ther. 313: 604–612 [DOI] [PubMed] [Google Scholar]
- 23.Jansen P. L., Strautnieks S. S., Jacquemin E., Hadchouel M., Sokal E. M., Hooiveld G. J., Koning J. H., De Jager-Krikken A., Kuipers F., Stellaard F., et al. 1999. Hepatocanalicular bile salt export pump deficiency in patients with progressive familial intrahepatic cholestasis. Gastroenterology. 117: 1370–1379 [DOI] [PubMed] [Google Scholar]
- 24.Yang J., Zhao X., Liu X., Wang C., Gao P., Wang J., Li L., Gu J., Yang S., Xu G. 2006. High performance liquid chromatography-mass spectrometry for metabonomics: potential biomarkers for acute deterioration of liver function in chronic hepatitis B. J. Proteome Res. 5: 554–561 [DOI] [PubMed] [Google Scholar]
- 25.Yin P., Wan D., Zhao C., Chen J., Zhao X., Wang W., Lu X., Yang S., Gu J., Xu G. 2009. A metabonomic study of hepatitis B-induced liver cirrhosis and hepatocellular carcinoma by using RP-LC and HILIC coupled with mass spectrometry. Mol. Biosyst. 5: 868–876 [DOI] [PubMed] [Google Scholar]
- 26.Tanaka N., Matsubara T., Krausz K. W., Patterson A. D., Gonzalez F. J. Disruption of phospholipid and bile acid homeostasis in mice with nonalcoholic steatohepatitis. Hepatology. Epub ahead of print. January 30, 2012; doi:10.1002/hep.25630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swain M. G., Maric M. 1996. Tumor necrosis factor-alpha stimulates adrenal glucocorticoid secretion in cholestatic rats. Am. J. Physiol. 270: G987–G991 [DOI] [PubMed] [Google Scholar]
- 28.Bonzo J. A., Patterson A. D., Krausz K. W., Gonzalez F. J. 2010. Metabolomics identifies novel Hnf1alpha-dependent physiological pathways in vivo. Mol. Endocrinol. 24: 2343–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato Y., Kubo Y., Iwata D., Kato S., Sudo T., Sugiura T., Kagaya T., Wakayama T., Hirayama A., Sugimoto M., et al. 2010. Gene knockout and metabolome analysis of carnitine/organic cation transporter OCTN1. Pharm. Res. 27: 832–840 [DOI] [PubMed] [Google Scholar]
- 30.Urakami Y., Kimura N., Okuda M., Inui K. 2004. Creatinine transport by basolateral organic cation transporter hOCT2 in the human kidney. Pharm. Res. 21: 976–981 [DOI] [PubMed] [Google Scholar]
- 31.Ciarimboli G., Lancaster C. S., Schlatter E., Franke R. M., Sprowl J. A., Pavenstadt H., Massmann V., Guckel D., Mathijssen R. H., Yang W., et al. 2012. Proximal tubular secretion of creatinine by organic cation transporter OCT2 in cancer patients. Clin. Cancer Res. 18: 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green R. M., Lo K., Sterritt C., Beier D. R. 1999. Cloning and functional expression of a mouse liver organic cation transporter. Hepatology. 29: 1556–1562 [DOI] [PubMed] [Google Scholar]
- 33.Urakami Y., Okuda M., Masuda S., Saito H., Inui K. I. 1998. Functional characteristics and membrane localization of rat multispecific organic cation transporters, OCT1 and OCT2, mediating tubular secretion of cationic drugs. J. Pharmacol. Exp. Ther. 287: 800–805 [PubMed] [Google Scholar]
- 34.Wikoff W. R., Nagle M. A., Kouznetsova V. L., Tsigelny I. F., Nigam S. K. 2011. Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J. Proteome Res. 10: 2842–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eraly S. A., Vallon V., Vaughn D. A., Gangoiti J. A., Richter K., Nagle M., Monte J. C., Rieg T., Truong D. M., Long J. M., et al. 2006. Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J. Biol. Chem. 281: 5072–5083 [DOI] [PubMed] [Google Scholar]
- 36.Bergwerk A. J., Shi X., Ford A. C., Kanai N., Jacquemin E., Burk R. D., Bai S., Novikoff P. M., Stieger B., Meier P. J., et al. 1996. Immunologic distribution of an organic anion transport protein in rat liver and kidney. Am. J. Physiol. 271: G231–G238 [DOI] [PubMed] [Google Scholar]
- 37.Cattori V., van Montfoort J. E., Stieger B., Landmann L., Meijer D. K., Winterhalter K. H., Meier P. J., Hagenbuch B. 2001. Localization of organic anion transporting polypeptide 4 (Oatp4) in rat liver and comparison of its substrate specificity with Oatp1, Oatp2 and Oatp3. Pflugers Arch. 443: 188–195 [DOI] [PubMed] [Google Scholar]
- 38.Cattori V., Hagenbuch B., Hagenbuch N., Stieger B., Ha R., Winterhalter K. E., Meier P. J. 2000. Identification of organic anion transporting polypeptide 4 (Oatp4) as a major full-length isoform of the liver-specific transporter-1 (rlst-1) in rat liver. FEBS Lett. 474: 242–245 [DOI] [PubMed] [Google Scholar]
- 39.Meng L. J., Wang P., Wolkoff A. W., Kim R. B., Tirona R. G., Hofmann A. F., Pang K. S. 2002. Transport of the sulfated, amidated bile acid, sulfolithocholyltaurine, into rat hepatocytes is mediated by Oatp1 and Oatp2. Hepatology. 35: 1031–1040 [DOI] [PubMed] [Google Scholar]
- 40.Tobin K. A., Steineger H. H., Alberti S., Spydevold O., Auwerx J., Gustafsson J. A., Nebb H. I. 2000. Cross-talk between fatty acid and cholesterol metabolism mediated by liver X receptor-alpha. Mol. Endocrinol. 14: 741–752 [DOI] [PubMed] [Google Scholar]
- 41.Patel D. D., Knight B. L., Soutar A. K., Gibbons G. F., Wade D. P. 2000. The effect of peroxisome-proliferator-activated receptor-alpha on the activity of the cholesterol 7 alpha-hydroxylase gene. Biochem. J. 351: 747–753 [PMC free article] [PubMed] [Google Scholar]
- 42.Marrapodi M., Chiang J. Y. 2000. Peroxisome proliferator-activated receptor alpha (PPARalpha) and agonist inhibit cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J. Lipid Res. 41: 514–520 [PubMed] [Google Scholar]
- 43.Kok T., Bloks V. W., Wolters H., Havinga R., Jansen P. L., Staels B., Kuipers F. 2003. Peroxisome proliferator-activated receptor alpha (PPARalpha)-mediated regulation of multidrug resistance 2 (Mdr2) expression and function in mice. Biochem. J. 369: 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hossain M. A., Tsujita M., Gonzalez F. J., Yokoyama S. 2008. Effects of fibrate drugs on expression of ABCA1 and HDL biogenesis in hepatocytes. J. Cardiovasc. Pharmacol. 51: 258–266 [DOI] [PubMed] [Google Scholar]
- 45.Chinetti G., Lestavel S., Bocher V., Remaley A. T., Neve B., Torra I. P., Teissier E., Minnich A., Jaye M., Duverger N., et al. 2001. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat. Med. 7: 53–58 [DOI] [PubMed] [Google Scholar]
- 46.Rakhshandehroo M., Sanderson L. M., Matilainen M., Stienstra R., Carlberg C., de Groot P. J., Muller M., Kersten S. 2007. Comprehensive analysis of PPARalpha-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007: 26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakhshandehroo M., Knoch B., Muller M., Kersten S. 2010. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 61: 393–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyata M., Tozawa A., Otsuka H., Nakamura T., Nagata K., Gonzalez F. J., Yamazoe Y. 2005. Role of farnesoid X receptor in the enhancement of canalicular bile acid output and excretion of unconjugated bile acids: a mechanism for protection against cholic acid-induced liver toxicity. J. Pharmacol. Exp. Ther. 312: 759–766 [DOI] [PubMed] [Google Scholar]
- 49.Wang R., Lam P., Liu L., Forrest D., Yousef I. M., Mignault D., Phillips M. J., Ling V. 2003. Severe cholestasis induced by cholic acid feeding in knockout mice of sister of P-glycoprotein. Hepatology. 38: 1489–1499 [DOI] [PubMed] [Google Scholar]
- 50.Wang L., Lee Y. K., Bundman D., Han Y., Thevananther S., Kim C. S., Chua S. S., Wei P., Heyman R. A., Karin M., et al. 2002. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell. 2: 721–731 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.