Abstract
Glycosylphosphatidylinositol (GPI) anchor biosynthesis takes place in the endoplasmic reticulum (ER). After protein attachment, the GPI anchor is transported to the Golgi where it undergoes fatty acid remodeling. The ER exit of GPI-anchored proteins is controlled by glycan remodeling and p24 complexes act as cargo receptors for GPI anchor sorting into COPII vesicles. In this study, we have characterized the lipid profile of mammalian cell lines that have a defect in GPI anchor biosynthesis. Depending on which step of GPI anchor biosynthesis the cells were defective, we observed sphingolipid changes predominantly for very long chain monoglycosylated ceramides (HexCer). We found that the structure of the GPI anchor plays an important role in the control of HexCer levels. GPI anchor-deficient cells that generate short truncated GPI anchor intermediates showed a decrease in very long chain HexCer levels. Cells that synthesize GPI anchors but have a defect in GPI anchor remodeling in the ER have a general increase in HexCer levels. GPI-transamidase-deficient cells that produce no GPI-anchored proteins but generate complete free GPI anchors had unchanged levels of HexCer. In contrast, sphingomyelin levels were mostly unaffected. We therefore propose a model in which the transport of very long chain ceramide from the ER to Golgi is regulated by the transport of GPI anchor molecules.
Keywords: ceramides, glycolipids, lipidomics, mass spectrometry, sphingolipid, transport, glucosylceramide
Lipid anchoring of proteins to the outer leaflet of the plasma membrane is essential for cellular function and development (1). One prominent lipid anchor is a complex glycolipid called glycosylphosphatidylinositol (GPI). The GPI anchor has the core structure phosphatidylinositol (PI)-glucosamine (GlcN)-(Mannose)3-phosphoethanolamine (EtN-P), which is conserved among all species. After biosynthesis, the GPI anchor is attached posttranslationally to the newly generated C terminus of certain eukaryotic proteins destined for anchoring thereby tethering the protein to the membrane surface by the glycolipid moiety. GPI-anchored proteins can be released from the cell surface by phosphatidylinositol specific phospholipases and this cleavage event can induce major conformational changes on the GPI-anchored protein itself (2).
At least three organelles, the endoplasmic reticulum (ER), Golgi, and peroxisomes, are involved in the biosynthesis and remodeling of the GPI anchor. The biosynthesis is initiated on the outer side of the ER membrane. After the first two reactions, the GPI anchor precursor is flipped and biosynthesis continues on the luminal side of the ER where the diacyl chains of phosphatidylinositol are then replaced by alkyl-acyl chains. This step is impaired in mutants of the peroxisomal alkyl phospholipid biosynthesis pathway (3).
After protein attachment, the GPI anchor undergoes complex remodeling that begins in the ER with the removal of the inositol-linked acyl chain (4) and the remodeling of the GPI glycan part (5). Glycan remodeling is crucial for sorting GPI-anchored proteins into ER exit sites and their subsequent ER to Golgi transport (6). In mammalian cells, remodeling of the GPI anchor is then continued in the Golgi where the unsaturated fatty acid of the GPI anchor is replaced by a saturated fatty acid chain (7). From the Golgi compartment, GPI-anchored proteins are transported to the plasma membrane where they are thought to associate preferentially with glycosphingolipids and cholesterol to be enriched in lipid-ordered microdomains. Lipid remodeling is likely to be important for this association because unremodeled GPI-anchored proteins, which carry unsaturated fatty acids, are no longer enriched in detergent resistant membrane fractions (7). Treatments that deplete either membrane cholesterol or sphingolipids also disrupt the association of GPI-anchored proteins with detergent-resistant membranes (DRMs) further supporting the notion that specialized domains are critical for the correct localization of this subset of proteins (8, 9).
Importantly, trafficking of GPI-anchored proteins is affected by alterations in sphingolipids and sterols. In yeast, the ER-to-Golgi transport of GPI-anchored proteins is rapidly reduced by inhibition of de novo sphingolipid biosynthesis without affecting the transport of soluble or transmembrane proteins (10, 11). However, GPI-anchored proteins might play an important role in the transport of membrane proteins such as Tat2 and Fur4p, which are no longer associated with DRMs and are retained in the ER in cells that are deficient at an early stage of GPI anchor biosynthesis (12). In mammalian cells, it was shown that inhibition of sphingolipid biosynthesis affects apical targeting of GPI-anchored proteins in Madin-Darby canine kidney (MDCK) cells (13) and sorting of the axonal GPI-anchored protein Thy-1 in primary hippocampal neurons (14). However unlike yeast, ER-to-Golgi transport of GPI-anchored proteins in mammalian cells does not depend on de novo sphingolipid biosynthesis (15).
An important characteristic of sphingolipid trafficking is the coexistence of at least two different ceramide transport pathways, a major ATP and cytosol-dependent pathway and a minor ATP or cytosol-independent pathway (16). Evidence for two different ceramide transport pathways was first obtained with the isolation of the Chinese hamster ovary (CHO) mutant cell line LY-A that shows a defect in SM but not in HexCer biosynthesis (17) and the subsequent identification of a ceramide transport protein called CERT (18). Two pathways for ceramide transport also exist in yeast (19).
As changes in lipid composition affect GPI-anchored proteins, we asked whether a lack of GPI-anchored proteins together with the abnormal accumulation of GPI anchor intermediates would affect the lipid profile of mammalian cells. To address this question, we made use of a series of mutant CHO cell lines (1) that have defects along the GPI anchor biosynthesis pathway. We determined the lipid profile of the GPI anchor deficient cells using a lipidomics approach. Lipidomics has emerged in the era of genomics and proteomics as a rapidly expanding research field due to recent advances in mass spectrometry and bioinformatics. Here, we applied tandem mass spectrometry coupled with multiple reaction monitoring to detect and quantify over 850 phospho- and sphingo-lipids from GPI anchor deficient cells. Using the same crude lipid extracts, we analyzed complex glycosphingolipids by a nontargeted mass spectrometry approach and also investigated the sterol composition of each sample by GC-MS.
Materials and Methods
Chemicals and lipid standards
DLPC 12:0/12:0 (850335), PE 17:0/14:1 (PE31:1, LM-1104), PI 17:0/14:1 (PI31:1, LM-1504), PS 17:0/14:1 (PS31:1, LM-1304), C17:0 ceramide (860517), C12:0 SM (860583) and Glucosyl C8:0 Cer (860540) were used as internal lipid standards and were purchased from Avanti Polar Lipids Inc. (Alabaster, AL). Ergosterol was used as sterol standard and was purchased from Fluka (Buchs, Switzerland).
Methyl tert-butyl ether (MTBE) was from Fluka (Buchs). Methyl amine (33% in absolute ethanol) was from Sigma Aldrich (Steinheim, Germany). HPLC-grade chloroform was purchased from Acros (Geel, Belgium), LC-MS grade methanol and LC-MS grade ammonium acetate were from Fluka. LC-MS grade water was purchased from Biosolve (Valkenswaard, The Netherlands).
Cell culture and transfection
All CHO cell lines used in this study were from the laboratory of Taroh Kinoshita (1). Cells of the F21 background stably express the GPI anchor marker proteins CD59 and DAF and twelve proteins involved in GPI anchor biosynthesis (20) whereas cells of the C311 background express four proteins of the GPI anchor biosynthesis pathway in addition to the markers CD59 and DAF (21–23). Presence of those plasmids was verified by antibiotic resistance of cells to G418, hygromycin B and blasticidin S (F21 series) or resistance of cells to G418, hygromycin B and puromycin (C311 series). For lipid extraction, cells were maintained in Ham's F-12 medium (Invitrogen) supplemented with 10% fetal calf serum (FCS) and 1% PS (penicillin (50 U/ml) and streptomycin (50 U/ml), Invitrogen). HeLa cells were maintained in DMEM (Invitrogen) with 10% FCS supplemented with 1% PS. All cells were grown at 37°C and 5% CO2.
HeLa cells were transiently transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Human ON-TARGETplus SMARTpool siRNA for PIG-L (L-011953-01-0005), DPM3 (L-017492-02-0005), PIG-X (L-013784-02-0005), PIG-F (L-011753-01-0005), PIG-O (L-008728-01-0005), PIG-U (L-017428-00-0005), PGAP1 (L-008110-01-0005), PGAP5 (L-008547-01-0005), p23 (L-003718-00-0005), p24 (L-008074-01-0005), CERT (L-012101-00-0005) and control scrambled siRNA (ON-TARGETplus nontargeting Pool D-001810-10-05) were purchased from Thermo Scientific. GeneSolution siRNA against PGAP2 (1027416) was purchased from Qiagen.
Stable CHO transfectants expressing human PIG-F or PIG-U respectively together with the Venus-FLAG-CD59 construct were established by transfection with Lipofectamine 2000 (Invitrogen) followed by selection with Zeocin (300 μg/ml, Invitrogen) for 2 weeks. Single clones were isolated, expanded and analyzed for CD59-Venus surface expression as a marker for restored GPI anchor biosynthesis.
Plasmids
Using the EcoRI / NotI restriction sites human PIG-F and PIG-U were subcloned from pMEori-PIG-F and pME-PIG-U (20) into the pcDNATM3.1/Zeo(+) mammalian expression vector (Invitrogen). Constructs were verified by sequencing. Vector pME-puro-Venus-FLAG-CD59 (15) was obtained from the laboratory of Reika Watanabe (University of Geneva, Switzerland).
RNA isolation and quantitative RT-PCR
Total RNA was isolated from HeLa cells 72 h after transfection using the RNeasy MINI kit (Qiagen) according to the manufacturer's instructions. RNA was converted into cDNA using random hexamers and Superscript II reverse transcriptase (Invitrogen). qRT-PCR was carried out on a BIO-RAD iCycler machine (BioRad, Hercules, CA) with the ABsoluteTMQPCR SYBR Green reagent (Thermo Scientific). Results were normalized against TBP expression. All primers except for PIG-U, PGAP1, and PGAP5 were designed using the NCBI Primer BLAST web tool. The following primers were used: PIGL.For 5′-GGGTGCTCTGTGCTCACGCT-3′, PIGL.Rev 5′-TG GCTTTCTTGGCCTGTGCCA-3′ DPM3.For 5′-GGCCACTGCCC GCCTACTTG-3′, DPM3.Rev 5′-GTCGGCTCGGGCCTCCTGTA-3′ PIGX.For 5′-GCTCTGACGCCGGCATAAGGG-3′, PIGX.Rev 5′-GA C GGCAGGTGTGCAAGTCCTC-3′ PIGF.For 5′-GCCGCCCGT C G TACCTGATG-3′, PIGF.Rev 5′-TGGCTAGCTAACTCTCCCT CC CG-3′ PIGO.For 5′-CACCACCATGCAGCGCCTCA-3′, PIGO.Rev 5′-CGCCTTCCTGCACTGGTGAGC-3′ PGAP2.For 5′-GCTG GAGTGTACACCATCTTTGCC-3′, PGAP2.Rev 5′-CCGAAGTCCC AC CAGGCCGT-3′ CERT.For 5′-AGGCTGTCATCACACCTCAC GA-3′, CERT.Rev 5′-AGCCATGTCGACGCAAGCTGG-3′ p23.For 5′-TGCGCAGCCACCTCAAGATCAC-3′, p23.Rev 5′-CGCCCTGTT C CCTTGCTCTCA-3′ p24.For 5′-TCGACGTGGAGATTACAGG A CCA-3′, p24.Rev 5′-TGGAGTCATGGTGGACATCCGGT-3′ TBP.For 5′-CCGAATATAATCCCAAGCGGT-3′, TBP.Rev 5′ AAATCAGTGCCGTGGTTCGT-3′. For PIG-U, PGAP1, and PGAP5 predesigned primersets from Qiagen (QuantiTect Primer Assay) were purchased. Relative CHOP and BiP mRNA levels were measured against TBP expression by qRT-PCR using primer: CHOP.For 5′-AGAACCA GGAAACGGAAACAGA-3′ CHOP.Rev 5′-TCTCCTTCATGCGCT GCTTT-3′ BiP.For 5′-TGTTCAACCAATTATCAGCAAACTC-3′ BiP.Rev 5′-TTCTGCTGTATCCTCTTCACCAGT-3′ (24). The efficiency of each primer set was determined to be between 90 and 100%.
Lipid extraction protocols
Lipid extracts were prepared using the MTBE protocol (25). Briefly, 2.5 × 106 cells were resuspended in 100 μl water. The cell suspension was transferred into a 2 ml Eppendorf tube. Three hundred and sixty microliters methanol and a mix of internal standards was added (400 pmol DLPC, 1000 pmol PE31:1, 1000 pmol PI31:1, 3300 pmol PS31:1, 2500 pmol C12SM, 500 pmol C17Cer and 100 pmol C8GC). Samples were vortexed and 1.2 ml of MTBE was added. Samples were placed for 10 min on a multitube vortexer at 4°C (Lab-tek International, Christchurch, New Zealand) followed by an incubation for 1 h at room temperature (RT) on a shaker. Phase separation was induced by addition of 200 μl MS-grade water. After 10 min of incubation at RT samples were centrifuged at 1,000 g for 10 min. The upper (organic) phase was transferred into a 13 mm glass tube (Corning) with a Teflon-lined cap and the lower phase was reextracted with 400 μl of a MTBE/MeOH/H2O mixture (10:3:1.5). Samples were vortexed, incubated for 10 min at RT, and centrifuged for 10 min at 1000 g. The upper phase was collected and the combined organic phases were dried in a CentriVap Vacuum Concentrator (Labconco, MO). In total, 1,500 μl of organic phase was recovered from each samples and split into three parts. One part was treated by alkaline hydrolysis to enrich for sphingolipids and the other two aliquots were used for glycerophospholipid and sterol analysis, respectively.
Glycerophospholipids were deacylated according to the method by Clarke (26). Briefly, 1 ml freshly prepared monomethylamine reagent [methylamine/H2O/n-butanol/methanol at 5/3/1/4 (v/v)] was added to the dried lipid extract and then incubated at 53°C for 1 h in a water bath. Lipids were cooled to RT and then dried in a CentriVap Vacuum Concentrator. For desalting, the dried lipid extracts were resuspended in 300 μl water-saturated n-butanol. The extracts were sonicated and then extracted with 150 µl H2O. The organic phase was collected, and the aqueous phase was rec xextracted twice with 300 µl water-saturated n-butanol. The organic phases were pooled and dried in a CentriVap Vacuum Concentrator.
Determination of total phosphorus
The dried glycerophospholipid extract was resuspended in 250 μl chloroform/methanol (1:1) and 100 μl were placed into a 13 mm disposable pyrex tube. The solvent was completely evaporated to avoid inhibition of the reaction by organic solvents. 0, 2, 5, 10, 20 μl of a 3 mM KH2PO4 standard solution were placed into separate pyrex tubes. To each tube, 20 μl of water and 140 μl of 70% perchloric acid were added. Tubes were heated at 180°C for 1 h in a hood. Tubes were removed from the heat block and kept at RT for 5 min. Then 800 μl of freshly prepared water/1.25% NH4Molybdate (50 mg/4 ml water)/1.67% ascorbic acid (100 mg/6 ml water) in the ratio of 5:2:1 were added. Tubes were heated at 100°C for 5 min with a marble on each tube to prevent evaporation during heating. Tubes were removed from the block and cooled at RT for 5 min. One hundred microliters of each sample was then transferred into a 96-well microplate and the absorbance at 820 nm was measured.
Phospho-and sphingo-lipid analysis by tandem mass spectrometry
Tandem mass spectrometry for the identification and quantification of phospho- and sphingo-lipid molecular species was performed using multiple reaction monitoring with a TSQ Vantage Triple Stage Quadrupole Mass Spectrometer (Thermo Scientific) equipped with a robotic nanoflow ion source, Nanomate HD (Advion Biosciences, Ithaca, NY). Each individual ion dissociation pathway was optimized with regard to collision energy. Lipid concentrations were calculated relative to the relevant internal standards as described in (27) and then normalized to the total phosphate content of each total lipid extract to adjust for difference in cell size, membrane content, and extraction efficiency.
Cellular ceramide glycosylation assay
The cellular ceramide glycosylation assay was performed as described previously (28). Briefly, HeLa cells were treated with siRNA against PIG-F, PIG-O, or PIG-U for 70 h. Scrambled (SCR) siRNA was used as control. Cells were then switched to 950 μl of 1% BSA DMEM medium containing 10 μM NBD C6-ceramide for 2 h. Cells were then rinsed with ice-cold PBS, scraped, and pelleted. Sphingolipids were extracted using the MTBE/methylamine protocol. Samples were resolved on partisil HPTLC plates with fluorescent indicator (Whatman). To distinguish GalCer and GlcCer the HPTLC plates were impregnated with borate as described previously (28). After dipping the plates into a 1% aqueous sodium tetraborate solution, the HPTLC plates were activated at 120°C for 30 min. Sphingolipids were resolved with the solvent system of chloroform/MeOH/water (100:30:4). Fluorescent lipids were visualized under UV exposure. NBD C6-Cer and NBD C6-Cer complexed to BSA were purchased from Invitrogen, NBD C6-GlcCer and NBD C6-GalCer were purchased from Matreya. Bands were quantified with the Image J software and values were calculated as percentage of input (NBD C6-Cer).
Determination of GM3 levels
Individual GM3 species were detected by high resolution mass spectrometry on the LTQ Orbitrap XL linear ion trap (Thermo Scientific). Sphingolipid-enriched extracts were infused at a low flow rate using the TriVersa NanoMate robotic ESI source (Advion Biosciences) equipped with a standard ESI chip (Advion Biosciences). Samples were analyzed in negative ion mode. Individual GM3 species were identified by their parental mass combined with fragmentation. Product ions of m/z 290 were obtained from HCD fragmentation of the GM3 precursor ions. These ions correspond to Neu5Ac fragments obtained after cleavage of the glycosidic bond.
Sterol analysis by GC-MS
Extracts were analyzed by GC-MS as described (29). Briefly, samples were injected into a VARIAN CP-3800 gas chromatograph equipped with a Factor Four Capillary Column VF-5ms 15 m × 0.32 mm i.d. DF = 0.10 and analyzed by a Varian 320 MS triple quadrupole with electron energy set to – 70 eV at 250°C. Samples were applied with the column oven at 45°C, held for 4 min, then raised to 195°C (20°C/min). Sterols were eluted with a linear gradient from 195 to 230°C (4°C / min), followed by raising to 320°C (10°C / min). Finally, the column temperature was raised to 350°C (6°C / min) to elute sterol esters. Cholesterol and cholesterol esters were identified by their retention times (compared with standards) and fragmentation patterns, which were compared with the NIST library.
Statistical analyses
All results are representative of at least three independent experiments. Statistical analyses were performed using an unpaired Student's t-test. Differences were considered significant for P < 0.05 (*), P < 0.01 (**), and P < 0.005 (***).
Results
The goal of this study was to establish the lipid profile of cells that have a defect in GPI anchor biosynthesis. CHO GPI anchor mutants have been very useful in the past to understand the GPI biosynthesis pathway and have allowed cloning of the majority of genes involved in this process (1). We focused on CHO mutants that had either the F21 or the C311 genetic background. The F21 series includes cell lines that are defective in Dol-P-Mannose synthase (DPM3), GPI mannosyltransferase I (PIG-X), ethanolamine phosphate transferase II and III (PIG-F), and GPI lipid remodeling (PGAP2). From the C311 series, we analyzed cells defective in ethanolamine phosphate transferase III (PIG-O) and GPI transamidase (PIG-U).
Defects in GPI anchor biosynthesis lead to changes in HexCer levels
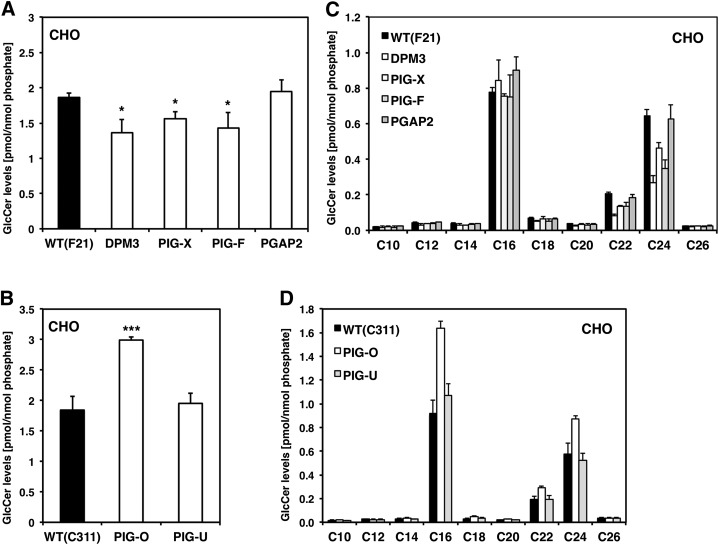
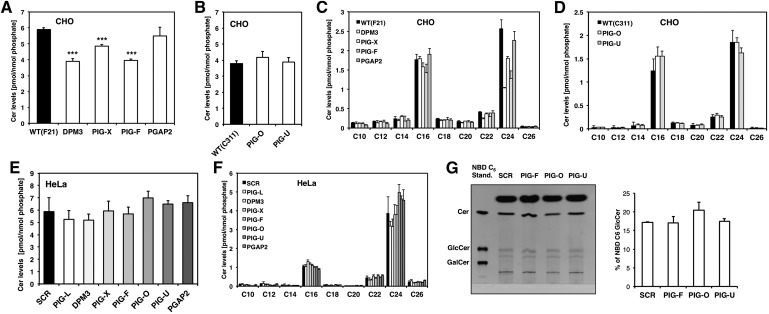
As can be seen in Fig. 1 we observed changes in HexCer levels in CHO mutant cells that have a defect in GPI anchor biosynthesis. Because CHO cells do not possess endogenous GalCer (30) the changes in HexCer are due to a difference in their glucosylceramide (GlcCer) levels. In detail, we observed a downregulation of total GlcCer levels in a subset of GPI anchor mutants (DPM3, PIG-X, and PIG-F) that generate short truncated GPI anchor intermediates. To our surprise, we did not see an effect in GPI anchor mutant PIG-U cells (Fig. 1B). Because PIG-U mutant cells (PA16.1) have no expression of the GPI marker protein CD59 and only 1% remaining DAF expression (supplementary Table I), this indicates that the observed GlcCer changes are not due to a general absence of GPI-anchored proteins (23). Lipid remodeling in the Golgi also did not influence GlcCer levels as we did not observe a change in PGAP2 deficient cells (Fig. 1A), which have normal GPI anchor biosynthesis in the ER but greatly reduced surface expression levels of CD59 and DAF (31) due to rapid secretion (supplementary Table 1). In contrast, the GPI anchor mutant PIG-O (22), which shows a great reduction but not a complete deficiency in the surface expression of GPI-anchored proteins, displayed a strong increase in GlcCer-levels (supplementary Table I, Fig. 1B).
Fig. 1.
Glucosylceramide profile of GPI anchor deficient CHO cells. A: Total GlcCer levels in F21 (wild-type) and GPI anchor mutants DPM3, PIG-X, PIG-F, and PGAP2. B: Total GlcCer levels in C311 (wild-type) and GPI anchor mutant cell lines PIG-O and PIG-U. C: Individual GlcCer profile of GPI anchor mutants from the F21 series and (D) mutants from the C311 series.
When GlcCers were analyzed in detail, we observed an effect concerning the chain length of individual glucosylceramide species. Very long chain GlcCers with a fatty acid chain length of C22 and C24 were strongly decreased in the GPI anchor mutants DPM3, PIG-X, and PIG-F whereas GlcCer species with a fatty acid chain length between C10 and C20 were mostly unaffected (Fig. 1C). In contrast, PIG-O mutant cells showed a general upregulation of all glucosylceramide species regardless of their chain length (Fig. 1D).
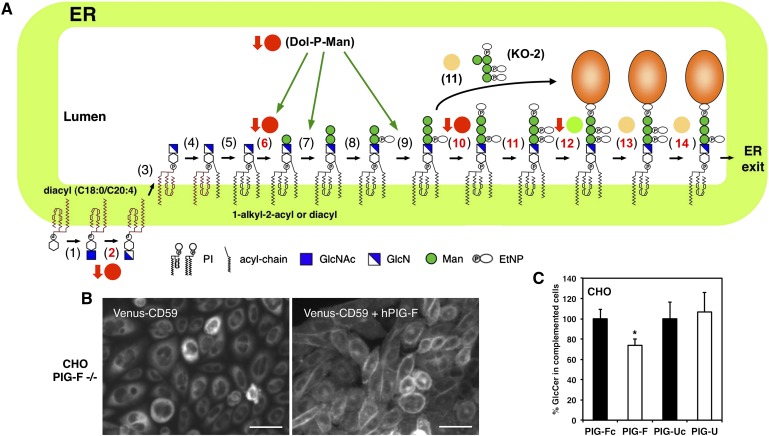
We next focused on the GPI anchor mutants PIG-F and PIG-U, which showed different phenotypes in our lipid analysis. Both GPI anchor mutants cannot generate GPI-anchored proteins and accumulate free GPI anchor intermediates that are similar in structure (Fig. 2A). However, PIG-U mutants are deficient in one subunit of the GPI transamidase and accumulate otherwise a functional free GPI anchor. PIG-F mutants on the other hand lack the regulatory subunit of the EtN-P transferases II and III and consequently have no EtN-P on both Man2 and Man3. The EtN-P on Man2 is later recognized and removed by PAGP5, a step which is necessary for efficient incorporation of GPI-anchored proteins into ER exit sites (5). PIG-F and PIG-U mutants belong to different series of CHO mutant cells (F21 and C311 respectively) and carry plasmids, which encode for several GPI anchor biosynthesis enzymes and GPI anchor marker proteins (20, 23). To test if the observed GlcCer changes are due to the defect in GPI anchor biosynthesis or if they are connected to their genetic background, we stably complemented both PIG-F and PIG-U mutant cells with the corresponding wild-type gene. In order to monitor restored GPI anchor biosynthesis we stably coexpressed Venus tagged CD59. As can be seen in Fig. 2B, the GPI-anchored protein CD59 localizes to the cell surface in PIG-F complemented cells whereas in the PIG-F deficient control cells, CD59 does not reach the cell surface. We then analyzed the lipid profile from both uncomplemented and complemented cells. As can be seen in Fig. 2C, GlcCer levels returned to wild-type in the complemented PIG-F cells indicating that the decrease in GlcCer levels was due to their defect in GPI anchor biosynthesis. As expected, we did not observe any changes in PIG-U cells after complementation (Fig. 2C).
Fig. 2.
Complementation of the GPI anchor mutants PIG-F and PIG-U. (A) Biosynthetic pathway of mammalian GPI-APs (1). GPI-N-acetylglucosamine transferase (2), GlcNAc-PI deN-acetylase (PIG-L) (3), Flippase (4), Inositol acyltransferase (5), PI remodeling enzyme (6), GPI-MT I (PIG-M/PIG-X) (7), GPI-MT II (8), EtN-P transferase I (9), GPI-MT III (10), EtN-P transferase III (PIG-O/PIG-F) (11), EtN-P transferase II (PIG-G/PIG-F) (12), GPI transamidase (PIG-K, GAA1, PIG-S, PIG-T, PIG-U) (13), Inositol deacylase (PGAP1) (14), EtN-P Phosphoesterase (PGAP5). Red circles, decreased levels of HexCer; green circles, unchanged levels of HexCer; yellow circles, elevated levels of HexCer. Red arrows indicate a reduction in cholesterol ester levels. B: Left panel: PIG-F mutant CHO cells (CHO PIG-F −/−) stably expressing Venus-CD59. Right panel: PIG-F mutant CHO cells (CHO PIG-F −/−) stably expressing Venus-CD59 and human PIG-F. Bar = 10 μm. C: Relative GlcCer levels in the GPI anchor mutants PIG-F and PIG-U and in the complemented PIG-Fc and PIG-Uc cells. GlcCer levels were calculated as percentage of PIG-Fc and PIG-Uc respectively. *P < 0.05 uncomplemented versus complemented cells.
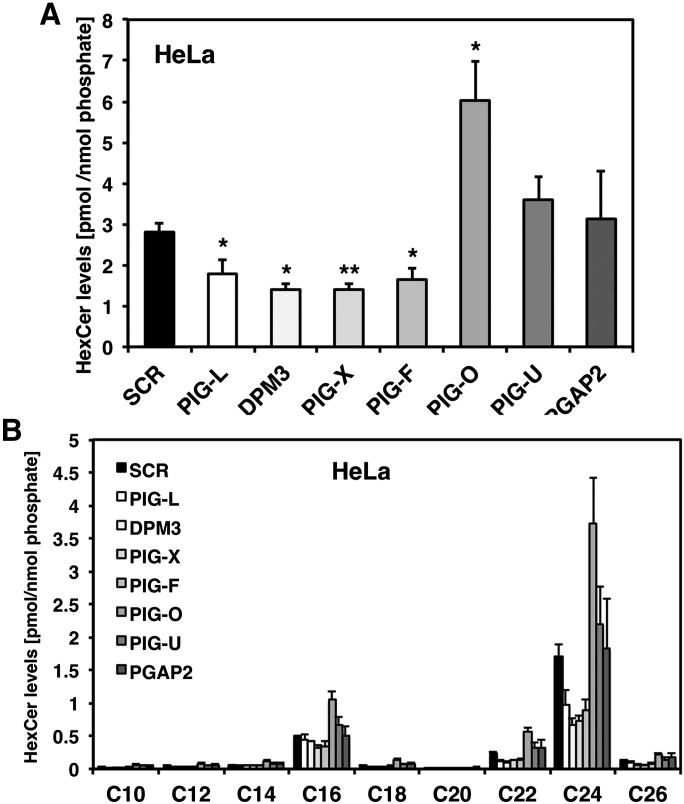
We next wanted to see whether the observed sphingolipid changes are CHO cell specific or if they can be reproduced in another mammalian, preferentially human cell line. In addition, we were interested if a transient knockdown of GPI anchor biosynthesis leads to the same sphingolipid changes as a genetic mutation. To test these two parameters, we transiently silenced PIG-L, DPM3, PIG-X, PIG-F, PIG-O, PIG-U, and PGAP2 expression in HeLa cells by siRNA. The gene silencing efficiency was confirmed by quantitative RT-PCR (supplementary Fig. I). Hela cells possess ceramide galactosyltransferase activity (32) therefore their HexCer levels might be composed of GlcCer and GalCer species. When analyzed by HPTLC, HeLa cells showed higher levels of GlcCer than GalCer (supplementary Fig. II). As can be seen in Fig. 3A we found a decrease in total HexCer levels in PIG-L, DPM3, PIG-X, and PIG-F but not in PIG-U or PGAP2 deficient cells. Again, very long chain C22 and C24 HexCer species were strongly affected whereas C16 HexCer was only slightly decreased (Fig. 3B). Because HeLa cells have a higher percentage of very long chain C22 and C24 HexCer species than CHO cells, the decrease in total HexCer was more pronounced. A specific decrease of very long chain HexCer species was also observed for the less abundant sphinganine containing dihydroglycosylceramides (DHHexCer, supplementary Fig. III). Interestingly, PIG-O depleted cells showed again a strong increase in total HexCer levels and a general upregulation of all HexCer species similar to what had been observed for CHO cells (Fig. 3A, B).
Fig. 3.
HexCer profile of GPI anchor deficient HeLa cells. A: Total HexCer levels of HeLa cells treated with siRNA against PIG-L, DPM3, PIG-X, PIG-F, PIG-O, PIG-U, or PGAP2, respectively. Scrambled (SCR) siRNA was used as control. Cells were harvested 72 h after transfection. B: Individual HexCer levels of the GPI anchor-deficient HeLa cells. *P < 0.05 and **P < 0.01 SCR versus siRNA treated cells.
Decrease in HexCer affects downstream glycosphingolipids
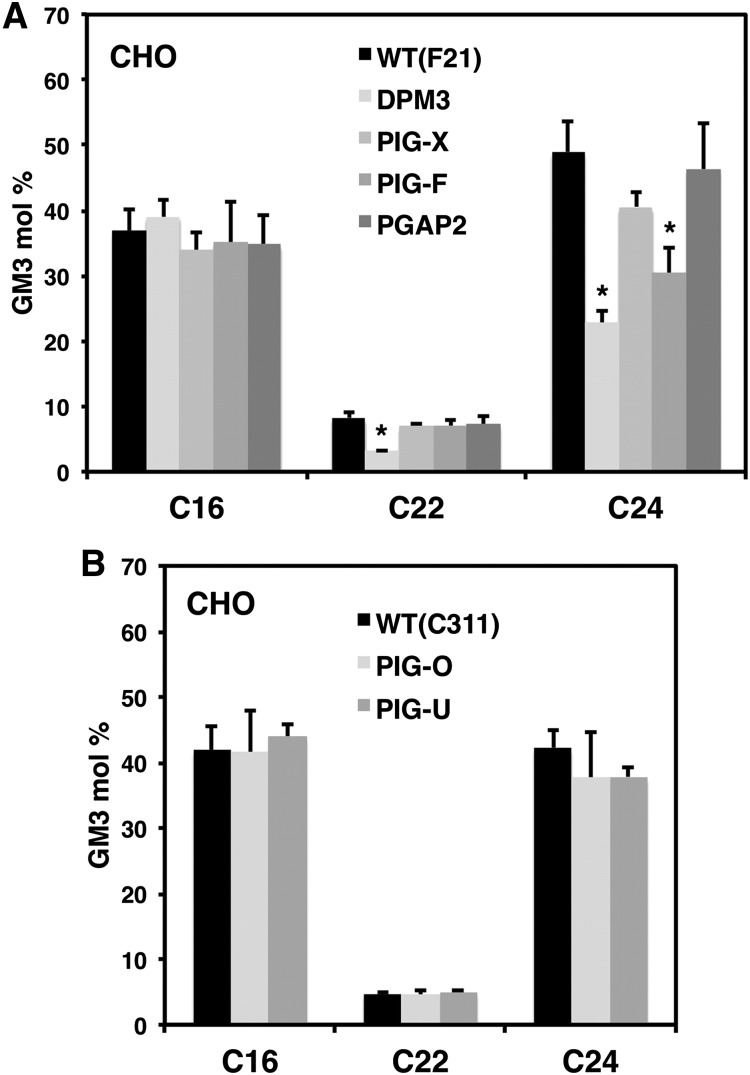
We next analyzed if the observed decrease in GlcCer levels in CHO cells also affects downstream glycosphingolipids such as GM3. The glycosphingolipid profile of CHO cells is rather simple as they only synthesize GlcCer, lactosylceramide and the ganglioside NeuAcα2-3Galβ1-4Glcβ1-1Cer (GM3) (30). CHO cells accumulate high amounts of GM3 because they are unable to synthesize GM2 and other complex gangliosides such as GM1 and GD1a (33). We therefore measured GM3 levels in the GPI anchor mutant CHO cells. When individual GM3 species were analyzed by high-resolution mass spectrometry, we found a decrease of the very long chain GM3 species for DPM3, PIG-X, and PIG-F but not for PGAP2 deficient cells (Fig. 4A). GM3 levels also did not change in PIG-O and PIG-U deficient cells indicating that the increase in GlcCer levels that was observed in PIG-O deficient cells did not translate into an upregulation of GM3 (Fig. 4B). HeLa cells predominantly synthesize Gb3 and have low amounts of GM3 (34), which were below our quantification level.
Fig. 4.
GM3 levels of GPI anchor mutant CHO cells. A: Individual GM3 profile of wild-type (F21) and GPI anchor mutant cell lines DPM3, PIG-X, PIG-F and PGAP2. B: Individual GM3 profile of wild-type (C311) and GPI anchor mutant PIG-O and PIG-U cells. *P < 0.05 wild-type versus GPI anchor mutant.
Ceramide levels are affected by defects in GPI anchor biosynthesis in a cell line dependent manner
Because ceramides are direct precursors of monoglycosylated ceramides we analyzed if the observed effect on HexCer levels was reflected by a decrease in ceramide. We measured ceramide levels in the same set of GPI anchor mutant CHO cells and found that DPM3, PIG-X, and PIG-F cells had reduced ceramide levels whereas no change was observed for PGAP2 deficient cells (Fig. 5A). On the individual level very long chain ceramides, in particular C24 ceramide, were decreased (Fig. 5C). In PIG-U and PIG-O mutant cells, ceramide levels were unaffected (Fig. 5B, D). In HeLa cells, however, total ceramide levels were not decreased upon knockdown of PIG-L, DPM3, PIG-X, or PIG-F (Fig. 5E) and also on the individual level, there was no significant change in very long chain ceramides (Fig. 5F). This indicates that the decrease in very long chain ceramides could be cell line-specific or might be a compensatory effect of the genetic mutation. In addition, it also raises the possibility that reduced ceramide biosynthesis is not the only cause for the observed decrease in HexCer levels. We next tested if glycosylation of ceramide is affected in the GPI anchor deficient cells. It has been shown that glucosylceramide synthase localizes to the Golgi in HeLa cells (35) but it cannot be fully ruled out that some GlcCer synthase activity might also exist in the ER (36). To test if ceramide glycosylation is affected in the GPI anchor deficient cells, we performed an in vivo ceramide glycosylation assay (28). As can be seen in Fig. 5G, NBD-C6 ceramide was efficiently converted into GlcCer in PIG-F, PIG-O, and PIG-U depleted cells. There was, however, no synthesis of GalCer, which shows that ceramide galactosyltransferase activity is very low in HeLa cells (Fig. 5G). Relative to the total amount of input (NBD-C6 Cer) there was an increase in NBD-C6 GlcCer in the PIG-O depleted cells, which indicates more GlcCer synthase activity in PIG-O depleted cells.
Fig. 5.
Ceramide profile of GPI anchor-deficient cells. A: Total ceramide (Cer) levels in F21 wild-type and GPI anchor mutants DPM3, PIG-X, PIG-F, and PGAP2. B: Total Cer levels in C311 wild-type and GPI anchor mutant cell lines PIG-O and PIG-U. C: Individual Cer profile of GPI anchor mutants from the F21 series and (D) mutants from the C311 series. E: Total Cer and (F) individual Cer levels of HeLa cells treated with siRNA against PIG-L, DPM3, PIG-X, PIG-F, PIG-O, PIG-U, or PGAP2, respectively. Scrambled (SCR) siRNA was used as control. ***P < 0.005 wild-type versus GPI anchor mutant. G: Ceramide glycosylation assay: HeLa cells were treated with siRNA against PIG-F, PIG-O, and PIG-U, respectively. After 70 h, cells were incubated with NBD C6-Cer for 2 h after which sphingolipids were extracted. Lipid extracts were resolved on borate impregnated HPTLC plates and visualized under UV exposure. Standards were NBD C6-Cer, NBD C6-GlcCer, and NBD C6 GalCer. Bands were quantified with Image J software and values were calculated as percentage of input (NBD C6-Cer).
Very long chain sphingomyelin levels are partially affected in GPI anchor deficient cells
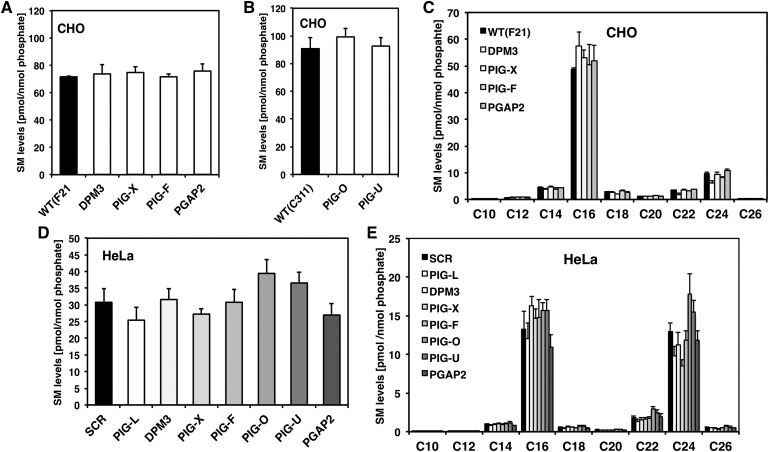
In contrast to the observed changes in HexCer and ceramide, total sphingomyelin levels were not affected in the same set of GPI anchor mutant CHO cells (Fig. 6A, B). Total sphingomyelin levels also did not change in both the PIG-F and the PIG-U complemented cells (supplementary Fig. IV). However, CHO cells have predominantly C16 sphingomyelin, which accounts for 70% of the total sphingomyelin and very long chain sphingomyelins were slightly reduced in CHO mutants that have a decrease in GlcCer levels (Fig. 6C). Total sphingomyelin levels were also not significantly changed in HeLa cells (Fig. 6D) but the detailed analysis showed that defects in GPI anchor biosynthesis have an effect on very long chain sphingomyelins (Fig. 6E). As there is much more sphingomyelin than HexCer in both CHO and HeLa cells, the absolute amounts of sphingomyelin that are affected could be in fact more than HexCers.
Fig. 6.
Sphingomyelin levels do not change in GPI anchor deficient cells. A: Total sphingomyelin (SM) levels of F21 wild-type and GPI anchor mutant cell lines DPM3, PIG-X, PIG-F, and PGAP2. B: Total SM levels of C311 wild-type and GPI anchor mutant cell lines PIG-O and PIG-U. C: Individual SM levels in the F21 series of GPI anchor mutants. D: Total SM levels of Hela cells treated with siRNA gainst PIG-L, DPM3, PIG-X, PIG-F, PIG-O, PIG-U, and PGAP2. Scrambled (SCR) siRNA was used as control. E: Individual SM profile of GPI anchor-deficient HeLa cells.
The ceramide transport protein CERT mediates transport of ceramide for sphingomyelin biosynthesis. However, CERT deficiency does not affect total GlcCer levels in CHO cells, indicating that there is an alternative ceramide transport pathway for the biosynthesis of GlcCer (18). In vitro CERT has a strong preference for the transport of ceramides that are C20 or shorter and does not efficiently transport very long chain ceramides (37). We silenced CERT in HeLa cells and observed a decrease in total sphingomyelin as expected (Fig. 7A). HexCer levels did not decrease and even modestly increased, which confirms the existence of different ceramide transport pathways (Fig. 7A). As shown in Fig. 7B, knockdown of CERT affected both C16 and C24 sphingomyelins, indicating that in vivo CERT is also required for the efficient transport of very long chain ceramides. This might explain why very long chain sphingomyelins are only partially affected in GPI anchor deficient cells. Individual HexCer levels showed a stronger increase in C16 than in C22 glycosylceramides (supplementary Fig. V).
Fig. 7.
CERT knockdown does not lead to a decrease in HexCer levels. A: Total sphingomyelin and HexCer levels of Hela cells treated with siRNA against the ceramide transport protein CERT. Scrambled (SCR) siRNA was used as control. B: Individual sphingomyelin profile of HeLa cells treated with siRNA against CERT. **P < 0.01 SCR versus siRNA knockdown.
Defects in GPI glycan remodeling or GPI anchor trafficking affect HexCer levels
We had noticed a strong global increase in HexCer levels for PIG-O deficient CHO cells, which was reproduced in HeLa cells during a transient knockdown of PIG-O. In PIG-O deficient cells, ethanolamine phosphate transferase III (PIG-F/GPI7) is active and cells can attach EtN-P onto mannose 2, which is important for glycan remodeling. Interestingly, PIG-O mutants are not completely deficient in the surface expression of GPI-anchored proteins and accumulate an intermediate, called KO-2 (Fig. 2A), which is competent for protein attachment but has an abnormal glycan structure (38). The glycan structure plays an important role for sorting of GPI-anchored proteins into ER exit sites (5, 6) and only glycan remodeled GPI-anchored proteins associate with p24 family proteins (6, 39). We therefore analyzed the lipid profile of cells in which the GPI remodeling enzymes PGAP1, PGAP5, or the p24 family members p23 or p24 were silenced respectively. As can be seen in Fig. 8A, total HexCer levels increased upon knockdown of PGAP1, PGAP5, p23, and in particular for p24. Individual HexCer levels showed that all HexCer species were affected equally (Fig. 8B). This result is in line with the observed increase of HexCer levels in PIG-O deficient cells and indicates that an abnormal GPI structure or a delay in ER to Golgi transport can lead to an increase in total HexCer levels. We also observed some increase in ceramide levels in particular for PGAP1 but sphingomyelin levels were not changed (supplementary Figs. VI , VII). In yeast, it has been shown that a defect in GPI anchor remodeling or trafficking results in ER stress and induces the upregulation of the unfolded protein response (UPR) (40–42). We assessed ER stress by measuring transcriptional activation of the UPR. The C/EBP homology protein (CHOP), which is involved in ER stress-mediated apoptosis was upregulated in the GPI anchor mutant PIG-O, the remodeling mutants PGAP1 and PGAP5, as well as in p23 and p24 knockdowns but not in PIG-F deficient cells (Fig. 8C). The ER chaperone BiP, was upregulated in PIG-O, PGAP5, and p23 depleted cells.
Fig. 8.
HexCer levels are affected by GPI glycan remodeling and involve the p24 family members. A: Total HexCer levels of Hela cells treated with siRNA against PGAP1, PGAP5, p23 or p24, respectively. Scrambled (SCR) siRNA was used as control. B: Detailed HexCer profile of the corresponding cells. C: Real time RT-PCR analysis of genes involved in UPR induction. HeLa cells were treated with siRNA against PIG-F, PIG-O, PGAP1, PGAP5, p23 and p24. SCR was used as control. Cells were harvested 72 h after transfection and total RNA was isolated. Relative levels of CHOP and BiP were assessed by qRT-PCR.
Cholesterol ester levels are reduced in cells that do not synthesize GPI-anchored proteins
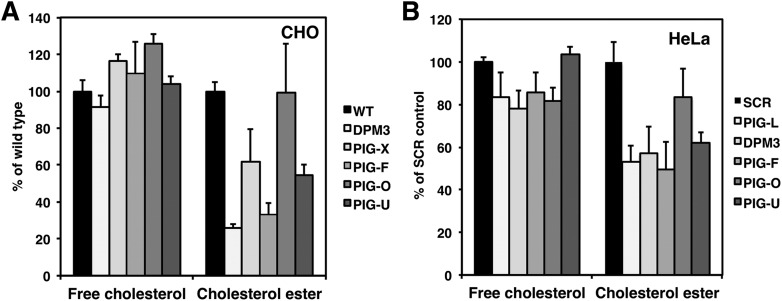
It has been previously reported that GPI-deficient CHO cells have cholesterol contents similar to wild-type CHO cells (43). Using our mass spectrometry approach, we measured the amount of free cholesterol and of cholesterol ester in the GPI anchor-deficient CHO and HeLa cells. As can be seen in Fig. 9A, free cholesterol levels are unchanged between wild-type and the GPI anchor mutants. We found, however, that all GPI anchor mutant cells including PIG-U had lower levels of cholesterol ester than the corresponding wild-type cells (Fig. 9A). PIG-O mutant cells, which are not completely deficient in the biosynthesis of GPI-anchored proteins, did not show a reduction in cholesterol esters. We then also analyzed the sterol composition of HeLa cells in which PIG-L, DPM3, PIG-F, PIG-O, or PIG-U had been silenced, respectively. Under these conditions, we observed a similar result: free cholesterol levels were mostly unchanged (Fig. 9B) whereas cholesterol ester levels were strongly reduced in all knockdowns except PIG-O (Fig. 9B). This result suggests that the observed effect on cholesterol ester is due to the inability to synthesize certain GPI-anchored proteins and is not caused by a specific GPI anchor intermediate that is generated.
Fig. 9.
Sterol levels in the GPI anchor deficient cells. A: Free cholesterol and cholesterol ester levels in in the GPI anchor deficient CHO cell lines DPM3, PIG-X, PIG-F, PIG-O, and PIG-U. Values were calculated as percentage of wild-type (F21 or C311 respectively). B: Free cholesterol and cholesterol ester levels in HeLa cells after a transient knockdown of the GPI anchor biosynthesis genes PIG-L, DPM3, PIG-F, PIG-O, and PIG-U respectively. Scrambled (SCR) siRNA was used as control.
In parallel to the observed changes in glycolipids and sterols we also analyzed the phospholipid content of each GPI anchor mutant by mass spectrometry. We measured the levels of phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylinositol (PI), and phosphatidylserine (PS). Among those glycerophospholipids we did not observe any major changes except for a mild increase of total PE in the GPI-anchor mutants DPM3, PIG-X, and PIG-O (supplementary Fig. VIII).
Discussion
In this study we show that GPI anchor biosynthesis mutants that generate incomplete GPI anchor intermediates have a strong decrease in very long chain C22 and C24 glucosylceramide levels, without greatly affecting long chain C16 glucosylceramides. In contrast, GPI transamidase-deficient cells (PIG-U) that synthesize complete anchors but do not attach them to proteins do not show this effect. This suggests that the GPI anchor, not necessarily attached to proteins, regulates very long chain GlcCer levels. Complementation experiments showed that these changes are specific to the GPI anchor biosynthesis and are not due to the complex genetic background of the GPI anchor mutant cells. siRNA silencing of GPI anchor biosynthesis genes in HeLa cells demonstrated that the observed effects on GlcCers are not restricted to one cell type and that the strategy of siRNA gene downregulation can be used to see changes in lipid profiles in a relatively short time period.
In addition to the decrease in GlcCer, we also observed a decrease in very long chain ceramides in the CHO GPI anchor mutants. However, this decrease was not observed in HeLa cells following a transient knockdown of the same GPI anchor biosynthesis genes (Fig. 5E). This implies that the decrease in very long chain HexCer is probably not just the consequence of reduced ceramide levels.
Defects in GPI anchor biosynthesis lead to the ER accumulation of GPI anchor intermediates such as GlcNAc-PI and GlcN-acyl-PI as well as the mannose containing GPI intermediates H2-H8 (1). Metabolic labeling with myo-[3H]inositol has shown that GPI anchor mutants such as DPM3 and PIG-X accumulate moderate amounts of the early GPI anchor intermediate GlcN-acyl-PI whereas PIG-U mutants show a high ER accumulation of all GPI anchor intermediates (20, 23). In particular, Man-GlcN-(acyl)-PI labels strongly in PIG-U deficient cells cultured with [3H]mannose, suggesting that GlcN-(acyl)-PI is also elevated in those cells (23). Because GlcCer levels were unaffected in PIG-U deficient cells, it is therefore unlikely that the ER accumulation of GPI anchor intermediates is causing an inhibition of certain ceramide synthases or that GlcCer synthase activity would be affected. Another possible reason for reduced ceramide and HexCer levels could be that ER to Golgi transport of ceramide is linked to the transport of GPI-anchored proteins and free GPI anchor molecules. If very long chain ceramides are not transported efficiently from the ER and are locally accumulating this could lead to a feedback inhibition of certain ceramide synthases and subsequently to a decrease in HexCers. The degree of feedback inhibition might be different in HeLa versus CHO cells.
In contrast to HexCer, sphingomyelin levels were mostly unaffected in the GPI anchor-deficient cells. When analyzed in detail, we found that CHO cells have primarily C16 sphingomyelin (Fig. 6C). In contrast, HeLa cells have a much higher content of very long chain sphingomyelins (C24 sphingomyelin is about 45% of total sphingomyelin) and very long chain sphingomyelin levels were mildly affected in HeLa cells (Fig. 6E). The major fraction of ceramide in mammalian cells is transported to the Golgi in a nonvesicular manner via the ceramide transport protein CERT and mutants in CERT affect mainly sphingomyelin levels, which suggested an alternative mechanism of transport of ceramide destined for GlcCer biosynthesis (18). CERT has a preference for ceramide species with an acyl chain length of C20 or less and transports C22 and C24 ceramide with greatly reduced efficiency in vitro (37). However, our results show that knockdown of CERT led to a global decrease of all sphingomyelin species and also greatly reduced C24 sphingomyelin (Fig. 7B). We therefore conclude that in vivo the transport of very long chain ceramides from ER to Golgi for sphingomyelin biosynthesis is regulated by CERT. Interestingly, ER to Golgi trafficking of GPI-anchored proteins is normal in CERT deficient cells (44). Taken together, these data support a model in which GPI anchor molecules regulate the transport of very long chain ceramides destined for GlcCer and to some degree also for sphingomyelin biosynthesis.
A common feature among the mutants that show decreased levels of very long chain GlcCer is the accumulation of GPI anchor precursors that have incomplete glycan structures. The GPI anchor mutants PIG-L, DPM3, and PIG-X produce very short truncated GPI anchor intermediates (GlcNAc-PI, GlcN-(acyl)-PI) that are unlikely to leave the ER. The mutant PIG-F however generates a GPI anchor intermediate that possesses an almost complete glycan structure but still has reduced GlcCer levels (Fig. 2A). PIG-F is the regulatory subunit of the GPI EtN-P transferase II (PIG-O/PIG-F) and III (GPI7/PIG-F) (45). Consequently, both EtN-P transferases are not active in PIG-F deficient cells (38) and mutant cells do not attach EtN-P onto mannose 2 or 3 (46). Because EtN-Ps are important for glycan remodeling and ER to Golgi transport, we speculate that this abnormal glycan structure makes it impossible for the PIG-F GPI anchor intermediate to exit the ER.
In contrast, PIG-U deficient cells, which make a complete GPI anchor but lack the enzymatic activity to transfer the free GPI anchor to a protein, have unchanged levels of HexCer. Interestingly, it has been reported that cells generate large pools of free nonproteins linked GPI anchors that exit the ER and are transported to the cell surface (47, 48). Although there is no direct evidence that free GPI anchors can be remodeled in vivo, it has been shown that free GPI anchors are substrates for remodeling enzymes in vitro (5).
In contrast to the mutants PIG-L, DPM3, PIG-X, and PIG-F, we observed a strong increase in HexCer levels for the GPI anchor mutant PIG-O. This was observed in both the CHO cell lines as well as HeLa cells under siRNA silencing conditions. PIG-O mutants generate intermediates with a similar GPI structure as PIG-F. However, the ethanolamine phosphate transferase II, which is a complex of GPI7 and PIG-F is active in PIG-O deficient cells (45). Ethanolamine phosphate transferase II adds EtN-P onto mannose 2, which is important for glycan remodeling by PGAP5 (5). Interestingly, PIG-O mutants are not completely deficient in the surface expression of GPI-anchored proteins (38). The surface expression of PIG-O deficient cells is due to a minor GPI anchor intermediate called KO-2 that is competent for protein attachment. KO-2 has an abnormal glycan structure with most likely four mannoses and EtN-P on Man1 and Man3 (38). An additional EtN-P on mannose 2 might be transitory and is a potential substrate for PGAP5. Because of this abnormal glycan structure, we speculate that PIG-O deficient cells might have a defect in sorting of free or protein anchored GPIs into ER exit sites. The ER exit of GPI-anchored proteins is controlled by glycan remodeling and p24 complexes act as cargo receptors for GPI anchor sorting into COPII vesicles (6, 39, 49). In agreement with this model, we observed a global increase in HexCer levels in the remodeling mutants PGAP1 and PGAP5 as well as for the p24 family proteins. However,we did not detect an increase in GM3 levels in PIG-O depleted cells, which shows that a global increase in HexCer does not necessarily translate into an increase in GM3 and most likely follows a different mechanism. In yeast, it has been shown that the unfolded protein response is highly activated in the GPI remodeling mutants (40–42). Here, we show that ER stress markers such as CHOP are upregulated in the GPI anchor remodeling and trafficking mutants including PIG-O but not in PIG-F deficient cells (Fig. 8C). Recently, we have shown that ceramide levels are increased in response to induction of the unfolded protein response (27). Because HexCer synthesis protects against Cer-induced stress in mammalian cells (50, 51), we speculate that the ER stress response might cause the observed increase in HexCer levels in the GPI anchor remodeling mutants.
Finally, we did not observe any sphingolipid changes for the GPI anchor mutant PGAP2, which has normal GPI anchor biosynthesis in the ER but greatly reduced surface expression levels of GPI-anchored proteins due to secretion (31). Sorting of GPI-anchored proteins into ER exit sites and ER to Golgi trafficking is not affected in the PGAP2 deficient cells.
Concerning the sterol composition of the GPI anchor-deficient cells we observed no effect on free cholesterol levels but found a decrease in cholesterol ester (CE) levels in certain GPI anchor-deficient cells (Fig. 9). There was, however, no correlation between the sterol and the HexCer profile since PIG-U deficient cells showed a similar reduction of CE levels as PIG-F deficient cells. In contrast, PGAP2 and PIG-O deficient cells, which synthesize GPI-anchored proteins had almost wild-type levels of CEs. We therefore speculate that the decrease in CE is due to the absence of certain GPI-anchored proteins that affect biosynthesis or degradation of CEs. In addition, uptake of cholesterol from the medium, which contains FCS, might be affected in cells that lack GPI-anchored proteins.
GPI-anchored proteins have been postulated to segregate into sphingolipid enriched lipid ordered micro domains (52). In yeast, it has been shown that ongoing ceramide synthesis is required for GPI-anchored protein transport from the ER to the Golgi compartment, linking the two processes (10, 11). In mammalian cells, however, de novo sphingolipid biosynthesis is not required to transport mammalian GPI-anchored proteins from the ER to the Golgi (15, 53). However, this does not exclude the possibility that also in mammalian cells ER to Golgi transport of ceramide is linked to the transport of GPI-anchored proteins. It might be possible that under sphingolipid depleted conditions GPI-anchored proteins are trafficked normally but that in the absence of GPI-anchored proteins very long chain ceramides are not transported properly.
We present here a model whereby the levels of very long chain GlcCer are correlated to the biosynthesis of GPI anchor molecules. We propose that very long chain GlcCer levels and to some extent also very long chain sphingomyelin levels are linked to GPI biosynthesis to ensure that proportional amounts of each lipid class are synthesized and transported.
Supplementary Material
Acknowledgments
The authors are grateful to Morihisa Fujita and Yusuke Maeda (Osaka University, Japan) for critical reading, plasmids, and cell lines. We thank Adrien Schmid (EPFL, Switzerland), Nicolas Guex (University of Lausanne, Switzerland), and Isabelle Riezman (University of Geneva, Switzerland) for mass spectrometry support and Reika Watanabe (University of Geneva, Switzerland) for plasmids and discussion.
Footnotes
Abbreviations:
- CE
- cholesterol ester
- Cer
- ceramide
- CHO
- Chinese hamster ovary
- ER
- endoplasmic reticulum
- EtN-P
- ethanolamine-phosphate
- GlcCer
- glucosylceramide
- GPI
- glycosylphosphatidylinositol
- HexCer
- hexylceramide
- PC
- phosphatidylcholine
- PE
- phosphatidyle-thanolamine
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
This project was supported by the Swiss National Science Foundation (HR) and the Swiss SystemsX.ch initiative, evaluated by the Swiss National Science Foundation.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of supplementary data, eight figures, and one table.
REFERENCES
- 1.Maeda Y., Ashida H., Kinoshita T. 2006. CHO glycosylation mutants: GPI anchor. Methods Enzymol. 416: 182–205 [DOI] [PubMed] [Google Scholar]
- 2.Bütikofer P., Malherbe T., Boschung M., Roditi I. 2001. GPI-anchored proteins: now you see ‘em, now you don't. FASEB J. 15: 545–548 [DOI] [PubMed] [Google Scholar]
- 3.Kanzawa N., Maeda Y., Ogiso H., Murakami Y., Taguchi R., Kinoshita T. 2009. Peroxisome dependency of alkyl-containing GPI-anchor biosynthesis in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 106: 17711–17716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka S., Maeda Y., Tashima Y., Kinoshita T. 2004. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279: 14256–14263 [DOI] [PubMed] [Google Scholar]
- 5.Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., Kinoshita T. 2009. GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell. 139: 352–365 [DOI] [PubMed] [Google Scholar]
- 6.Fujita M., Watanabe R., Jaensch N., Romanova-Michaelides M., Satoh T., Kato M., Riezman H., Yamaguchi Y., Maeda Y., Kinoshita T. 2011. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 194: 61–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeda Y., Tashima Y., Houjou T., Fujita M., Yoko-o T., Jigami Y., Taguchi R., Kinoshita T. 2007. Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol. Biol. Cell. 18: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder R. J., Ahmed S. N., Zhu Y., London E., Brown D. A. 1998. Cholesterol and sphingolipid enhance the Triton X-100 insolubility of glycosylphosphatidylinositol-anchored proteins by promoting the formation of detergent-insoluble ordered membrane domains. J. Biol. Chem. 273: 1150–1157 [DOI] [PubMed] [Google Scholar]
- 9.Hanada K., Nishijima M., Akamatsu Y., Pagano R. E. 1995. Both sphingolipids and cholesterol participate in the detergent insolubility of alkaline phosphatase, a glycosylphosphatidylinositol-anchored protein, in mammalian membranes. J. Biol. Chem. 270: 6254–6260 [DOI] [PubMed] [Google Scholar]
- 10.Horvath A., Sutterlin C., Manning-Krieg U., Movva N. R., Riezman H. 1994. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 13: 3687–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe R., Funato K., Venkataraman K., Futerman A. H., Riezman H. 2002. Sphingolipids are required for the stable membrane association of glycosylphosphatidylinositol-anchored proteins in yeast. J. Biol. Chem. 277: 49538–49544 [DOI] [PubMed] [Google Scholar]
- 12.Okamoto M., Yoko-o T., Umemura M., Nakayama K., Jigami Y. 2006. Glycosylphosphatidylinositol-anchored proteins are required for the transport of detergent-resistant microdomain-associated membrane proteins Tat2p and Fur4p. J. Biol. Chem. 281: 4013–4023 [DOI] [PubMed] [Google Scholar]
- 13.Mays R. W., Siemers K. A., Fritz B. A., Lowe A. W., van Meer G., Nelson W. J. 1995. Hierarchy of mechanisms involved in generating Na/K-ATPase polarity in MDCK epithelial cells. J. Cell Biol. 130: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledesma M. D., Simons K., Dotti C. G. 1998. Neuronal polarity: essential role of protein-lipid complexes in axonal sorting. Proc. Natl. Acad. Sci. USA. 95: 3966–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivier A. S., Castillon G. A., Michon L., Fukasawa M., Romanova-Michaelides M., Jaensch N., Hanada K., Watanabe R. 2010. Exit of GPI-anchored proteins from the ER differs in yeast and mammalian cells. Traffic. 11: 1017–1033 [DOI] [PubMed] [Google Scholar]
- 16.Yamaji T., Kumagai K., Tomishige N., Hanada K. 2008. Two sphingolipid transfer proteins, CERT and FAPP2: their roles in sphingolipid metabolism. IUBMB Life. 60: 511–518 [DOI] [PubMed] [Google Scholar]
- 17.Hanada K., Hara T., Fukasawa M., Yamaji A., Umeda M., Nishijima M. 1998. Mammalian cell mutants resistant to a sphingomyelin-directed cytolysin. Genetic and biochemical evidence for complex formation of the LCB1 protein with the LCB2 protein for serine palmitoyltransferase. J. Biol. Chem. 273: 33787–33794 [DOI] [PubMed] [Google Scholar]
- 18.Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 426: 803–809 [DOI] [PubMed] [Google Scholar]
- 19.Funato K., Riezman H. 2001. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J. Cell Biol. 155: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashida H., Hong Y., Murakami Y., Shishioh N., Sugimoto N., Kim Y. U., Maeda Y., Kinoshita T. 2005. Mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I. Mol. Biol. Cell. 16: 1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong Y., Kang J. Y., Kim Y. U., Shin D. J., Choy H. E., Maeda Y., Kinoshita T. 2005. New mutant Chinese hamster ovary cell representing an unknown gene for attachment of glycosylphosphatidylinositol to proteins. Biochem. Biophys. Res. Commun. 335: 1060–1069 [DOI] [PubMed] [Google Scholar]
- 22.Hong Y., Ohishi K., Inoue N., Kang J. Y., Shime H., Horiguchi Y., van der Goot F. G., Sugimoto N., Kinoshita T. 2002. Requirement of N-glycan on GPI-anchored proteins for efficient binding of aerolysin but not Clostridium septicum alpha-toxin. EMBO J. 21: 5047–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong Y., Ohishi K., Kang J. Y., Tanaka S., Inoue N., Nishimura J., Maeda Y., Kinoshita T. 2003. Human PIG-U and yeast Cdc91p are the fifth subunit of GPI transamidase that attaches GPI-anchors to proteins. Mol. Biol. Cell. 14: 1780–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oslowski C. M., Urano F. 2011. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 490: 71–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., Schwudke D. 2008. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke N. G., Dawson R. M. 1981. Alkaline O leads to N-transacylation. A new method for the quantitative deacylation of phospholipids. Biochem. J. 195: 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epstein S., Kirkpatrick C. L., Castillon G. A., Muniz M., Riezman I., David F. P., Wollheim C. B., Riezman H. 2011. Activation of the unfolded protein response pathway causes ceramide accumulation in yeast and INS-1E insulinoma cells. J. Lipid Res. 53: 412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta V., Patwardhan G. A., Zhang Q. J., Cabot M. C., Jazwinski S. M., Liu Y. Y. 2010. Direct quantitative determination of ceramide glycosylation in vivo: a new approach to evaluate cellular enzyme activity of glucosylceramide synthase. J. Lipid Res. 51: 866–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan X. L., Riezman I., Wenk M. R., Riezman H. 2010. Yeast lipid analysis and quantification by mass spectrometry. Methods Enzymol. 470: 369–391 [DOI] [PubMed] [Google Scholar]
- 30.van der Bijl P., Strous G. J., Lopes-Cardozo M., Thomas-Oates J., van Meer G. 1996. Synthesis of non-hydroxy-galactosylceramides and galactosyldiglycerides by hydroxy-ceramide galactosyltransferase. Biochem. J. 317: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tashima Y., Taguchi R., Murata C., Ashida H., Kinoshita T., Maeda Y. 2006. PGAP2 is essential for correct processing and stable expression of GPI-anchored proteins. Mol. Biol. Cell. 17: 1410–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaji T., Nishikawa K., Hanada K. 2010. Transmembrane BAX inhibitor motif containing (TMBIM) family proteins perturbs a trans-Golgi network enzyme, Gb3 synthase, and reduces Gb3 biosynthesis. J. Biol. Chem. 285: 35505–35518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosales Fritz V. M., Daniotti J. L., Maccioni H. J. 1997. Chinese hamster ovary cells lacking GM1 and GD1a synthesize gangliosides upon transfection with human GM2 synthase. Biochim. Biophys. Acta. 1354: 153–158 [DOI] [PubMed] [Google Scholar]
- 34.Okuda T., Nakayama K. 2008. Identification and characterization of the human Gb3/CD77 synthase gene promoter. Glycobiology. 18: 1028–1035 [DOI] [PubMed] [Google Scholar]
- 35.Halter D., Neumann S., van Dijk S. M., Wolthoorn J., de Maziere A. M., Vieira O. V., Mattjus P., Klumperman J., van Meer G., Sprong H. 2007. Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. J. Cell Biol. 179: 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohyama-Koganeya A., Sasamura T., Oshima E., Suzuki E., Nishihara S., Ueda R., Hirabayashi Y. 2004. Drosophila glucosylceramide synthase: a negative regulator of cell death mediated by proapoptotic factors. J. Biol. Chem. 279: 35995–36002 [DOI] [PubMed] [Google Scholar]
- 37.Kumagai K., Yasuda S., Okemoto K., Nishijima M., Kobayashi S., Hanada K. 2005. CERT mediates intermembrane transfer of various molecular species of ceramides. J. Biol. Chem. 280: 6488–6495 [DOI] [PubMed] [Google Scholar]
- 38.Hong Y., Maeda Y., Watanabe R., Inoue N., Ohishi K., Kinoshita T. 2000. Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem. 275: 20911–20919 [DOI] [PubMed] [Google Scholar]
- 39.Takida S., Maeda Y., Kinoshita T. 2008. Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochem. J. 409: 555–562 [DOI] [PubMed] [Google Scholar]
- 40.Castillon G. A., Aguilera-Romero A., Manzano-Lopez J., Epstein S., Kajiwara K., Funato K., Watanabe R., Riezman H., Muniz M. 2011. The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol. Biol. Cell. 22: 2924–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S., et al. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 323: 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Promlek T., Ishiwata-Kimata Y., Shido M., Sakuramoto M., Kohno K., Kimata Y. 2011. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell. 22: 3520–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abrami L., Fivaz M., Kobayashi T., Kinoshita T., Parton R. G., van der Goot F. G. 2001. Cross-talk between caveolae and glycosylphosphatidylinositol-rich domains. J. Biol. Chem. 276: 30729–30736 [DOI] [PubMed] [Google Scholar]
- 44.Fukasawa M., Nishijima M., Hanada K. 1999. Genetic evidence for ATP-dependent endoplasmic reticulum-to-Golgi apparatus trafficking of ceramide for sphingomyelin synthesis in Chinese hamster ovary cells. J. Cell Biol. 144: 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shishioh N., Hong Y., Ohishi K., Ashida H., Maeda Y., Kinoshita T. 2005. GPI7 is the second partner of PIG-F and involved in modification of glycosylphosphatidylinositol. J. Biol. Chem. 280: 9728–9734 [DOI] [PubMed] [Google Scholar]
- 46.Puoti A., Conzelmann A. 1993. Characterization of abnormal free glycophosphatidylinositols accumulating in mutant lymphoma cells of classes B, E, F, and H. J. Biol. Chem. 268: 7215–7224 [PubMed] [Google Scholar]
- 47.Baumann N. A., Vidugiriene J., Machamer C. E., Menon A. K. 2000. Cell surface display and intracellular trafficking of free glycosylphosphatidylinositols in mammalian cells. J. Biol. Chem. 275: 7378–7389 [DOI] [PubMed] [Google Scholar]
- 48.Singh N., Liang L. N., Tykocinski M. L., Tartakoff A. M. 1996. A novel class of cell surface glycolipids of mammalian cells. Free glycosyl phosphatidylinositols. J. Biol. Chem. 271: 12879–12884 [DOI] [PubMed] [Google Scholar]
- 49.Bonnon C., Wendeler M. W., Paccaud J. P., Hauri H. P. 2010. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J. Cell Sci. 123: 1705–1715 [DOI] [PubMed] [Google Scholar]
- 50.Niimura Y., Moue T., Takahashi N., Nagai K. 2010. Modification of sphingoglycolipids and sulfolipids in kidney cell lines under heat stress: activation of monohexosylceramide synthesis as a ceramide scavenger. Glycobiology. 20: 710–717 [DOI] [PubMed] [Google Scholar]
- 51.Uchida Y., Murata S., Schmuth M., Behne M. J., Lee J. D., Ichikawa S., Elias P. M., Hirabayashi Y., Holleran W. M. 2002. Glucosylceramide synthesis and synthase expression protect against ceramide-induced stress. J. Lipid Res. 43: 1293–1302 [PubMed] [Google Scholar]
- 52.Fujita M., Umemura M., Yoko-o T., Jigami Y. 2006. PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol. Biol. Cell. 17: 5253–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sutterwala S. S., Creswell C. H., Sanyal S., Menon A. K., Bangs J. D. 2007. De novo sphingolipid synthesis is essential for viability, but not for transport of glycosylphosphatidylinositol-anchored proteins, in African trypanosomes. Eukaryot. Cell. 6: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.