Abstract
Dysregulation of ceramide synthesis has been associated with metabolic disorders such as atherosclerosis and diabetes. We examined the changes in lipid homeostasis and gene expression in Huh7 hepatocytes when the synthesis of ceramide is perturbed by knocking down serine pal mitoyltransferase subunits 1, 2, and 3 (SPTLC123) or dihydroceramide desaturase 1 (DEGS1). Although knocking down all SPTLC subunits is necessary to reduce total ceramides significantly, depleting DEGS1 is sufficient to produce a similar outcome. Lipidomic analysis of distribution and speciation of multiple lipid classes indicates an increase in phospholipids in SPTLC123-silenced cells, whereas DEGS1 depletion leads to the accumulation of sphingolipid intermediates, free fatty acids, and diacylglycerol. When cer amide synthesis is disrupted, the transcriptional profiles indicate inhibition in biosynthetic processes, downregulation of genes involved in general endomembrane traffi cking, and upregulation of endocytosis and endosomal recycling. SPTLC123 silencing strongly affects the expression of genes involved with lipid metabolism. Changes in amino acid, sugar, and nucleotide metabolism, as well as vesicle trafficking between organelles, are more prominent in DEGS1-silenced cells. These studies are the first to provide a direct and comprehensive understanding at the lipidomic and transcriptomic levels of how Huh7 hepatocytes respond to changes in the inhibition of ceramide synthesis.
Keywords: serine palmitoyl transferase, dihydroceramide desaturase, ceramide, sphingolipids, human hepatocytes, lipidomic, microarray, LDL receptor, apoB secretion
Dyslipidemia leading to cardiovascular and metabolic diseases remains a major issue in many developed countries. Oversupply from diets and poor processing of lipids at a cellular level can promote greater risk toward metabolic disorders. Recent studies have become more focused on the function and regulation of different lipid classes, including sphingolipids, to understand their contribution to diseases. For instance, sphingolipids, such as ceramides, are highly elevated in the serum, liver, and muscle of type 2 diabetic patients (1) and insulin-resistant rodents (2). Besides being a component of lipid rafts and cell membranes, sphingolipids have been reported to function as mediators of cell growth, cell differentiation, and cell death (3–7). De novo ceramide biosynthesis starts with the first and rate-limiting step to condense serine and palmitoyl-CoA by an enzyme complex called serine palmitoyltransferase (SPTLC) producing 3-ketosphinganine (Fig. 1A). During the next step, 3-ketosphinganine is reduced by 3-ketosphinganine reductase to generate sphinganine. Depending on the tissue distribution, a certain family member of ceramide synthase (CerS) preferentially uses a certain chain-length fatty acyl-CoA to convert sphinganine to dihydroceramide (dhCer), which is then desaturated by dihydroceramide desaturase (DEGS) to yield a unique subspecies of ceramide.
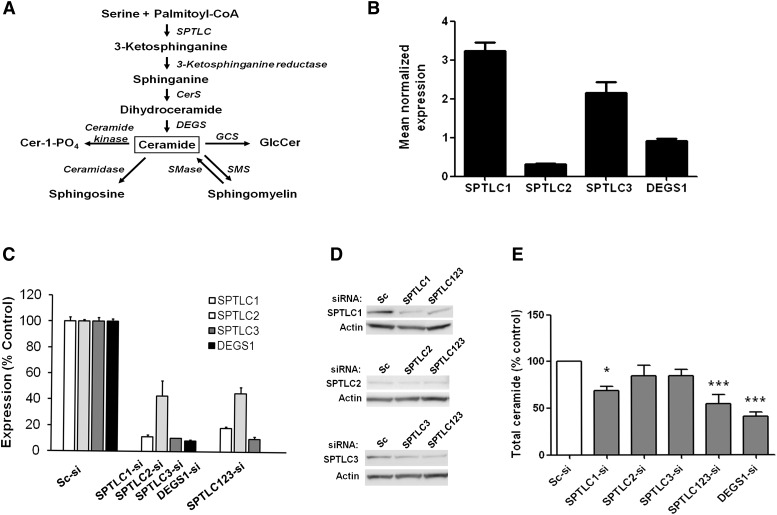
Fig. 1.
Characterization of SPTLC and DEGS1 gene expression and silencing effects at mRNA, protein, and ceramide levels in Huh7 cells. (A) A schematic diagram representing the de novo ceramide biosynthesis pathway shows SPTLC as the first enzymatic reaction and DEGS1-desaturating reaction to convert dihydroceramide to ceramide, which can be converted to other sphingolipids. (B) SPTLC1, SPTLC2, SPTLC3, and DEGS1 mRNA expression in Huh7 cells. Transcriptional level of each SPTLC subunit and DEGS1 in the Huh7 cells was quantified and normalized to cyclophillin A mRNA levels as described in Materials and Methods. (C) Silencing SPTLC and DEGS1 mRNA expression by siRNA after 72 h of treatment. Transcriptional level of SPTLC1 (white bar), SPTLC2 (light gray bar), SPTLC3 (dark gray bar), and DEGS1 (black bar) obtained from the cells transfected with SPTLC1, SPTLC2, SPTLC3, DEGS1, or combination of SPTLC123 siRNA and compared with scramble siRNA (Sc-si). The expression of each gene was normalized to PPIA. (D) A Western blot analysis was performed with antibodies against SPTLC1, SPTLC2, and SPTLC3 protein to confirm the knockdown effects at the protein level. β-actin was used to ensure equal protein loading in each lane. (E) Total level of major ceramides analyzed from the whole-cell lysate of SPTLC- and DEGS1-silenced cells after 72 h of siRNA transfection. Data are represented as a percent of Sc-si control from three to five independent experiments (mean ± SEM, *P < 0.05, ***P < 0.001).
Currently, the major identified SPTLC subunits, SPTLC1, SPTLC2, and SPTLC3, are differentially expressed in human tissues (8). Both SPTLC2 and SPTLC3 subunits contains pyridoxal phosphate binding motif, but each appears to have a different substrate preference as shown in the overexpressed human embryonic kidney cells (9). Whereas palmitate-CoA is the predominant substrate for SPTLC2, mysristoyl and lauryl-CoA are preferential substrates for SPTLC3, resulting in a different chain length of the sphingoid base. Another gene family (ssSPT) with two small subunits has recently been discovered and is believed to be associated with the major SPTLC subunits to form a large enzyme complex (10). Multiple reports indicate that using myriocin to reduce SPTLC activities decreases the production of ceramide and is suggested to prevent the formation or slow the progression of atherosclerotic lesions, improve plasma lipid profiles, and prevent the onset of diabetes in rodents (11–13). Not only has inhibiting SPTLC activity by myriocin shown to be beneficial but also a small interfering RNA (siRNA) against SPTLC1 stimulates cholesterol efflux in macrophages by activating ATP cassette binding transporter 1 (ABCA1) (14). In addition, depletion of SPTLC in human macrophages shows a reduction in palmi tate-induced proinflamatory cytokine secretion of tumor necrosis factor (TNF)-α and interleukin (IL)-1β (15). A recent genome-wide association study in European populations established a strong correlation between SPTLC3 single nucleotide polymorphisms and circulating sphingolipid levels (16).
The inhibition of DEGS1 is hypothesized to benefit diabetes and cancer. DEGS1 is a protein that contains three histidine consensus motifs, a hallmark of the membrane fatty acid desaturase group. A study carried out by Holland et al. reported that heterozygous knockout mice are more sensitive to insulin and have normal glucose tolerance (13). In addition, silencing of DEGS1 in human neuroblastoma cells decreases ceramide synthesis and inhibits cell growth and cell-cycle progression through the dephosphorylation of the retinoblastoma protein (17).
Regardless of several reports describing various aspects of SPTLC and DEGS, the global changes in transcriptional regulation and lipid homeostasis responding to specific and targeted knockdown of these enzymes have not been closely examined. As liver is a major site of lipid production and homeostasis, a human hepatocarcinoma cell line, Huh7, is used as a model in this study. We utilize siRNA as a tool to knock down SPTLC or DEGS1 and to investigate the changes in lipids and gene expression as a result of ceramide reduction. Our initial hypothesis is that the changes in lipid profiles and transcriptional regulations will differ between the inhibition of SPTLC and DEGS1, although the depletion of either enzyme yields a similar reduction in the ceramide levels.
MATERIALS AND METHODS
Cell culture and siRNA transfection
Human hepatocarcinoma (Huh7) cells were obtained from Dr. Yi Luo (Pfizer Inc., Groton, CT) (18) and maintained at 70–80% confluence in the growing media consisting of high-glucose DMEM (Life Technologies, Carlsbad, CA), 10% fetal bovine serum (BSA, Sigma-Aldrich, St. Louis, MO), 2 mM L-glutamine, 10 mM hepes, and 1% penicillin-streptomycin (Life Technologies) at 37°C, 5% CO2. Huh7 cells were seeded at 60,000 cells/well in a 12-well plate or 6 millions cells/500 cm2 plate overnight. Next day, Silencer® Select siRNA (Life Technologies) against SPTLC123 or DEGS1 gene and siLentFect™ lipid reagent (Bio-Rad, Hercules, CA) was prepared in OptiMEM according to the manufacturer protocol. Silencer® Select scramble #1 was used as a nontargeting control. The siRNA-lipid complex was added to each well to yield the final concentration of 20 nM siRNA and incubated for 24 h at 37°C, 5% CO2. Media was replaced with the fresh-growing media, and cells were incubated for another 24 h before the conditioned media (DMEM + 1% FBS + fatty acid free BSA) was added to the cells and incubated for another 24 h prior to harvest.
Transcriptional expression analysis
In the QuantiGene 2.0 assay, cells were harvested by washing twice with Dulbecco's phosphate-buffered saline (D'PBS) and lysed with QuantiGene sample processing buffer (Affymetrix, Fremont, CA) according to manufacturer protocol. Corresponding gene-specific probes were purchased from Affymetrix. The QuantiGene 2.0 assay was used to detect mRNA expression of the gene of interest, and each relative light value was normalized to cyclophillin A (PPIA) expression. The expression of target genes from CRE analysis (supplementary File VI) was performed using Taqman® Gene Expression method. Total RNA of siRNA-transfected cells was isolated using RNeasy Mini Kit (Qiagene, Valencia, CA). Quantitative PCR was performed according to the manufacturing protocol, in a 7900HT Sequence Detection System (Life Technologies) using reagents and gene specific Taqman® assays (Life Technologies) as indicated in the table in supplementary File VI. Relative mRNA levels were determined using the comparative Ct method and normalized to PPIA reference gene.
Western blot analysis
Cells were washed with D'PBS and lysed in 1× RIPA buffer (Sigma-Aldrich, St. Louis, MO), proteinase inhibitor cocktail and phosphatase inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). The supernatant was recovered by centrifugation, and the protein concentration was quantified in the BCA assay. Protein lysates were subjected to SDS-PAGE and Western blot analysis. Antibodies purchased commercially were against human SPTLC1 (mouse monoclonal, Santa Cruz Biotechnology, Santa Cruz, CA), SPTLC2 (rabbit polyclonal, Abcam, Cambridge, MA), SPTLC3 (goat polyclonal, Santa Cruz Biotechnology), β-actin (mouse monoclonal, Sigma-Aldrich), and LDLR (goat polyclonal, R and D Systems, Minneapolis, MN).
Cell proliferation, caspase 3/7 assay, and SPT activity
Cells were seeded and transfected with siRNA according to the transfection protocol described above and left in the growth media for 48 h. At 24 h before harvest, the growing media was replaced with the conditioned media. The transfected cells were subjected to cell proliferation or caspase 3/7 assay at the end of 24 h conditioned media treatment. Cell proliferation was performed using CyQUANT Direct Cell Proliferation assay kit (Life Technologies), and the Caspase 3/7 assay was performed using CellEvent Caspase-3/7 Green Detection Reagent (Life Technologies) following the manufacturer's protocol. Fluorescence was measured using SpectraMax Gemini EM plate reader (Molecular Devices, Sunnyvale, CA).
For the SPT activity, a radiolabeled 14C-serine was added to the siRNA-transfected cells that were grown in the conditioned media for 4 h before harvest. Cells were harvested with trypsin and washed three times with cold D'PBS. Lipid was extracted using Blight-Dye method (19) and spotted on a Baker-flex silica plate (Avantor Performance Materials, Phillipsburg, NJ) with a marker. The plate was run in a closed chamber with chloroform-methanol solvent. The signal was quantified with a scintillation detector.
Microarray hybridizations and bioinformatic analyses
Total RNA isolation and hybridizations to the human genome U133 Plus 2.0 Affymetrix array chip were performed by Gene Logic. All .CEL files from the microarray experiment have been submitted to GEO (http://www.ncbi.nlm.nih.gov/geo/) and can be identified with the accession ID GSE28059 (20). All data is minimum information about a microarray experiment (MIAME) compliant. First, .CEL files were normalized using robust multi-array analysis (RMA) (21). Then, pair-wise comparison between the different siRNA groups was performed using a simple linear model to calculate P values, followed by Benjamini and Hochberg false discovery rate (FDR) correction. Prior to bioin formatic analysis, Affymetrix probe sets designated with a “_x” were removed because they potentially lacked gene specificity. All remaining probes were annotated with information provided by NetAffx (Affymetrix), Gene Entrez ID (NCBI), Ingenuity Pathway Analysis (IPA; Ingenuity Sytems), and Ariadne Pathway Studios tools (MedScan Technology). For ease of comparison, bioinformatic analysis was performed so that all the treatment groups were compared with the scrambled siRNA treatment. An expression difference between treatments for a probe was considered significant if the fold-change was ≥1.5 and the FDR-adjusted P value ≤ 0.05. Hierarchical clustering was performed with Spotfire Decision Site 9.0 (http://www.spotfire.com) using a UPGMA clustering method, cosine similarity measure, and an input rank-order function. Gene set enrichment analysis (GSEA) was performed with DAVID Bioinformatics Resources 6.7, in which gene sets were considered significant if the enrichment score was greater than 2.0 (22, 23). Pathway and network analysis was performed using IPA tools (Ingenuity Systems) and Ariadne Pathway Studios tools (MedScan Technology).
Causal reasoning engine analysis
Causal reasoning engine (CRE) is an algorithm to infer upstream molecular mechanisms consistent with observed expression changes (24). Causal reasoning algorithms can be viewed as a form of gene set enrichment in which gene sets are defined by their common response to a defined molecular perturbation (e.g., inhibition of TNF activity). Two metrics to quantify the significance of an upstream hypothesis with respect to the experimental data were considered: The enrichment P value treats each set of downstream response genes as one gene set and does not take the direction of regulation into account. The correctness P value also accounts for upregulation and downregulation in the experimentally observed regulations as well as the literature-derived changes downstream of the molecular perturbation. To ensure statistical significance under a null model of randomly reassigning upregulated and downregulated transcripts to arbitrary transcript nodes, we recomputed this score 1,000 times under the null model and computed approximate P values. If the real score observed in our data was always greater than in any randomly assigned run, we noted a P value < 0.001.
The user of CRE applies cutoffs for both metrics of the automatically derived hypotheses. Then the user manually interprets the hypotheses for validity and context. For the purpose of this study, hypotheses were limited to those that are one step away from the observed transcriptional data and are reasonable for the Huh7 model used in the experiment. In addition, the nominal enrichment and correctness P values were limited based on a threshold of <0.01. Ultimately, we focused on the top 20 hypotheses and note that most of them have significant enrichment P values even under the conservative Bonferroni correction for multiple testing (number of tests is ∼3,500 in both cases; resulting Bonferroni threshold is 0.05 / 3,500 = 1.4e−05).
Lipid profiling
To analyze ceramides from the whole-cell lysate, Huh7 cells were lyzed with a homogenizer and immediately run on the high performance liquid chromatography (HPLC) system. Chromatography was achieved using a Phenomenex Synergi Polar-RP (2.0 × 150 mm, 4 μm, 80A) column. The injection volume of cell lysate was 30 μl with the flow rate at 0.220 ml/min. The HPLC system was coupled to an Applied Biosystems API 4000 mass spectrometer operated in positive multiple reaction monitoring (MRM) mode. The transition was monitored for ceramide C16:0, C18:0, C20:0, and C24:1. The quantity of ceramide was normalized with total protein and calculated as a percentage of the scrambled siRNA-treated sample.
Subspecies of purified sphingolipids were analyzed at Lipidomics Core Facility of the Medical University of South Carolina. Briefly, to extract sphingolipids, cell pellets were sonicated twice for 30 s each in 2 ml of isopropanol-water-ethyl acetate (30:10:60, v/v). After vortex and spin down at 4,000 g for 10 min, supernatants were pooled, dried, and then further extracted following the Bligh-Dye extraction method (19). Sphingolipids were measured by high-performance liquid chromatography-tandem mass spectrometry on a Thermo Finnigan (Waltham, MA) TSQ 7000 triple quadrupole mass spectrometer operating in a MRM positive ionization mode as previously described (25).
Other lipid classes, including triacylglycerol, diacylglycerol, glycerolphospholipids, free cholesterol, cholesteryl ester, free fatty acids, cardiolipin, and lysophosphatidylcholine, were analyzed at Lipomics Technologies, West Sacramento, CA. The lipids from cell pellets were extracted in the presence of authentic internal standards by the method of Folch et al. (26) using chloroform:methanol (2:1 v/v). Individual lipid classes within each extract were separated by liquid chromatography (Agilent Technologies model 1100 series). Each lipid class was transesterified in 1% sulfuric acid in methanol in a sealed vial under a nitrogen atmosphere at 100°C for 45 min. The resulting fatty acid methyl esters were extracted from the mixture with hexane containing 0.05% butylated hydroxytoluene and were prepared for gas chromatography by sealing the hexane extracts under nitrogen. Fatty acid methyl esters were separated and quantified by capillary gas chromatography (Agilent Technologies model 6890) equipped with a 30 m DB 88MS capillary column (Agilent Technologies) and a flame ionization detector.
Lipid bioinformatic analyses
Mass values for lipids were transformed to a ratio relative to scrambled siRNA by comparing each treatment with the average mass for the scrambled siRNA treatment. Statistic analysis was performed according to the following steps unless indicated otherwise. Treatment groups and lipid ratios were statistically compared by two-way ANOVA to calculate P values, followed by a Bonferroni FDR correction using GraphPad Prism version 5.01 for Windows (GraphPad Software, San Diego, CA). A difference between treatments for a lipid amount was considered significant if fold-change ≥ 1.5 (transformation of the calculated ratio) and the FDR-adjusted P value ≤ 0.05. Significant lipids were annotated with information provided by PubChem (NCBI), IPA tools (Ingenuity Sytems), and the Human Metabolome Database (27).
ApoB ELISA
Media from siRNA-treated cells was recovered from each well before harvest. Cells were washed with D'PBS three times before lysing with cell lysis buffer (50 mM HEPES, 1 mM EDTA, 1% NP40, 120 mM NaCl, 1% glycerol) and centrifugation. The lysate was then subjected to BCA assay for protein quantification. ApoB ELISA was performed as described previously (18).
Data plotting and statistical analysis
Data was plotted using Microsoft Excel or GraphPad Prism software. Unless stated in the legend of each figure or table, a basic statistic analysis was performed using one-way ANOVA. A P value < 0.05 compared with scrambled siRNA treatment was considered as significant.
RESULTS
SPTLC and DEGS1 expression in Huh7 cells
We determined the mRNA levels of the three major SPTLC subunits (SPTLC1, SPTLC2, and SPTLC3) and DEGS1 in Huh7 cells. Relative to cyclophillin A, the major SPTLC subunits expressed in this cell line are SPTLC1 and SPTLC3 (Fig. 1B). In agreement with a previous study (8), the expression of SPTLC2 was noticeably low (∼10-fold lower than SPTLC1 and ∼7-fold lower than SPTLC3). The expression of DEGS1 was also detectable in Huh7 cells, although at a lower level than SPTLC1 or SPTLC3.
DEGS1 and the three subunits of SPTLC were downregulated using siRNA without affecting cell morphology or viability as determined by the trypan blue staining method. There was no significant difference on mRNA knockdown 24, 48, or 72 h post transfection (data not shown). At 72 h post transfection, DEGS1, SPTLC1, and SPTLC3 mRNA levels were reduced by more than 75%. SPTLC2 mRNA was decreased by only ∼50% (Fig. 1C). Similar knockdown was obtained by using a different set of siRNA oligonucleotides (data not shown).
At the protein level, the silencing effects of SPTLC siRNA were detected at 72 h post transfection (Fig. 1D). As expected, the level of SPTLC1 and SPTLC3 protein were dramatically reduced. It was more difficult to visually observe the decrease in SPTLC2 protein compared with other SPTLC subunits. Synchronizing knockdown of three SPTLC subunits yielded a reduced expression similar to knocking down an individual subunit. We were not able to specifically and conclusively detect DEGS1 protein in the whole-cell lysate from scrambled or DEGS1 siRNA-treated cells despite testing all commercially available DEGS1 antibodies. Nevertheless, we believe that the synthesis of DEGS1 was significantly decreased as shown by the changes in the levels of mRNA and different classes of sphingolipids (see Figs. 1C, E and 2A, B, D and Table 1).
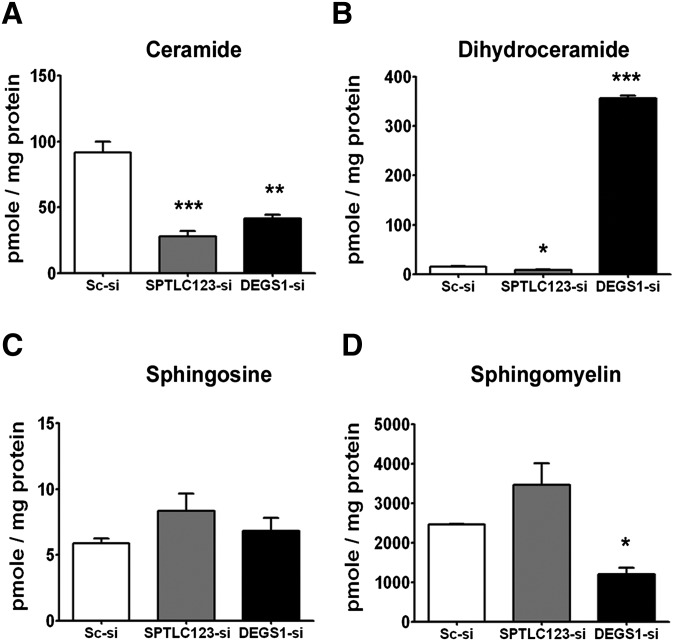
Fig. 2.
The effects of SPTLC123 or DEGS1 silencing on the level of different sphingolipids. Total purified (A) ceramide, (B) dihydroceramide, (C) sphingosine, and (D) sphingomyelin from siRNA-treated Huh7 cells were analyzed at Lipidomics Core Facility and normalized to the total protein. The complete data of each sphingolipid is indicated in supplementary File II. Data are plotted as mean ± SEM (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).
TABLE 1.
Significant changes in lipid species in SPTLC123 or DEGS1 siRNA-treated cells
| Lipid Class | Lipid Species | Scrambled siRNA | SPTLC123 siRNA | SPTLC123 P | DEGS1 siRNA | DEGS1 P | SPTLC123 versus DEGS1 P |
| Ceramide | Cer 16:0 | 1.000 | −1.469 | NS | −3.408 | P < 0.01 | NS |
| Ceramide | Cer 24:0 | 1.000 | −4.485 | P < 0.001 | −2.560 | P < 0.001 | P < 0.001 |
| Ceramide | Cer 24:1 | 1.000 | −3.428 | P < 0.001 | −2.152 | P < 0.001 | P < 0.001 |
| Ceramide | Cer 26:1 | 1.000 | −8.185 | P < 0.001 | −3.194 | P < 0.01 | NS |
| Dihydroceramide | dhCer 14:0 | 1.000 | −1.799 | NS | 10.072 | P < 0.001 | P < 0.001 |
| Dihydroceramide | dhCer 16:0 | 1.000 | −1.497 | NS | 12.207 | P < 0.001 | P < 0.001 |
| Dihydroceramide | dhCer 18:0 | 1.000 | −1.371 | NS | 8.332 | P < 0.001 | P < 0.001 |
| Dihydroceramide | dhCer 20:0 | 1.000 | 1.268 | NS | 10.512 | P < 0.001 | P < 0.001 |
| Dihydroceramide | dhCer 22:0 | 1.000 | −1.867 | NS | 25.166 | P < 0.001 | P < 0.001 |
| Dihydroceramide | dhCer 22:1 | 1.000 | −4.376 | NS | 9.928 | P < 0.001 | P < 0.001 |
| Dihydroceramide | dhCer 24:0 | 1.000 | −3.427 | NS | 67.891 | P < 0.001 | P < 0.001 |
| Dihydroceramide | dhCer 24:1 | 1.000 | −3.847 | NS | 27.561 | P < 0.001 | P < 0.001 |
| Dihydroceramide | dhCer 26:0 | 1.000 | −1.680 | NS | 25.921 | P < 0.001 | P < 0.001 |
| Dihydroceramide | dhCer 26:1 | 1.000 | −4.689 | NS | 78.955 | P < 0.001 | P < 0.001 |
| Sphinganine | dhSph | 1.000 | −1.041 | NS | 7.490 | P < 0.001 | P < 0.001 |
| Sphingomyelin | SM 16:0 | 1.000 | 1.461 | P < 0.001 | −2.546 | P < 0.001 | P < 0.001 |
| Cardiolipin | CL 16:0 | 1.000 | 1.236 | P < 0.001 | 1.195 | P < 0.001 | NS |
| Cardiolipin | CL 16:1n7 | 1.000 | 1.273 | P < 0.001 | −1.144 | P < 0.01 | P < 0.001 |
| Cardiolipin | CL 18:1n7 | 1.000 | 1.210 | P < 0.001 | −1.038 | NS | P < 0.001 |
| Cholesteryl ester | CE 16:0 | 1.000 | 1.201 | P < 0.05 | 1.429 | P < 0.001 | P < 0.01 |
| Free fatty acid | FFA 16:0 | 1.000 | 1.026 | NS | 1.578 | P < 0.001 | P < 0.001 |
| Free fatty acid | FFA 18:1n9 | 1.000 | −1.156 | NS | 1.878 | P < 0.001 | P < 0.001 |
| Diacylglycerol | DAG 16:0 | 1.000 | −1.413 | NS | 1.613 | P < 0.01 | P < 0.001 |
| Triacylglycerol | TAG 16:1n7 | 1.000 | −1.273 | NS | −2.645 | P < 0.05 | NS |
| Triacylglycerol | TAG 18:1n7 | 1.000 | −1.322 | NS | −2.152 | P < 0.01 | NS |
| Triacylglycerol | TAG 18:1n9 | 1.000 | −1.657 | P < 0.001 | −2.414 | P < 0.001 | P < 0.01 |
| Lysophosphatidylcholine | LYPC 16:0 | 1.000 | 1.703 | P < 0.01 | 1.607 | P < 0.05 | NS |
| Phosphatidylcholine | PC 16:0 | 1.000 | 1.330 | P < 0.001 | −1.016 | NS | P < 0.001 |
| Phosphatidylcholine | PC 16:1n7 | 1.000 | 1.441 | P < 0.001 | −1.283 | NS | P < 0.001 |
| Phosphatidylcholine | PC 18:1n7 | 1.000 | 1.299 | P < 0.01 | −1.334 | NS | P < 0.001 |
| Phosphatidylcholine | PC 18:1n9 | 1.000 | 1.118 | P < 0.01 | −1.097 | NS | P < 0.001 |
| Phosphatidylethanolamine | PE 20:4n6 | 1.000 | 1.434 | P < 0.05 | 1.248 | NS | NS |
| Phosphatidylserine | PS 18:0 | 1.000 | 1.113 | P < 0.01 | 1.059 | NS | NS |
| Phosphatidylserine | PS 20:4n6 | 1.000 | 1.205 | NS | −1.160 | NS | P < 0.05 |
Lipid levels were determined by LC/MS in Huh7 cells treated with scrambled, SPTLC123, or DEGS1 siRNA. The table summarizes lipid species across the different siRNA treatments are expressed as fold-change relative to scrambled siRNA treatment. Statistically significant differences are represented in bold (P ≤ 0.05 after Bonferroni correction). All 289 lipids compared in this study containing the actual values can be found in supplementary File II.
NS, nonsignificant.
Even though there may be a substrate specificity of SPTLC2 and SPTLC3 subunits in Huh7 cells as seen in Hek293 cells (9), for the main scope of this study, we focused on the effects of downregulating SPTLC and DEGS1 in ceramide synthesis; therefore, the levels of sphingoid base were not measured. We examined the changes in ceramide levels 72 h post siRNA transfection. Using mass spectrometry, C16:0, C18:0, C20:0, and C24:1 were detected as major ceramide subspecies. Interestingly, the level of ceramide was not altered when the SPTLC2 or SPTLC3 subunit was individually silenced, but it was significantly reduced when all subunits were knocked down (Fig. 1E). Consistent with the findings observed at mRNA and protein levels, the residual SPT activity in SPTLC123-silenced cells was approximately 30% compared with scrambled siRNA cells (supplementary Fig. I-A). Although SPTLC1 knockdown caused a significant decrease in ceramide compared with scrambled siRNA-treated control, silencing all three SPTLC subunits led to further reduction of total ceramide levels. DEGS1 siRNA-treated cells showed a decrease in ceramides similar to that observed in cells when all three SPTLC subunits were knocked down. This finding suggests that silencing DEGS1 is sufficient to significantly decrease ceramide levels, whereas all SPTLC subunits need to be silenced to achieve a similar effect.
We also evaluated whether SPTLC123 and DEGS1 siRNA knockdown affected cell proliferation and apoptosis. Although a significant decrease in cell proliferation was observed in the DEGS1-silenced cells, no changes were observed in SPTLC123-silenced cells (supplementary Fig. I-B). There was no evidence suggesting an effect on apoptosis in either SPTLC123 or DEGS1-silenced cells (supplementary Fig. I-C).
Effects of SPTLC123 or DEGS1 knockdown on sphingolipids levels
Because silencing all three SPTLC subunits at a time or silencing DEGS1 led to a significant reduction in ceramide levels, we continued our studies using SPTLC123 and DEGS1 siRNA-treated Huh7 cells. The levels of sphingolipids extracted from the silenced cells were analyzed at the Lipidomics Core Facility of the Medical University of South Carolina using a high performance liquid chromatography-tandem mass spectrometer (Fig. 2A–D, Table 1, and supplementary File II). From this analysis, we confirmed the significant reduction of total ceramide levels in both samples compared with the scrambled siRNA-treated control (Fig. 2A). Fold-change differences of sphingolipid subspecies relative to the scrambled siRNA treated group are shown in Table 1. Levels for all lipid species are shown in supplementary File II. Specifically, the major ceramide subspecies that were substantially reduced (at least 2-fold in both SPTLC123 and DEGS1 siRNA-treated cells) are very long chained ceramides C24:0, C24:1, and C26:1. In addition, DEGS1 depletion reduced C16:0 ceramide by 3.4-fold compared with the nontargeting control.
We observed more than 20-fold accumulation of dhCer when DEGS1 was knocked down. Conversely, dhCer decreased in the SPTLC123 siRNA-treated cells (Fig. 2B). All dhCer subspecies were increased following DEGS1 knockdown but to a different degree (Table 1). C24:0 and C26:0, very long chain fatty acid dhCer, were increased over 60-fold in DEGS1-treated cells. Sphinganine, a sphingolipid intermediate produced upstream of dhCer, increased more than 7-fold in DEGS1 siRNA-treated cells. No change in sphingosine levels was observed in either SPTLC123- or DEGS1-silenced cells (Fig. 2C). Sphingomyelin and its C16:0 subspecies were reduced substantially in DEGS1-silenced cells. On the contrary, there was a slight increasing trend, but not significant, in sphingomyelin and C16:0 subspecies in SPTLC123 siRNA-treated cells (Fig. 2D and Table 1).
Changes in lipid classes at the subspecies level in SPTLC123- and DEGS1-silenced cells
As treating Huh7 cells with SPTLC123 or DEGS1 siRNA substantially affects lipid levels in the sphingolipid pathway, there is a strong possibility that other lipid synthesis pathways could be altered. We next assessed the subspecies of cholesteryl ester (CE), cardiolipin (CL), diacylglycerol (DAG), free cholesterol (FC), free fatty acid (FFA), lysophophatidylcholine (LYPC), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS) and triacylglycerol (TAG). Although the total level of each lipid class was not altered, there were substantial changes at the subspecies level (Table 1 and supplementary File II). For example, the C16:0, C16:1n7, C18:1n7, and C18:1n9 PC significantly increased in the SPTLC123-silenced cells. A slight increase in C18:0 PS and C20:4n6 PE was also observed in the SPTLC123 siRNA-treated cells. Interestingly, silencing DEGS1 did not increase phospholipids as seen in the SPTLC123-depleted cells; rather, there was a slightly reducing trend, especially in the PC subspecies.
TAG subspecies C16:1n7, C18:1n7, and C18:1n9 were reduced, but the C16:0 DAG and C16:0 and C18:1n9 FFA were elevated in DEGS1-silenced cells. The strong directional changes in TAG and DAG were not evident in SPTLC123 knockdown cells. However, we observed a slight but significant increase of C16:0, C16:1n7, and C18:1n7 cardiolipin in these cells. Although the majority of lipid changes is different between SPTLC123 and DEGS1 knockdown, there was an increase of C16:0 CE and LYPC in both groups. Interestingly, C16:0 is the most common lipid subspecies changed in both SPTLC123 and DEGS1 siRNA-treated cells, which is consistent with the role that palmitate plays as a primary substrate for ceramide biosynthesis.
Comparison of SPTLC123- and DEGS1-silencing effects in lipid synthesis pathways
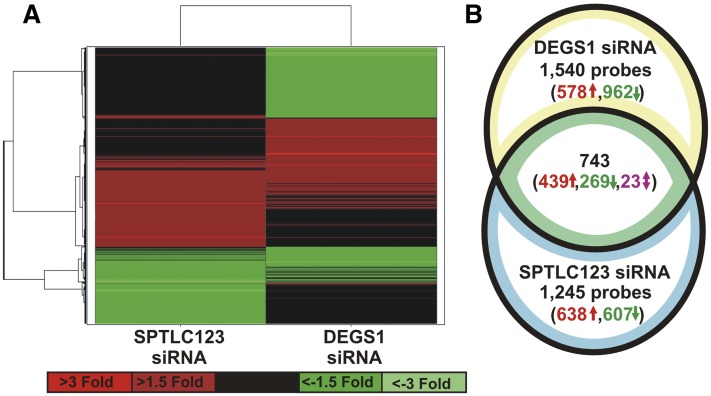
Global changes in gene expression were evaluated at 72 h after transfection with scrambled, SPTLC123, or DEGS1 siRNA. Statistical tests were performed so that the SPTLC123 siRNA and the DEGS1 siRNA-treated cells were compared with the scrambled siRNA treatment. Hierarchical clustering was performed with the statistically significant 3,528 probes with >1.5-fold change in at least one treatment using a UPGMA clustering method to identify gene expression clusters (Fig. 3A). The Venn diagram (Fig. 3B) highlights the extent of probe overlap between SPTLC123 and DEGS1 siRNA treatments. There were fewer overall expression changes in the SPTLC123 than in the DEGS siRNA-treated cells and fewer overlapping probes than the uniquely changed probes of each siRNA-treated group, suggesting a distinct pattern of gene expression between the two treatments.
Fig. 3.
Global changes of gene transcription in Huh7 cells treated with SPTLC123 or DEGS1 siRNA. (A) Affymetrix microarray gene expression analysis was performed with RNA from Huh7 cells treated with scrambled, SPTLC123, or DEGS1 siRNA as described under Materials and Methods. (B) A Venn diagram grouping the 3,528 probes on direction of change observed across both targeted siRNA treatments was presented. The number of upregulated probes is red with an up arrow, and the number of downregulated genes is green with a down arrow. The number of probes with direction disagreement is magenta with a bidirectional arrow. A complete table for all 3,528 probes compared in this study containing the fold change, adjusted P value, and further annotation can be found in supplementary File III.
To better understand the consequences of SPTLC123 or DEGS1 knockdown on lipid homeostasis, the lipid classes with statistical significance at a subspecies level and transcripts involved with the lipid synthesis pathways were studied. (See supplementary Fig. I-D, E for a simplified model highlighting changes in lipid levels.) When cells were treated with SPTLC123 siRNA, there was a marked reduction in ceramide and an increase in phospholipids. The corresponding changes in genes involved in PE synthesis, such as CDP-diacylglycerol synthase (CDS1), suggest that the increase of PE could be a result of the flux from DAG and ceramide. Using DAG to generate PE could lead to a reduction of substrates available for TAG production. An increase in PE appeared to flux to LYPC and PC, which is a substrate for sphingomyelin production. Also there was a significant increase in the expression of carnitine palmitoyltransferase 1 (CPT1) when SPTLC123 was silenced.
When cells were treated with DEGS1 siRNA, there were significant changes in sphingolipid intermediates, whereas changes in the expression of genes associated with lipid synthesis pathways was less pronounced. Knocking down DEGS1 significantly reduced ceramide and sphingomyelin and increased sphinganine and dhCer. The accumulation of sphinganine and dhCer is associated with a significant shift in lipid flux leading to the accumulation of FFA. The increase of DAG is consistent with the increased expression of 1-acylglycerol-3-P acyltransferase and diacylglycerol kinase. The expression of diacylglycerol acyltransferase is decreased, supporting the decrease in TAG levels observed in DEGS1-silenced hepatocytes.
The effects of SPTLC123 or DEGS1 knockdown on phosphatidylinositol (PI) or ganglioside synthesis were not addressed in this study because the levels of these lipids could not be accurately measured. Nevertheless, changes in the expression of genes involved in PI (PIP4K2A, PIP4K2B, PIP4K2C, PIK3AP1, PIK3R3, and PTEN) and ganglioside biosynthesis (glucosyl ceramide transferase or GCS), suggest that the inactivation of ceramide synthesis has an impact on PI and ganglioside biosynthesis. (See supplementary File III for exact gene expression changes.) To validate these changes, mRNA expression of GCS, DGAT2, and CDS1 were confirmed using QuantiGene 2.0 assay (data not shown).
Effect of SPTLC123 and DEGS1 knockdown on global gene expression
To gain insight into cellular processes that may be affected by SPTLC123 or DEGS1 knockdown, the transcripts with statistically significant changes in expression were interrogated by DAVID gene set enrichment analysis. Using a gene set enrichment score > 2 as the cutoff, both SPTLC123 and DEGS1 knockdown were statistically overrepresented for genes with functions associated with negative regulation of biosynthetic processes, endomembrane system, and translation (supplementary File IV). The clusters from DEGS1 siRNA-treated cells were more agreeable in a direction, either up or down, than were those from SPTLC123 knockdown cells (Table 2).
TABLE 2.
GSEA of SPTLC123- or DEGS1-silenced cells
| Rank | Cluster Key Term | Change | Score | Term |
| SPTLC123 siRNA GSEA | ||||
| 1 | Negative regulation of biosynthetic process | UP | 4.02 | 74 |
| 2 | Endomembrane system | MIXED | 3.28 | 87 |
| 3 | Organelle lumen | MIXED | 3.10 | 179 |
| 4 | Response to extracellular stimulus | MIXED | 2.95 | 35 |
| 5 | Transcription factor binding | UP | 2.91 | 71 |
| 6 | Blood vessel development | MIXED | 2.51 | 34 |
| 7 | Serpin | DOWN | 2.34 | 12 |
| 8 | Response to organic substance | MIXED | 2.29 | 90 |
| 9 | Translation regulation | MIXED | 2.04 | 13 |
| DEGS1 siRNA GSEA | ||||
| 1 | Endoplasmic reticulum | DOWN | 7.33 | 142 |
| 2 | Golgi apparatus | MIXED | 5.05 | 122 |
| 3 | Endomembrane system | DOWN | 4.72 | 114 |
| 4 | Negative regulation of transcription | UP | 3.45 | 58 |
| 5 | Organelle lumen | MIXED | 3.23 | 208 |
| 6 | Positive regulation of cell migration | UP | 2.98 | 20 |
| 7 | Vesicle | MIXED | 2.94 | 89 |
| 8 | Transcription repressor activity | UP | 2.66 | 52 |
| 9 | Cellular response to insulin stimulus | UP | 2.62 | 19 |
| 10 | Membrane fraction | DOWN | 2.55 | 96 |
| 11 | Regulation of translation | MIXED | 2.50 | 25 |
| 12 | Lysosome | DOWN | 2.48 | 26 |
| 13 | Pleckstrin homology-type | UP | 2.30 | 46 |
| 14 | Regulation of hydrolase activity | DOWN | 2.13 | 49 |
| 15 | Establishment of protein localization (transport) | DOWN | 2.09 | 102 |
| 16 | PDZ (synaptic scaffolding protein) | UP | 2.02 | 24 |
| SPTLC123 and DEGS1 overlap GSEA | ||||
| 1 | Negative regulation of cellular biosynthetic process | UP | 3.37 | 37 |
| 2 | Blood vessel development | MIXED | 2.48 | 18 |
| 3 | Carboxylic acid / lipid biosynthetic process | DOWN | 2.03 | 14 |
The clusters based on similarity from DAVID for the gene set enrichment analyses were included in the table if their enrichment score, represented as score in the table, was greater than 2 for each targeted siRNA as well as the overlapping probes that agreed in directionality. The overall change for each process was indicated as UP, DOWN, or MIXED if most (>60%) of the genes represented in the cluster were upregulated, downregulated, or had an undetermined directionality, respectively. The number of terms or genes for each cluster is noted. Gene lists and their directional change for the clusters from the DAVID GSEA can be found in supplementary File IV.
Genes clustered with negative regulation of biosynthetic processes were mostly upregulated for Huh7 cells treated with either SPTLC123 or DEGS1 siRNA, suggesting that cellular biosynthesis was inhibited for both treatments. However, there were more genes enriched for the endomembrane system, such as the endoplasmic reticulum (ER) or lysosome, that associates with clusters that were downregulated in the DEGS1 knockdown cells. This finding suggests that DEGS1 knockdown has a greater impact on endomembrane trafficking than does SPTLC123 knockdown. For the genes involved with the regulation of translation, expression levels were directionally variable in the cells treated with either SPTLC123 or DEGS1 siRNA. Interestingly, there were more changes in the gene clusters associated with negative regulation of transcription, positive regulation of cell migration, cytoskeleton modification/cell polarity, and cellular response to insulin stimulus in the DEGS1-silenced cells. (More information on the specific genes that changed and their expression for each cluster from the DAVID gene set enrichment can be found in supplementary File IV.)
To provide better clarity for all the cellular processes that may be affected by knocking down SPTLC123 or DEGS1, cellular processes of interest based on Ariadne Pathway Studios annotation were examined for changes and highlighted if their function was also represented in DAVID gene set enrichment (Table 3 and supplementary File V). We observed a greater directional effect on gene expression across all pathways in the DEGS1 knockdown cells compared with SPTLC123-silenced hepatocytes. Gene expression changes in most functions appeared to be downregulated, except for the endocytosis process, which was upregulated in both siRNA treatments. Nevertheless, expression of genes involved in protein folding and translation was upregulated in the SPTLC123 siRNA-treated cells but downregulated in the DEGS1 knock down cells.
TABLE 3.
Ariadne pathway analysis with directional changes of SPTLC123- or DEGS1-silenced cells
| Ariadne Process | SPTLC123 siRNA | DEGS1 siRNA |
| Adherens junction assembly | MIXED | UP |
| DNA recombination, repair, replication | DOWN | DOWN |
| Polymerase II transcription | MIXED | DOWN |
| Protein folding and translation | UP | DOWN |
| Nuclear export and import | MIXED | DOWN |
| Ion transport | MIXED | DOWN |
| Endocytosis | UP | UP |
| ER to Golgi transport | MIXED | DOWN |
| Fatty acid biosynthesis | DOWN | DOWN |
| A, D, E, Q, P, S, G, and T metabolism | DOWN | DOWN |
| Branched amino acid metabolism | DOWN | DOWN |
| Sugar (amino sugar, glucose, mannose) metabolism | MIXED | DOWN |
| Tricarboxylic acid cycle | DOWN | DOWN |
| Carbohydrate metabolism, oxidative phosphorylation, and ROS metabolism | MIXED | DOWN |
| Purine metabolism | MIXED | DOWN |
| Nucleotide (purine and pyrimidine) metabolism | MIXED | DOWN |
Pathways were included if there were at least 10 genes with a statistical change in any treatment and if there was a directional change in at least one treatment. No enrichment analysis was performed for these pathways. If the majority of transcripts in a pathway for a given siRNA treatment were upregulated, downregulated, or had an undetermined directionality; the box was indicated as UP, DOWN, or MIXED, respectively. Gene lists and their directional change for all Ariadne pathways can be found in supplementary File V.
ROS, reactive oxygen species.
Although the expression of genes associated with amino acid metabolism was downregulated in both DEGS1 and SPTLC123 knockdown cells, the effect from DEGS1 silencing was stronger than the SPTLC123 siRNA treatment, suggesting that DEGS1 knockdown reduces the processes involved with nitrogen metabolism, gluconeogenesis, or ketogenesis irrespective of their function in Huh7 cells. In addition, more genes associated with sugar metabolism were statistically downregulated in cells treated with DEGS1 siRNA than with SPTLC123 siRNA. The downregulation of genes associated with nucleotide metabolism suggests a decrease of nucleotide needs for DNA, RNA synthesis, or key cofactors, such as ATP or cyclic CTP. Genes associated with endocytosis and endosomal recycling processes were statistically upregulated for cells treated with either DEGS1 or SPTLC123 siRNA.
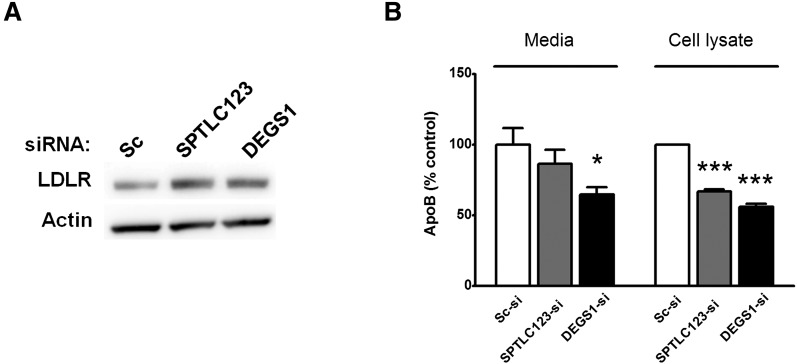
To validate the upregulation of the endocytosis pathway, we determined the levels of low density lipoprotein receptor (LDLR) protein as well as used cellular and secreted levels of apoB as markers of cellular cholesterol regulation and vesicular trafficking. Western blot analysis of LDLR (Fig. 4A) shows an increase in the LDLR protein level (∼1.5- to 2-fold compared with scrambled siRNA-treated control) in SPTLC123- or DEGS1-silenced hepatocytes. The cellular level of apoB was decreased significantly in DEGS1- and SPTCL123-silenced cells, whereas the reduction of apoB secreted to media was more pronounced in DEGS1 knockdown group (Fig. 4B).
Fig. 4.
LDLR and apoB levels in SPTLC123- or DEGS1-silenced Huh7 cells. (A) A Western blot representation from three independent experiments illustrated the changes in LDLR protein level. β-actin antibody was used on the same blot to verify an equal protein loading in each lane. (B) Secreted and cellular apoB levels were determine by ELISA as described under Materials and Methods. (mean ± SEM, n = 3, *P < 0.05, ***P < 0.001).
Finally, the CRE algorithm was used to infer possible upstream molecular mechanisms that could cause the observed gene expression changes following SPTLC123 or DEGS1 knockdown. The most functionally relevant CRE hypotheses are indicated in Table 4. (An inclusive list of CRE results can be found in supplementary File VI.) From the statistically significant gene expression changes observed with SPTLC123 or DEGS1 knockdown, the hypotheses generated from CRE suggested reduced responses to hypoxia and inflammation. Enzymes with transcriptional activity within the cell cycle/adhesion function, such as GTPase HRas (HRAS), transforming growth factor (TGF)B1, and protein C-ets1 (ETS1) were predicted to be less active in DEGS1-silenced cells. Interestingly, two members of the hepatic nuclear factor (HNF) family were differentially predicted to be downregulated in the knockdown cells, with HNF4A in the SPTLC123-silenced cells and HNF1A in the DEGS1-silenced cells. To confirm these predictions, five HNF4A- and five HNF1A-target genes were selected from the CRE predictions, and their expression levels were measured in SPTLC123- and DEGS1-silenced cells. In agreement with the CRE predictions, changes in mRNA levels of HNF4- and HNF1-target genes were demonstrated via qPCR (supplementary File VI).
TABLE 4.
CRE hypotheses of SPTLC123 or DEGS1 knockdown cells
| Rank | CRE Hypothesis | Direction | Function | Enrichment | Correctness | C | I | A | T |
| SPTLC123 siRNA | |||||||||
| 1 | Response to hypoxia | − | Response to O2 | 4.0e−21 | <0.001 | 87 | 59 | 0 | 146 |
| 2 | Methylprednisolone | + | Inflammation | 3.4e−11 | 0.019 | 32 | 15 | 0 | 57 |
| 3 | EPAS1 | − | Response to O2 | 1.2e−09 | <0.001 | 26 | 12 | 0 | 38 |
| 4 | STAT4 | − | Inflammation | 6.9e−04 | <0.001 | 17 | 5 | 0 | 22 |
| 7 | Lipopolysaccharide | − | Inflammation | 8.9e−06 | <0.001 | 52 | 41 | 0 | 99 |
| 11 | RELA | − | Inflammation | 5.4e−06 | 0.002 | 17 | 8 | 0 | 26 |
| 12 | Hypoxia | − | Response to O2 | 3.8e−04 | <0.001 | 16 | 7 | 0 | 23 |
| 13 | IL6 | − | Inflammation | 5.3e−08 | <0.001 | 31 | 22 | 0 | 54 |
| 19 | STAT3 | − | Inflammation | 4.0e−06 | <0.001 | 22 | 14 | 0 | 36 |
| 20 | HNF4A | − | Liver | 8.5e−05 | <0.001 | 17 | 9 | 0 | 26 |
| DEGS1 siRNA | |||||||||
| 1 | Response to hypoxia | − | Response to O2 | 1.0e−12 | <0.001 | 85 | 52 | 0 | 137 |
| 2 | Lipopolysaccharide | − | Inflammation | 2.7e−06 | <0.001 | 65 | 42 | 6 | 113 |
| 3 | HNF1A | − | Liver | 1.0e−09 | <0.001 | 29 | 7 | 1 | 37 |
| 4 | XBP1 | − | ER stress | 9.7e−10 | <0.001 | 26 | 5 | 0 | 31 |
| 5 | Response to oxidative stress | − | Response to O2 | 3.0e−06 | <0.001 | 29 | 8 | 0 | 37 |
| 6 | PPARG | − | Adipocyte | 1.7e−03 | <0.001 | 26 | 8 | 2 | 36 |
| 7 | HRAS | − | Cell cycle/ adhesion | 1.9e−05 | 0.004 | 29 | 13 | 0 | 42 |
| 9 | IL6 | − | Inflammation | 3.9e−06 | <0.001 | 34 | 20 | 0 | 54 |
| 11 | Bacterial infection | − | Inflammation | 7.0e−06 | <0.001 | 25 | 11 | 0 | 36 |
| 12 | TGFB1 | − | Cell cycle/ adhesion | 2.5e−09 | <0.001 | 52 | 39 | 6 | 97 |
| 13 | ETS1 | − | Cell cycle/ adhesion | 6.9e−05 | <0.001 | 22 | 9 | 0 | 31 |
| 17 | EPAS1 | − | Response to O2 | 3.5e−06 | <0.001 | 23 | 11 | 0 | 34 |
| 18 | HIF1A | − | Response to O2 | 2.5e−08 | <0.001 | 31 | 20 | 2 | 53 |
| 19 | NFE2L2 | − | Response to O2 | 4.4e−06 | <0.001 | 31 | 20 | 0 | 51 |
| 20 | VHL | + | Response to O2 | 9.3e−06 | <0.001 | 13 | 2 | 1 | 16 |
The top 20 most functionally relevant CRE hypotheses for each siRNA treatment were examined. The direction of the hypothesis is provided as a +, representing active/present, or a –, representing inactive/absent. Correctness and enrichment P values are computed as described in Materials and Methods. A single-letter column represents the statistically significant genes from the microarray gene expression profile for each treatment that is in the correct fold-change direction for the hypothesis (C), incorrect fold-change direction for the hypothesis (I), ambiguous fold-change based on the current literature for the hypothesis (A), and total genes for that hypothesis in the dataset (T). A complete hypotheses list for each targeted siRNA treatment and evidence matrix for the genes supporting these hypotheses can be found in supplementary File VI.
DISCUSSION
Ceramide is a class of sphingolipids that contributes to membrane components and functions as an intracellular signaling molecule (3, 28, 29). SPTLC is the first key enzyme involved in the rate-limiting step of ceramide biosynthesis. Multiple reports indicate that inhibiting SPTLC activity with RNAi (30) or myriocin (31) in rodents or cell lines or by genetic modulation in mice reduces ceramide levels, leading to an improvement in lipid profiles as well as prevention of atherosclerosis and onset of diabetes (12, 13, 32). In addition, the heterozygous DEGS1 knockout mice have demonstrated enhanced insulin sensitivity and are refractory to dexamethasone-induced insulin resistance (13). As the disturbance of ceramide production has been achieved successfully through an inhibition of SPTLC or DEGS1 enzyme, in this study, we used human hepatocarcinoma cell line Huh7, which has a comparable SPTLC expression to HepG2 and human liver tissue (8), as a model to study global changes of gene expression and lipid homeostasis as a result of the inhibition of ceramide synthesis.
Using targeted siRNA against three major SPTLC subunits or DEGS1, we were able to substantially decrease ceramide levels by downregulating two critical steps within the ceramide biosynthetic pathway. At the message level, the knockdown effect is more pronounced with SPTLC1, SPTLC3, and DEGS1 than with SPTLC2 siRNA; nevertheless, knocking down SPTLC1, DEGS1, or all three SPTLC subunits led to a significant reduction of total ceramides. Neither SPTLC2- nor SPTLC3-silenced cells showed a decrease in ceramides compared with the nontargeting control. It is possible that these two subunits can be dynamically interchangeable within a SPT enzyme complex to replenish the ceramide pool, although the substrate specificity to produce a different chained length of sphingoid base of ceramide is unique for SPTLC2 and SPTLC3, as reported from the findings in overexpressed Hek293 cells (9, 33). Beside the effects of siRNA on ceramide levels, we observed a decreased cell proliferation in the DEGS1-silenced cells but not in the SPTLC123 siRNA-treated cells. This observation is in agreement with several reports in the literature demonstrating cell-cycle growth arrest caused by an inhibition of DEGS1 (17, 34, 35). It is unlikely that the low level of ceramide will lead to cell growth arrest, as the level of ceramide is significantly decreased in SPTLC123- and DEGS1-silenced cells, but only DEGS1 knockdown cells decreased cell proliferation. Rather, the accumulation of lipid intermediates and/or changes in cellular processes that are unique to DEGS1-silenced cells are most likely responsible for this phenotype. No evidence of cell death or apoptosis was observed in either SPTLC123- or DEGS1-silenced cells.
The synchronized knockdown of all SPTLC subunits or DEGS1 has a marked impact on the global lipidomic and gene expression profiles. First, the sphingolipid profile of SPTLC123 and DEGS1 siRNA-treated cells appeared to be noticeably different, especially the dhCer, sphinganines, and sphingomyelin levels. Silencing DEGS1 but not SPTLC123 led to a pronounced increase in dhCer. This finding is consistent with studies in the DEGS1-deficient mice and cells in which the activity of DEGS1 was inhibited using celecoxib (13, 17, 35, 36). Not only does dhCer accumulate significantly but also intermediates upstream of dhCer, such as sphinganine, increase dramatically. This finding indicates that the attenuation of DEGS1 function does not inhibit the upstream synthesis of intermediates in the pathway; rather, the synthesis continues, causing both dhCer and sphinganine to build up. Interestingly, the level of sphingomyelin (but not sphingosine) is significantly reduced in DEGS1 siRNA-treated cells, whereas neither changes in SPTLC123 knockdown cells. Possibly, sphingomyelin from DEGS1-depleted cells continues to salvage and replenish the ceramide pool, which can then be converted to other classes of sphingolipids, such as ganglioside or ceramide-1-phosphate (Cer-1-PO4). Additional quantifications of gangliosides and Cer-1-PO4 are necessary to confirm this hypothesis. On the other hand, reducing ceramide levels in SPTLC123 siRNA-treated cells did not significantly alter the level of sphingolipids downstream of ceramide, suggesting that other pathways are involved in maintaining sphingomyelin and sphingosine levels. Taken together, these findings demonstrate that even though the cellular levels of ceramide are approximately the same, through depleting the enzymes responsible for different reactions in the biosynthetic pathway, the total levels and speciation of sphingolipids can be dramatically different.
The depletion of ceramide through SPTLC123 or DEGS1 siRNA treatment on lipid subspecies and gene expression was modeled (supplementary Fig. I-F, G). Dramatic changes in lipid synthesis and gene expression of cellular functions that are unique to SPTLC123 and DEGS1 knockdown were revealed. These figures are the first model combining changes in lipid subspecies and gene expression that highlights how Huh7 cells respond and adapt to the disruption of ceramide synthesis. Previous studies focused on the effects of altering SPTLC functions on cholesterol efflux (14), apoE secretion (37), necrosis (38), and inflammation (39). The changes observed in lipid synthesis clearly indicate that inhibiting SPTLC123 by siRNA-mediated knockdown is sufficient to reduce ceramide synthesis and alter certain subspecies (mainly C16:0) of other lipid classes, especially phospholipids. Most of these lipid changes would be considered beneficial to the physiology of the cell (4, 40–42). In addition to the effects on lipids, other cellular processes are affected in SPTLC123-silenced Huh7 hepatocytes. First, GSEA indicates the enrichment of genes involved in the regulation of biosynthetic process. Ariadne pathway analysis also highlights the downregulation of lipid and amino acid biosynthesis, suggesting that the knockdown of SPTLC123 leads to the utilization of different substrates for energy or autophagy (43). An increase in the uptake of nutrients while reducing the synthesis of lipids is supported by the increased endosomal flux. Second, CRE analysis for the SPTLC123-silenced cells also indicated a decrease in inflammation, suggesting that the decreases of various lipid classes functioning as energy substrate, membrane constituents, and signaling regulators do not result in increasing cellular stress through an inflammatory pathway (44, 45). Third, Huh7 hepatocytes treated with SPTLC123 siRNA exhibit a decrease in hypoxic function driven by alterations in genes involved with fatty acid metabolism. Finally, decreases in HNF4A, a transcription factor associated with maturity-onset diabetes, is predicted in SPTLC123-silenced hepatocytes. In these cells, we observed and confirmed the changes in the expression level by qPCR of HNF4A-target genes, including apoM and apoC3, two genes involved in lipid metabolism (46, 47).
Similarly, previous studies of altering the level of DEGS1 in cells focused on selected processes, such as metabolic stress (13), oxidative stress (36, 48), and apoptosis (49), as well as cell cycle and cancer (17, 50, 51), which correspond well to our findings. The changes observed in lipid synthesis indicates that the inhibition of DEGS1 is sufficient to reduce both ceramide and sphingomyelin synthesis, while increasing the cellular levels of sphinganine and dhCer. Unlike the beneficial lipid changes associated with SPTLC123 knockdown (4, 40, 41), the accumulation of FFA and DAG in DEGS1 knockdown could be detrimental to the cell (52). In addition to the effects on lipids, DEGS1 knockdown significantly alters cellular functions, more so than SPTLC123 knockdown. The downregulation in almost all of the metabolic biosynthesis pathways suggests either the use of different substrates for energy or entering senescence. Our findings, together with the cell proliferation assay, confirm that DEGS1 knockdown hepatocytes undergo a reduction in cell replication. Several potential mechanisms, such as the decrease in membrane lipid synthesis, utilization of all biosynthetic substrates, or related effects on multiple signaling cascades, could contribute to the effects of cell replication. Additional studies are needed to determine this hypothesis. Consistent with our findings, several published studies indicate that DEGS1 plays a role in the cell cycle and apoptosis (17, 34, 35, 48–51). Our studies also indicate that cell-to-cell adhesion and organelle/vesicle migration is strongly affected when DEGS1 is knocked down. Downregulation in expression of genes involved with trafficking from the ER to the Golgi and upregulation of endocytosis process is consistent with an increased autophagy in DEGS1-silenced hepatocytes. In addition, DEGS1-silenced cells exhibit an increase of cholesterol influx through an elevation of LDLR protein and a reduction of cholesterol efflux through the cellular content and secretion of apoB-containing lipoproteins. Finally, CRE analysis also predicted a decrease in the activity of HNF1A, a transcription factor associated with maturity-onset diabetes, in DEGS1 knockdown hepatocytes, possibly driven by genes involved with steroid metabolism, inflammation, and secreted factors, including C8B and HSD17B2 (47, 53, 54). Despite the dramatic changes in the various lipids and other cellular processes, there is no strong evidence for cellular stress through inflammation, hypoxia, or endoplasmic reticulum stress in DEGS1-silenced Huh7 cells (44, 45, 55).
Taken together, the findings from our studies indicate that there are significant differences between knocking down the levels of SPTLC123 or DEGS1 in Huh7 hepatocytes with respect to lipid synthesis and alterations in cellular function. These differences in modulating the ceramide/sphingolipid synthesis pathway suggest that there is a therapeutic potential for different diseases depending upon which enzyme in the pathway is affected. The reduction of ceramide and TAG, with an increase in phospholipids, upregulation of endocytosis, as well as decreased inflammation and hypoxia, add more supporting evidence that the inhibition of the three SPTLC subunits is best suited for the treatment of diseases associated with lipid metabolism, such as lipotoxicity, insulin resistance, heart failure, and atherosclerosis (11, 30, 32, 34, 37, 56–58). Meanwhile, the accumulation of FFA and DAG, downregulation of most biosynthetic pathways, upregulation of endocytosis, downregulation of vesicle trafficking, and inhibition of cell-cycle progression, cytoskeleton modification, inflammation, and hypoxia provide supporting evidence that the inhibition of DEGS1 is better suited as a therapeutic mechanism for treating cancer (17, 49–51).
Irrespective of the therapeutic potential, this study provides a comprehensive insight to the disruption of ceramide synthesis and highlights major adaptive changes at both lipidomic and transcriptomic levels.
Supplementary Material
Acknowledgments
The authors acknowledge the contribution of Steven S. Gernhardt for ceramide quantification using mass spectrometry.
Footnotes
Abbreviations:
- CE
- cholesteryl ester
- CL
- cardiolipin
- CRE
- causal reasoning engine (algorithm)
- DAG
- diacylglycerol
- DEGS1
- dihydroceramide desaturase 1
- dhCER
- dihydroceramide
- ER
- endoplasmic reticulum
- ETS1
- protein C-ets1
- FC
- free cholesterol
- FDR
- false discovery rate
- GSEA
- gene set enrichment analysis
- HNF
- hepatic nuclear factor
- HRAS
- GTPase HRas
- LDLR
- LDL receptor
- LYPC
- lysophophatidylcholine
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- SPTLC
- serine palmitoyltransferase
- SPTLC123
- SPTLC subunits 1, 2, 3
- TAG
- triacylglycerol
- TGFB1
- transforming growth factor B1
All studies were funded by Pfizer Inc.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one fi gure and five files.
REFERENCES
- 1.Haus J. M., Kashyap S. R., Kasumov T., Zhang R., Kelly K. R., Defronzo R. A., Kirwan J. P. 2009. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes. 58: 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holland W. L., Summers S. A. 2008. Sphingolipids, insulin resistance, and metabolic disease: new insights from in vivo manipulation of sphingolipid metabolism. Endocr. Rev. 29: 381–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannun Y. A., Obeid L. M. 2002. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 277: 25847–25850 [DOI] [PubMed] [Google Scholar]
- 4.Kolesnick R. 2002. The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J. Clin. Invest. 110: 3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hannun Y. A. 1994. The sphingomyelin cycle and the second messenger function of ceramide. J. Biol. Chem. 269: 3125–3128 [PubMed] [Google Scholar]
- 6.Hannun Y. A., Obeid L. M. 1995. Ceramide: an intracellular signal for apoptosis. Trends Biochem. Sci. 20: 73–77 [DOI] [PubMed] [Google Scholar]
- 7.Hannun Y. A., Obeid L. M. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150 [DOI] [PubMed] [Google Scholar]
- 8.Hornemann T., Richard S., Rutti M. F., Wei Y., von Eckardstein A. 2006. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J. Biol. Chem. 281: 37275–37281 [DOI] [PubMed] [Google Scholar]
- 9.Hornemann T., Penno A., Rutti M. F., Ernst D., Kivrak-Pfiffner F., Rohrer L., von Eckardstein A. 2009. The SPTLC3 subunit of serine palmitoyltransferase generates short chain sphingoid bases. J. Biol. Chem. 284: 26322–26330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han G., Gupta S. D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R. H., Jr., Harmon J. M., Dunn T. M. 2009. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. USA. 106: 8186–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park T. S., Rosebury W., Kindt E. K., Kowala M. C., Panek R. L. 2008. Serine palmitoyltransferase inhibitor myriocin induces the regression of atherosclerotic plaques in hyperlipidemic ApoE-deficient mice. Pharmacol. Res. 58: 45–51 [DOI] [PubMed] [Google Scholar]
- 12.Glaros E. N., Kim W. S., Quinn C. M., Jessup W., Rye K. A., Garner B. 2008. Myriocin slows the progression of established atherosclerotic lesions in apolipoprotein E gene knockout mice. J. Lipid Res. 49: 324–331 [DOI] [PubMed] [Google Scholar]
- 13.Holland W. L., Brozinick J. T., Wang L. P., Hawkins E. D., Sargent K. M., Liu Y., Narra K., Hoehn K. L., Knotts T. A., Siesky A., et al. 2007. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 5: 167–179 [DOI] [PubMed] [Google Scholar]
- 14.Tamehiro N., Zhou S., Okuhira K., Benita Y., Brown C. E., Zhuang D. Z., Latz E., Hornemann T., von Eckardstein A., Xavier R. J., et al. 2008. SPTLC1 binds ABCA1 to negatively regulate trafficking and cholesterol efflux activity of the transporter. Biochemistry. 47: 6138–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Håversen L., Danielsson K. N., Fogelstrand L., Wiklund O. 2009. Induction of proinflammatory cytokines by long-chain saturated fatty acids in human macrophages. Atherosclerosis. 202: 382–393 [DOI] [PubMed] [Google Scholar]
- 16.Hicks A. A., Pramstaller P. P., Johansson A., Vitart V., Rudan I., Ugocsai P., Aulchenko Y., Franklin C. S., Liebisch G., Erdmann J., et al. 2009. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 5: e1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraveka J. M., Li L., Szulc Z. M., Bielawski J., Ogretmen B., Hannun Y. A., Obeid L. M., Bielawska A. 2007. Involvement of dihydroceramide desaturase in cell cycle progression in human neuroblastoma cells. J. Biol. Chem. 282: 16718–16728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y., Shelly L., Sand T., Chang G., Jiang X. C. 2010. Identification and characterization of dual inhibitors for phospholipid transfer protein and microsomal triglyceride transfer protein. J. Pharmacol. Exp. Ther. 335: 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- 20.Barrett T., Troup D. B., Wilhite S. E., Ledoux P., Rudnev D., Evangelista C., Kim I. F., Soboleva A., Tomashevsky M., Edgar R. 2007. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 35: D760–D765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 4: 249–264 [DOI] [PubMed] [Google Scholar]
- 22.Huang da W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- 23.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4: P3. [PubMed] [Google Scholar]
- 24.Chindelevitch L., Ziemek D., Enayetallah A., Randhawa R., Sidders B., Brockel C., Huang E. S. 2012. Causal reasoning on biological networks: interpreting transcriptional changes. Bioinformatics. 28: 1114–1121 [DOI] [PubMed] [Google Scholar]
- 25.Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. 2006. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 39: 82–91 [DOI] [PubMed] [Google Scholar]
- 26.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509 [PubMed] [Google Scholar]
- 27.Wishart D. S., Knox C., Guo A. C., Eisner R., Young N., Gautam B., Hau D. D., Psychogios N., Dong E., Bouatra S., et al. 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 37: D603–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merrill A. H., Jr. 2002. De novo sphingolipid biosynthesis: a necessary, but dangerous, pathway. J. Biol. Chem. 277: 25843–25846 [DOI] [PubMed] [Google Scholar]
- 29.Merrill A. H., Jr., Schmelz E. M., Dillehay D. L., Spiegel S., Shayman J. A., Schroeder J. J., Riley R. T., Voss K. A., Wang E. 1997. Sphingolipids--the enigmatic lipid class: biochemistry, physiology, and pathophysiology. Toxicol. Appl. Pharmacol. 142: 208–225 [DOI] [PubMed] [Google Scholar]
- 30.Watson M. L., Coghlan M., Hundal H. S. 2009. Modulating serine palmitoyl transferase (SPT) expression and activity unveils a crucial role in lipid-induced insulin resistance in rat skeletal muscle cells. Biochem. J. 417: 791–801 [DOI] [PubMed] [Google Scholar]
- 31.Park T. S., Panek R. L., Mueller S. B., Hanselman J. C., Rosebury W. S., Robertson A. W., Kindt E. K., Homan R., Karathanasis S. K., Rekhter M. D. 2004. Inhibition of sphingomyelin synthesis reduces atherogenesis in apolipoprotein E-knockout mice. Circulation. 110: 3465–3471 [DOI] [PubMed] [Google Scholar]
- 32.Hojjati M. R., Li Z., Jiang X. C. 2005. Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim. Biophys. Acta. 1737: 44–51 [DOI] [PubMed] [Google Scholar]
- 33.Hornemann T., Wei Y., von Eckardstein A. 2007. Is the mammalian serine palmitoyltransferase a high-molecular-mass complex? Biochem. J. 405: 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devlin C. M., Lahm T., Hubbard W. C., Van Demark M., Wang K. C., Wu X., Bielawska A., Obeid L. M., Ivan M., Petrache I. 2011. Dihydroceramide-based response to hypoxia. J. Biol. Chem. 286: 38069–38078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiffmann S., Sandner J., Schmidt R., Birod K., Wobst I., Schmidt H., Angioni C., Geisslinger G., Grosch S. 2009. The selective COX-2 inhibitor celecoxib modulates sphingolipid synthesis. J. Lipid Res. 50: 32–40 [DOI] [PubMed] [Google Scholar]
- 36.Idkowiak-Baldys J., Apraiz A., Li L., Rahmaniyan M., Clarke C. J., Kraveka J. M., Asumendi A., Hannun Y. A. 2010. Dihydroceramide desaturase activity is modulated by oxidative stress. Biochem. J. 427: 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., Li Y., Chakraborty M., Fan Y., Bui H. H., Peake D. A., Kuo M. S., Xiao X., Cao G., Jiang X. C. 2009. Liver-specific deficiency of serine palmitoyltransferase subunit 2 decreases plasma sphingomyelin and increases apolipoprotein E levels. J. Biol. Chem. 284: 27010–27019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta E., Ohira T., Matsue K., Ikeda Y., Fujii K., Ohwaki K., Osuka S., Hirabayashi Y., Sasaki M. 2009. Analysis of development of lesions in mice with serine palmitoyltransferase (SPT) deficiency -Sptlc2 conditional knockout mice. Exp. Anim. 58: 515–524 [DOI] [PubMed] [Google Scholar]
- 39.Suzuki H., Riley R. T., Sharma R. P. 2007. Inducible nitric oxide has protective effect on fumonisin B1 hepatotoxicity in mice via modulation of sphingosine kinase. Toxicology. 229: 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi S. S., Diehl A. M. 2008. Hepatic triglyceride synthesis and nonalcoholic fatty liver disease. Curr. Opin. Lipidol. 19: 295–300 [DOI] [PubMed] [Google Scholar]
- 41.Simha V., Garg A. 2009. Inherited lipodystrophies and hypertriglyceridemia. Curr. Opin. Lipidol. 20: 300–308 [DOI] [PubMed] [Google Scholar]
- 42.Wymann M. P., Schneiter R. 2008. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9: 162–176 [DOI] [PubMed] [Google Scholar]
- 43.Rautou P. E., Mansouri A., Lebrec D., Durand F., Valla D., Moreau R. 2010. Autophagy in liver diseases. J. Hepatol. 53: 1123–1134 [DOI] [PubMed] [Google Scholar]
- 44.He G., Karin M. 2011. NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell Res. 21: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tacke F., Luedde T., Trautwein C. 2009. Inflammatory pathways in liver homeostasis and liver injury. Clin. Rev. Allergy Immunol. 36: 4–12 [DOI] [PubMed] [Google Scholar]
- 46.Watt A. J., Garrison W. D., Duncan S. A. 2003. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology. 37: 1249–1253 [DOI] [PubMed] [Google Scholar]
- 47.Ellard S., Colclough K. 2006. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity-onset diabetes of the young. Hum. Mutat. 27: 854–869 [DOI] [PubMed] [Google Scholar]
- 48.Goldkorn T., Ravid T., Khan E. M. 2005. Life and death decisions: ceramide generation and EGF receptor trafficking are modulated by oxidative stress. Antioxid. Redox Signal. 7: 119–128 [DOI] [PubMed] [Google Scholar]
- 49.Stiban J., Fistere D., Colombini M. 2006. Dihydroceramide hinders ceramide channel formation: implications on apoptosis. Apoptosis. 11: 773–780 [DOI] [PubMed] [Google Scholar]
- 50.Kandyba A. G., Kobliakov V. A., Kozlov A. M., Nagaev I. Y., Shevchenko V. P., Dyatlovitskaya E. V. 2002. Dihydroceramide desaturase activity in tumors. Biochemistry (Mosc.). 67: 597–599 [DOI] [PubMed] [Google Scholar]
- 51.Zhou W., Ye X. L., Sun Z. J., Ji X. D., Chen H. X., Xie D. 2009. Overexpression of degenerative spermatocyte homolog 1 up-regulates the expression of cyclin D1 and enhances metastatic efficiency in esophageal carcinoma Eca109 cells. Mol. Carcinog. 48: 886–894 [DOI] [PubMed] [Google Scholar]
- 52.van Herpen N. A., Schrauwen-Hinderling V. B. 2008. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol. Behav. 94: 231–241 [DOI] [PubMed] [Google Scholar]
- 53.Jung D., Kullak-Ublick G. A. 2003. Hepatocyte nuclear factor 1 alpha: a key mediator of the effect of bile acids on gene expression. Hepatology. 37: 622–631 [DOI] [PubMed] [Google Scholar]
- 54.Armendariz A. D., Krauss R. M. 2009. Hepatic nuclear factor 1-alpha: inflammation, genetics, and atherosclerosis. Curr. Opin. Lipidol. 20: 106–111 [DOI] [PubMed] [Google Scholar]
- 55.Malhi H., Kaufman R. J. 2011. Endoplasmic reticulum stress in liver disease. J. Hepatol. 54: 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z., Park T. S., Li Y., Pan X., Iqbal J., Lu D., Tang W., Yu L., Goldberg I. J., Hussain M. M., et al. 2009. Serine palmitoyltransferase (SPT) deficient mice absorb less cholesterol. Biochim. Biophys. Acta. 1791: 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebert E. C. 2006. Hypoxic liver injury. Mayo Clin. Proc. 81: 1232–1236 [DOI] [PubMed] [Google Scholar]
- 58.Kleemann R., Verschuren L., van Erk M. J., Nikolsky Y., Cnubben N. H., Verheij E. R., Smilde A. K., Hendriks H. F., Zadelaar S., Smith G. J., et al. 2007. Atherosclerosis and liver inflammation induced by increased dietary cholesterol intake: a combined transcriptomics and metabolomics analysis. Genome Biol. 8: R200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.