Abstract
Ubiquitination appears to be involved in virus particle release from infected cells. Free ubiquitin (Ub), as well as Ub covalently bound to a small fraction of p6 Gag, is detected in mature HIV particles. Here we report that the p6 region in the Pr55Gag structural precursor polyprotein binds to Tsg101, a putative Ub regulator that is involved in trafficking of plasma membrane-associated proteins. Tsg101 was found to interact with Gag in (i) a yeast two-hybrid assay, (ii) in vitro coimmunoprecipitation by using purified Pr55Gag and rabbit reticulocyte lysate-synthesized Tsg101, and (iii) in vivo in the cytoplasm of COS cells transfected with gag. The PTAPP motif [or late (L) domain] within p6, which is required for release of mature virus from the plasma membrane, was the determinant for binding Pr55Gag. The N-terminal region in Tsg101, which is homologous to the Ubc4 class of Ub-conjugating (E2) enzymes, was the determinant of interaction with p6. Mutation of Tyr-110 in Tsg101, present in place of the active-site Cys that binds Ub in E2 enzymes, and other residues unique to Tsg101, impaired p6 interaction, indicating that features that distinguish Tsg101 from active E2 enzymes were important for binding the viral protein. The results link L-domain function in HIV to the Ub machinery and a specific component of the cellular trafficking apparatus.
Keywords: p6, virus assembly
The Pr55Gag protein of the HIV type 1 (HIV-1) contains all of the information required for transport to assembly sites on the plasma membrane, association with genomic RNA, and release into extracellular space (reviewed in ref. 1). However, although Gag is sufficient for viral assembly, cellular proteins are likely to facilitate the process. Several cellular proteins can be recovered from purified virions, suggesting proximity to the assembling particle (2, 3). Others interact directly with Pr55Gag (e.g., ref. 4). Interaction with still others is implied, because Gag contains posttranslational modifications (e.g., refs. 5–8). There are now reports that the region in Gag required for release of mature particles, the late (L) domain (9–11), directs the interaction of the protein with the ubiquitination machinery (12–16). In this study, HIV-1 Pr55Gag was used as bait in a yeast two-hybrid screen and identified Tsg101, the product of a mammalian tumor susceptibility gene, tsg101 (17), as a cellular protein that interacts with HIV-1 Gag. On the basis of its sequence and recent studies, Tsg101 is a ubiquitin (Ub)-conjugating E2 enzyme variant (UEV) protein involved in regulation of intracellular trafficking, transcriptional regulation, and cell cycle control (18–22). UEV proteins lack the critical Cys residue essential for conjugation and transfer of Ub to protein substrates or Ub-ligating (E3) enzymes (23, 24). They are highly conserved in evolution and constitute a family of proteins structurally related to, but distinct from, E2 enzymes. Here, we demonstrate that Tsg101 interacted specifically with the p6 region of HIV-1 Gag both in vitro and in the cytoplasm of transfected cells. Two highly conserved Pro residues in the L domain within p6 were critical for Tsg101 binding. Moreover, the altered Ub-binding site in the UEV domain in Tsg101, as well as other residues unique to Tsg101, were determinants of interaction with Gag. These results implicate a specific component of the cellular trafficking machinery in virus budding and maturation.
Materials and Methods
Plasmid Construction.
Oligonucleotides and procedures used for PCR and mutagenesis to construct GAL4-hybrids for expression in yeast, Tsg101 for in vitro expression, and Pr55GagΔp6 for expression in mammalian cells can be found in Table 1 and Supplemental Materials and Methods, which are published as supplemental data on the PNAS web site, www.pnas.org.
Two-Hybrid Assay.
The Pr55Gag–Tsg101 interaction was identified by a yeast two-hybrid screen by using a human B-cell library (25). Vectors pGBT9 and pGAD424 encoding Pr55Gag or Tsg101 sequences as GAL4 activation and binding domain fusion proteins were transformed into Saccharomyces cerevisiae Y153 by using procedures previously described (26). Briefly, interactions were detected by using a selection for Trp and Leu prototrophy followed by quantitative assay for LacZ activation. True positives were confirmed by demonstrating that they failed to interact with vectors carrying no insert or vectors carrying nonspecific genes (lamin). Proteins were identified after automated sequencing and matching of the DNA to a protein sequence in the database. Mapping of the interacting domain was performed by using vectors encoding the DNA-binding or activation domain of the yeast GAL4 transcriptional activator protein fused to truncations, deletions, or point mutations of the proteins. Interactions were tested in both orientations: the text describes the interactions of the GAL4 DNA-binding domain-Gag or -p1-p6 fusion proteins with the GAL4 activation domain-Tsg101 fusion protein. Expression of all GAL4 fusions was checked by analysis of yeast cell extracts by SDS–gel electrophoresis and Western blotting with an antibody directed against the GAL4-binding domain (Upstate Biochemical, Lake Placid, NY) and GAL4 transactivation domain (Santa Cruz Biotechnology).
Cell Culture, Transfection, and Preparation of Cytoplasmic Extracts.
COS-1 cells were cultured in DMEM supplemented with FBS to 60% confluency at 37°C. The cells were transfected by using the FuGene 6 reagent (Roche Molecular Biochemicals) according to the instructions of the manufacturer. At 48 h posttransfection, the cells were harvested into the media and collected by centrifugation. The pelleted cells were washed with cold PBS, allowed to swell in hypotonic buffer (10 mM Tris, pH 7.4/1 mM MgCl2, 4°C) containing protease inhibitors, and disrupted with a Dounce homogenizer (type B pestle). The total lysate was spun for 10 min at 10,000 × g at 4°C to remove unbroken cells, nuclei, and mitochondria.
Immune Capture Assays.
For in vitro assay of Tsg101–Gag interaction, Tsg101 was expressed in rabbit reticulocyte lysate (RRL) from a pET3a-tsg101 construct in the presence of [35S]-Met (DuPont NEN) by using the TNT T7 Quick Coupled Transcription/Translation System (Promega). Recombinant Pr55Gag, produced by using the T7 RNA polymerase promoter and containing amino acids 1–10 of T7 gene 10 at the N terminus, was purified from an expression strain of Escherichia coli (BL21-DE3) as described (27). Protein A agarose beads (Pierce), prewashed with nondenaturating buffer [25 mM Tris, pH 7.4/150 mM NaCl/0.5 mM MgCl2/1 mM CaCl2/1% IGEPAL (Sigma)] containing protease inhibitors (Roche Molecular Biochemicals) were incubated with the appropriate antibody, washed again, and then preincubated with Gag. Radiolabeled Tsg101 was then added, and the mixture incubated further at 4°C in a rotating device for 60 min. The beads were washed, suspended in SDS/PAGE loading buffer, and heated at 95°C for 5 min. Cytoplasmic extracts, prepared as described above, were also examined for Tsg101–Gag interaction by using the same procedure, except that the extract and the antibody-coated Protein A beads were maintained in 10 mM Tris, pH 7.4/1 mM MgCl2.
Protein Detection.
Proteins were separated by electrophoresis through a 12.5% SDS/polyacrylamide gel. For detection of radiolabeled Tsg101 after electrophoresis, gels were fixed, incubated for 30 min in EN3HANCE (DuPont NEN) autoradiography enhancer for gel fluorography, and dried. Radioactive bands were visualized by using imaging film (BioMax, Kodak). Gels with nonradioactive samples were transferred to nitrocellulose and analyzed by Western blotting. The following antibodies, as specified in the text, were used: anti-capsid (CA)1 and -CA2 (rabbit polyclonal antibodies raised against native and denatured forms of the CA protein, respectively; refs. 28 and 29), anti-CA3 (mouse monoclonal antibody, NEN-DuPont), anti-p6 [rabbit polyclonal against the C-terminal 16 amino acids, S. Campbell, National Cancer Institute–Frederick Cancer Research and Development Center (NCI-FCRDC)]; antinucleocapsid (NC, goat polyclonal, A. Rein, NCI-FCRDC); anti-T7 (Novagen); anti-Tsg1011 (monoclonal, Santa Cruz Biotechnology); and anti-Tsg1012 (rabbit polyclonal, S. Cohen, Stanford University). Proteins were visualized by chemiluminescence with Lumi-Light (Roche Molecular Biochemicals).
Results
Pr55Gag Interacts with Tsg101 in the Two-Hybrid Assay.
HIV-1 Pr55Gag was used as bait in the yeast two-hybrid screen (26) to identify interacting proteins encoded in a cDNA library derived from human B cells (25). Approximately 2 million transformants were screened, and two positives were isolated. Sequencing and matching to recorded entries in the GenBank database identified one interacting partner as cyclophilin B, which had been previously found to interact with Pr55Gag (4). The other interacting protein was identified as the product of the human tumor susceptibility gene, Tsg101 (17, 30).
Identification of the Region in Pr55Gag That Interacts with Tsg101.
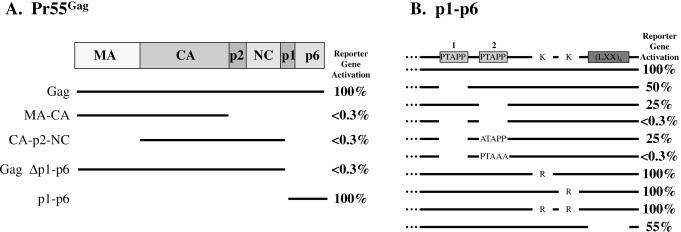
To localize the region of Pr55Gag required for interaction with Tsg101, plasmids encoding N- or C-terminally deleted-Pr55Gag fused to the DNA-binding domain of the yeast GAL4 protein were tested for LacZ reporter gene activation by using the two-hybrid assay. Western analysis, by using an antibody against the GAL4 DNA-binding domain in the fusion proteins showed that the mutated and wild-type proteins were all expressed at comparable levels (data not shown). As shown in Fig. 1A, the signal was lost on deletion of the p1-p6 region of Pr55Gag but was retained in plasmids encoding p1-p6. Thus, interaction with Tsg101 was determined by elements within p1-p6 of Pr55Gag.
Figure 1.
Identification of the region in Pr55Gag required for Tsg101 binding by using the two-hybrid assay. Reporter gene activation was quantified by determination of β-galactosidase units. (A) The interaction of Gag and p1-p6 with Tsg101. β-Galactosidase activity of Gag and p1-p6 was equivalent in two independent trials. (B) The interaction of p1-p6 with Tsg101 (taken as 100%) ranged from ≈10 to 30 β-galactosidase units in 20 independent trials. Negative interactions were equivalent to that obtained when p1-p6 was cotransformed with vector lacking Tsg101 (<0.3 units). The figure shows averaged values obtained for mutants in six independent trials as a percentage of the wild-type interaction ±1%.
The p6 region contains the conserved motifs P7T/SAP10P11 (numbering within p6 domain) and a repeating Leu sequence (LXX)4. The (LXX)4 motif is a critical determinant for Vpr binding, although it may not interact directly (reviewed in ref. 31). The PTAPP motif, or late (L) domain and, in particular, Pro10,11, is the determinant of mature virus release during the final stages of assembly (10, 11). The PTAPP motif overlaps a region (P5XP7) that is critical for efficient packaging of pol gene products into the assembled virus particle (32). The p6 region also contains Lys residues that are substrates for ubiquitination (8). To determine whether any of the above conserved motifs were important for Tsg101 binding, deletion and point mutations were engineered into a plasmid encoding the p1-p6 region of Pr55Gag, and protein–protein interactions were measured by using the two-hybrid assay. The p6 region from the pBH10 clone used for these studies contained two copies of the PTAPP motif (33), and each of these was deleted independently. Deletion of the first PTAPP motif (amino acids 455–459, numbering within Gag) reduced β-galactosidase activity to a level that was ≈50% that of wild-type protein binding (Fig. 1B). Deletion of the second PTAPP motif (amino acids 467–471), which is conserved among all HIV strains and most lentiviruses (34), reduced β-galactosidase activity to ≈25% that of wild-type protein binding. Deletion of both motifs (amino acids 455–459 and 467–471) reduced β-galactosidase activity to the level obtained when the vector lacked an insert (<0.3%). Deletion of the first PTAPP motif combined with substitution of Ala for Pro7 in the second motif reduced enzyme activity to 25% that of wild type. In the same context, substitution of Ala for Pro10,11 blocked the interaction completely. Substitution of Arg for either or both of the Lys residues that serve as substrates for Ub modification (8) gave wild-type β-galactosidase activity, indicating that the Ub substrates in p6 are not required for Tsg101 recognition. Deletion of the (LXX)4 repeat motif reduced β-galactosidase activity to ≈55% of the wild-type level. The same reduction was obtained when the first PTAPP motif was removed in its entirety. The more deleterious impairment caused by deletion of the entire second highly conserved PTAPP motif or by mutation of Pro10, 11 in this motif suggests that it is the major determinant of Tsg101 binding.
Tsg101 Binds to Pr55Gag in Vitro.
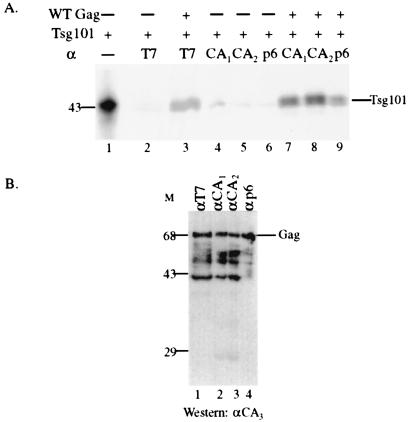
To confirm and extend our observations with the two-hybrid assay, in vitro coimmunoprecipitation studies were conducted by using unlabeled bacteriophage T7 protein-tagged recombinant Pr55Gag expressed in bacteria (27) and [35S]-Met-labeled mouse or human Tsg101 expressed in RRL. Mouse (a gift of S. Cohen, Stanford University) and human tsg101 (from the B cell library) are 94% identical and were used interchangeably in these assays. The radiolabeled protein shown in Fig. 2A (lane 1) migrated at the molecular mass expected for Tsg101 (391 amino acids, ≈43 kDa). The protein was detected consistently as a doublet, perhaps because of internal initiation at Met10. The protein was also sometimes detected as a doublet in cytoplasmic extracts. Tsg101 was captured by Protein A-coated beads on which antibodies directed against the T7 tag, CA, or p6 domains had been immobilized and preincubated with Pr55Gag (lanes 3 and 7–9). Beads not preincubated with Gag (lanes 2 and 4–6) or preincubated with Gag in the absence of antibody (data not shown) did not capture Tsg101. The anti-p6 antibody recognizes an antigenic site in the C-terminal 16 residues of the protein [including the (Leu-X-X)4 repeat]. The demonstration that the Pr55Gag bound to this anti-p6 antibody was still able to capture Tsg101 indicates that the Tsg101-binding region in p6 was exposed. This is consistent with the two-hybrid assay result, which showed that the C-terminal half of the p6 region was not a Tsg101-binding site. In competition assays, addition of a 5-fold molar excess of a peptide containing the PTAPP motif (ALQSRPEPTAPPEES) caused a 2.2-fold reduction in Tsg101 capture by Pr55Gag. The limited solubility of the peptide precluded testing at higher concentrations. In contrast, no change was detected with a 5-fold molar excess of a peptide containing the mutant LIAPP sequence, indicating that the effect of the PTAPP motif was specific (data not shown). Western analysis with an anti-CA monoclonal antibody confirmed the presence of full-length Pr55Gag on the beads coated with the anti-T7, -CA, and -p6 antibodies that captured Tsg101 (Fig. 2B, lanes 1–4). The results of the immune capture assay demonstrate that Tsg101 interacts specifically and stably with Pr55Gag in vitro through interaction with the L domain.
Figure 2.
Binding of Pr55Gag and Tsg101 in vitro. (A) Autoradiography to detect immune-captured radiolabeled Tsg101. Lane 1, Tsg101 synthesized in RRL. Lanes 2–9, determination of binding of radioactive Tsg101 to unlabeled Pr55Gag bound to anti-T7 (lane 3), anti-CA (lanes 7 and 8), or anti-p6 (lane 9) IgG immobilized on protein A beads. Antibodies are as defined in Materials and Methods. The amount of RRL used in lanes 2–9 was 5-fold greater than the amount used in lane 1. (B) Confirmation of the presence of Pr55Gag on the beads by Western analysis. A monoclonal antibody against an antigenic site in the CA domain was used to visualize the Gag proteins immunoprecipitated with the antibodies used in A. Molecular mass markers (kDa) are on the left.
Tsg101 Binds Pr55Gag in Vivo.
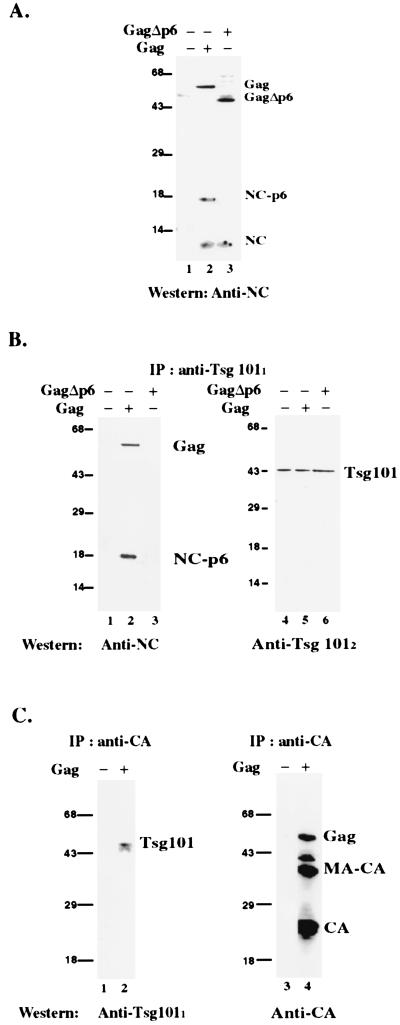
To determine whether Tsg101 and Gag associate during viral assembly, COS cells expressing Gag were examined for Tsg101–Gag complexes by coimmunoprecipitation assays. Plasmid pgp-RRE-r expresses the HIV-1 Gag and Gag-Pol polyproteins as well as Vif under the control of the simian virus 40 late promoter (35). Expression requires the Rev protein, which is provided in trans by expression of pCMV-rev (35). To ensure the specificity of the Tsg101–Gag interaction in the cytoplasm, a Gag mutant that lacked the p6 domain was included. Cytoplasmic extracts prepared from cells transfected with rev alone, rev, gag, and pol, or rev, gagΔp6, and pol were incubated with anti-Tsg101 mouse monoclonal antibody and the immunoprecipitate was examined for Gag-related proteins by Western blotting with a goat polyclonal antibody against the NC domain. First, the total cytoplasmic extract was examined (Fig. 3A). The anti-NC antibody recognized Pr55Gag, an 18-kDa NC-related protein, and NCp7 in the extract prepared from cells expressing the wild-type Gag protein (lane 2). The 18-kDa band was identified as NC-p6 on the basis of its reactivity with both anti-NC and anti-p6 (data not shown). The anti-NC antibody also recognized NCp7 and a protein that migrated at ≈49 kDa in the extract prepared from cells expressing the mutant (lane 3). The latter is the molecular mass expected for the Pr55GagΔp6 precursor protein. Consistent with this conclusion, the 49-kDa protein was not detected in extracts prepared from cells expressing Rev alone (lane 1) or the wild-type Gag protein (lane 2). Immunoprecipitation by using a monoclonal antibody against Tsg101 precipitated the wild-type Gag precursor and the NC-p6 protein but not NCp7 (Fig. 3B, lane 2) or Gag lacking the p6 domain (lane 3). Reprobing the same blot with another anti-Tsg101 antibody confirmed the presence of the cellular protein in the immunoprecipitates of all three extracts (lanes 4–6). In a reciprocal experiment, anti-CA antibody coimmunoprecipitated Tsg101 from extracts of cells expressing Gag (Fig. 3C, lane 2) but not extracts expressing Rev alone (lane 1). Reprobing the same blot with another anti-CA antibody confirmed the presence of Gag on the beads that captured Tsg101 (lane 4). Immunoprecipitation with an irrelevant antibody (rabbit anti-mouse IgG) did not precipitate Gag or Tsg101 (data not shown). The results indicate that the interaction between Tsg101 and Gag occurs in the cytoplasm of cells containing the viral protein and demonstrate that the p6 region of the Gag protein specifically directs the interaction.
Figure 3.
Coimmunoprecipitation of Pr55Gag and Tsg101 from cytoplasmic extracts. (A) Total cytoplasmic extract. Extracts were prepared from cells transfected with rev (lane 1), rev, gag, and pol (lane 2), or rev, gagΔp6, and pol (lane 3). (B) Immunoprecipitation with anti-Tsg101 monoclonal antibody by using extracts of cells transfected with rev (lane 1); rev, wild-type gag and pol (lane 2); or rev, gagΔp6, and pol (lane 3) as detected by the anti-NC polyclonal antibody. The blot was reprobed with an anti-Tsg101 polyclonal antibody to confirm the presence of Tsg101 in the immunoprecipitates (lanes 4–6). (C) Immunoprecipitation with anti-CA polyclonal antibody by using extracts from cells transfected with rev (lane 1) or rev, gag, and pol (lane 2) as detected by anti-Tsg101 monoclonal antibody. The blot was reprobed with anti-CA monoclonal antibody to confirm the presence of Gag in the immunoprecipitate (lanes 3 and 4). Molecular mass markers (kDa) are on the left.
Identification of the Region in Tsg101 That Interacts with Pr55Gag.
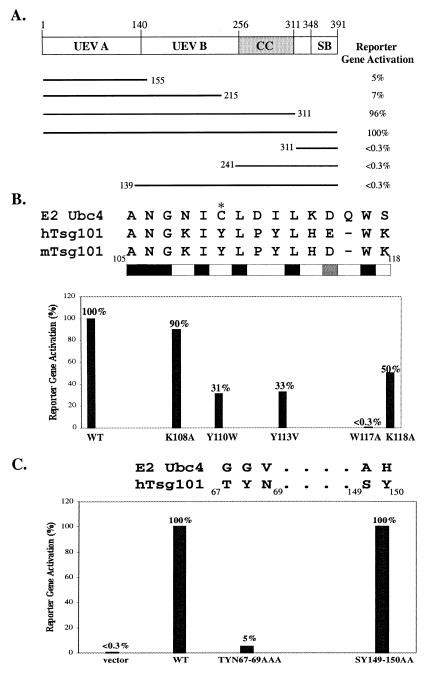
The Tsg101 protein contains an N-terminal E2-like (UEV) domain with homology to the Ub-conjugating (Ubc) 4 subgroup of E2 enzymes (23, 24). It also contains central Pro-rich and coiled-coil regions and a C-terminal steadiness box that controls its steady-state level (Fig. 4; ref. 36). On the basis of recent findings implicating Ub in Gag assembly (8, 12–16), it was of interest to determine which region of Tsg101 was recognized by the viral protein. To investigate this, the yeast two-hybrid system was used to localize the region in Tsg101 required for interaction with Pr55Gag. Plasmids encoding N- or C-terminally truncated Tsg101 fused to the activation domain of the GAL4 protein were assayed for the ability to bind p1-p6 fused to the GAL4 DNA-binding domain. As shown in Fig. 4A, assessment of reporter gene activation indicated that p6–Tsg101 interaction was maintained in fragments that retained the N-terminal 155 amino acids, but lost in the mutants that lacked this region, suggesting that the N-terminal region was the minimal determinant of binding. Western analysis showed comparable expression of the interacting and noninteracting fragments, thus supporting this conclusion (data not shown). Curiously, the N-terminal 155-, 215-, and 311-residue fragments bound p6 comparably in a qualitative assay, but the 311-residue fragment interacted to a significantly greater extent in the quantitative assay (Fig. 4A). This difference did not reflect a binding site downstream of amino acid 155, because a fragment extending from amino acid 139 to the C terminus of Tsg101 failed to interact. Moreover, no binding was detected in in vitro capture assays by using a hybrid protein comprised of glutathione S-transferase fused to Tsg101 residues 167–374 (data not shown). Optimal presentation of the p6-binding region in the N-terminal 155-residue fragment may require downstream sequences. In this regard, Tsg101 resembles class II E2 enzymes with a conserved catalytic core domain of ≈150 residues and an extra C-terminal extension attached to this core domain that is speculated to play a role in substrate recognition (37, 38).
Figure 4.
Identification of the region in Tsg101 required for Pr55Gag binding. Truncation (A) and substitution (B and C) mutants of Tsg101 were tested in the two-hybrid assay for interaction with the p1-p6 fusion protein. The figure shows averaged values as a percentage of the wild-type interaction with ±2–6% error. Notations are as in legend to Fig. 1.
In E2 enzymes including Ubc4, Ub is conjugated to an active-site Cys. In Tsg101, Tyr replaces this Cys residue. However, 8 of 14 residues flanking this Tyr are conserved in both Tsg101 and Ubc4 (refs. 23 and 24; Fig. 4B). To determine whether recognition by the p6 domain was because of amino acids unique to Tsg101, point mutations were engineered into the active-site homologue in the full-length protein, and interaction with p6 was tested in the two-hybrid assay. Western analysis by using an antibody against the GAD moiety in the fusion proteins showed that the mutated and wild-type proteins were all expressed at comparable levels (data not shown). Mutation of Trp117, which is conserved in all classes of E2 and UEV proteins and believed to demarcate the C terminus of the active site region (23, 39), eliminated binding completely, indicating that this residue is important in both Tsg101 and Ubc4. Consistent with the hypothesis that residues unique to Tsg101 determined L-domain interaction, substitution of three of the four nonconserved residues tested impaired p6 binding (Fig. 4B). Conservative substitution of Trp for Tyr110 reduced p6 interaction to 31% of wild type. Substitution of the hydrophobic amino acid Val for Tyr113 reduced binding to 33% of wild type. Substitution of Ala for Lys118 reduced binding to a lesser extent.
To further test the hypothesis, we examined residues in Tsg101 that are not conserved in E2 enzymes but align with regions previously shown to determine substrate recognition in the Ubc4 subgroup (23, 24, 40). Residues 49 and 125 in Ubc4 were found to determine the substrate specificity of structurally homologous but functionally distinct Ubc4 isoforms (40). Residue 49 aligns with Tsg101 Thr67 (23, 41) or Asn69 (24). Residue 125 aligns with Ser149 (24) or Tyr150 (41). We therefore substituted Ala for 69Thr-Tyr-Asn69 and 149Ser-Tyr150 and determined the effect on binding. As shown in Fig. 4C, mutation of 69Thr-Tyr-Asn69 reduced binding to 5% of the wild-type level. Mutation of 149Ser-Tyr150 had no effect. Western blotting confirmed that the mutants were expressed at wild-type levels (data not shown). The results are consistent with the conclusion that the interaction of Gag with Tsg101 is based on specific recognition and support the suggestion that the N-terminal E2-like domain of Tsg101 is the minimal determinant of p6 binding.
Discussion
In this report, we described the interaction of Tsg101 with HIV-1 Pr55Gag in vitro and in vivo. The N-terminal half of Tsg101, which contains the determinant of Gag binding, is homologous to Ub-conjugating E2 enzymes (23, 24, 37). The C-terminal half of Tsg101 has a coiled-coil domain that can interact with a cytoplasmic phosphoprotein, stathmin, implicated in microtubule dynamics (17), and contains a highly conserved sequence that regulates the steady-state level of the protein (35). On the basis of its structural features, Tsg101 has been speculated to be (i) a dominant-negative Ub regulator (23, 24); (ii) a transcriptional regulator (22); (iii) a regulator of the cell cycle (20, 21); and (iv) a regulator of membrane protein trafficking (18, 19). It is not clear whether these apparently diverse roles reflect independent or related functions of the protein. Furthermore, how participation in these functions may relate to Tsg101's role in the ubiquitination process is unknown.
The observation that Tsg101 interacts with HIV-1 Gag in mammalian cells suggests that the interaction is relevant to the viral life cycle. We noted that the interaction was much more efficient in cytoplasmic extracts than in vitro, perhaps suggesting a need for stabilizing cellular factors or a particular Gag assembly state. That the L-domain-containing p6 region of the protein is required for binding implicates the interaction in the late budding function. Our results indicate that deletion of the L-domain PTAPP motif prevents the interaction of Gag with Tsg101. This Pro-rich sequence is highly conserved in all lentiviruses except equine infectious anemia virus (34). It is duplicated in some isolates of HIV-1, HIV-2, and simian immunodeficiency virus. The human T-cell leukemia virus type 1 and the Mason–Pfizer monkey virus Gag proteins contain both the lentivirus motif [PT(S)AP] and the (P)PPPY motif, the functionally interchangeable avian retrovirus counterpart (9, 42). Studies indicating that the PY and PTAP motifs recruit the Ub machinery and that certain proteasome inhibitors cause alterations in viral particle budding similar to defects resulting from mutations in PTAPP and PY support the possibility that the cell's ubiquitination machinery is linked to viral assembly (12, 13, 16). However, it is unclear at this time whether this link reflects a direct or indirect involvement of Ub: on the one hand, mutation of the Lys residues in the p6 domain that are substrates for Ub modification has no apparent effect on virus assembly or release (15). On the other hand, covalent linkage of Ub to Gag was shown to rescue the defect in release caused by proteasome inhibitors (16). Perhaps other Lys residues serve as Ub substrates when the preferred sites in p6 are not available.
The notion that Tsg101 functions as a dominant-negative Ub regulator is based on the fact that Tsg101 lacks the active site Cys residue that conjugates Ub in active E2 enzymes (23, 24). The ubiquitination process requires the sequential action of two or three enzymes (reviewed in ref. 39). E1, a Ub-activating enzyme, binds Ub through a thioester bond, then transfers it to E2. E2 enzymes can function alone or in conjunction with E3 Ub-protein ligases to attach Ub to lysine residues in substrate proteins. Substrates modified by polyubiquitination are degraded by the proteasome; monoubiquitination serves as a signal for endocytosis (43). Although the active site Cys is not conserved in Tsg101, our results suggest that this region functions directly or indirectly in binding of the HIV-1 L domain. It is noteworthy that the residues in the altered Ub-binding site in Tsg101 that contribute to L-domain PTAPP binding are Tyr and Trp residues flanked by positively charged Lys residues (Fig. 4B). Aromatic amino acids flanked by charged residues are critical binding determinants for protein-binding motifs like SH3 and WW domains. SH3 domains bind Pro-rich sequences having the consensus PXXP, like PTAPP; WW domains interact with PPXY motifs (44, 45). Moreover, as the amino acids surrounding these aromatic residues in Tsg101 are conserved in active E2 enzymes, Tsg101 may maintain an E2-like ability to present Gagp6 to interacting E3 enzymes, as suggested for E2–Ub–E3 interacting complexes (46, 47). If so, the Tsg101–Gag–complex may associate with an active E3 enzyme to facilitate an event related to L-domain function. Furthermore, Tsg101 has been shown to function in membrane protein transport (18, 19). Its involvement in regulation of vesicles that are required for recycling of membrane-associated proteins (18, 48) may permit it to play a role in Gag trafficking to the site of particle maturation and release.
Our finding that HIV-1 Gag binds through the E2-like domain in Tsg101 suggests several hypotheses. The interaction of Gag with Tsg101 may be adventitious, based on Tsg101's similarity to active E2 enzymes. Alternatively, if Ub is required for assembly as suggested (12, 13, 16), Tsg101 may function as a cellular defense mechanism that prevents Gag interaction with active E2 enzymes. It is also possible that Tsg101 is recruited by the virus as a chaperone to block Gag polyubiquitination and subsequent degradation by the proteasome. This idea is supported by the fact that cyclin-specific E2 enzymes with Ser substituted for the active Cys are, in fact, dominant-negative inhibitors of cyclin destruction (49). Finally, Tsg101 may function like the yeast UEV Mms2 protein, which alters the function of interacting E3 proteins (50). Interestingly, the L domain of the Ebola virus matrix protein interacts with Nedd4, an E3 Ub protein ligase (51). The apparent conservation of L-domain interaction with cellular proteins that affect Ub modification suggests that the involvement of the Ub machinery is a highly conserved event in virus assembly and particle release both within and outside the retrovirus family.
Supplementary Material
Acknowledgments
We are grateful to I. Jayatilaka, M. Leussis, T. Ng, and A. Goff for excellent technical assistance. We thank Drs. S. Campbell, S. Cohen, A. Copeland, S. J. Elledge, S. Fields, W. H. Lee, A. Rein, and G. Zybarth for generously providing antibodies, plasmids, or peptides. We also thank L. Ehrlich, J. Konopka, and W. Parrish for helpful discussions. A.K. and J.L. were supported by National Institutes of Health (NIH) Grants CA38046 and CA52047; F.B. was supported by NIH Grant GM 58271 to C.A.C. and S. Scarlata. This work was supported by NIH Grant GM 48294 to C.A.C.
Abbreviations
- RRL
rabbit reticulocyte lysate
- CA
capsid
- NC
nucleocapsid
- L domain
late domain
- Ub
ubiquitin
- Ubc
Ub-conjugating
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Swanstrom R, Wills J W. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 263–334. [Google Scholar]
- 2.Arthur L O, Bess J W, Jr, Sowder R C I, Benveniste R E, Mann D L, Chermann J-C, Henderson L E. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 3.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D J, Sowder R C I, Chertova E N, Arthur L O, Henderson L E. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 5.Bryant M, Ratner L. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camaur D, Gallay P, Swingler S, Trono D. J Virol. 1997;71:6834–6841. doi: 10.1128/jvi.71.9.6834-6841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Göttlinger H G, Sodroski J G, Haseltine W A. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ott D E, Coren L V, Copeland T D, Kane B P, Johnson D G, Sowder R C, 2nd, Yoshinaka Y, Oroszlan S, Arthur L O, Henderson L E. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M, Orenstein J M, Martin M A, Freed E O. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert U, Ott D E, Chertova E N, Welker R, Tessmer U, Princiotta M F, Bennink J R, Krausslich H G, Yewdell J W. Proc Natl Acad Sci USA. 2000;97:13057–13062. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strack B, Calistri A, Accola M A, Palu G, Gottlinger H G. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogt V M. Proc Natl Acad Sci USA. 2000;97:12945–12947. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ott D E, Coren L V, Chertova E N, Gagliardi T D, Schubert U. Virology. 2000;278:111–121. doi: 10.1006/viro.2000.0648. [DOI] [PubMed] [Google Scholar]
- 16.Patnaik A, Chau V, Wills J W. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Cohen S N. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 18.Babst M, Odorizzi G, Estepa E J, Emr S D. Traffic. 2000;1:242–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- 19.Lemmon S K, Traub L M. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- 20.Xie W, Li L, Cohen S N. Proc Natl Acad Sci USA. 1998;95:1595–1600. doi: 10.1073/pnas.95.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong Q, Chen Y, Jones D, Lee W H. Cancer Res. 1998;58:2699–2702. [PubMed] [Google Scholar]
- 22.Sun Z, Pan J, Hope W X, Cohen S N, Balk S P. Cancer. 1999;86:689–696. doi: 10.1002/(sici)1097-0142(19990815)86:4<689::aid-cncr19>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 23.Koonin E V, Abagyan R A. Nat Genet. 1997;16:330–331. doi: 10.1038/ng0897-330. [DOI] [PubMed] [Google Scholar]
- 24.Ponting C P, Cai Y-D, Bork P. J Mol Med. 1997;75:467–469. [PubMed] [Google Scholar]
- 25.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 26.Bartel P L, Fields S. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 27.Ehrlich L S, Fong S, Scarlata S, Zybarth G, Carter C. Biochemistry. 1996;35:3933–3943. doi: 10.1021/bi952337x. [DOI] [PubMed] [Google Scholar]
- 28.Ebbets-Reed D. Molecular Microbiology. Stony Brook, NY: State University of New York; 1996. p. 209. [Google Scholar]
- 29.Ehrlich L S, Agresta B E, Carter C A. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Li X, Francke U, Cohen S N. Cell. 1997;88:143–154. doi: 10.1016/s0092-8674(00)81866-8. [DOI] [PubMed] [Google Scholar]
- 31.Frankel A D, Young J A. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Dettenhofer M, Yu X F. J Virol. 1999;73:4696–4704. doi: 10.1128/jvi.73.6.4696-4704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Nature (London) 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 34.Myers G, Korber B, Wain-Hobson S, Smith R F, Pavlakis G N. Human Retroviruses and AIDS. Los Alamos, NM: Los Alamos National Laboratory; 1995. [Google Scholar]
- 35.Smith A J, Cho M I, Hammarskjold M L, Rekosh D. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng G H, Lih C J, Cohen S N. Cancer Res. 2000;60:1736–1741. [PubMed] [Google Scholar]
- 37.Jentsch S, Seufert W, Sommer T, Reins H A. Trends Biochem Sci. 1990;15:195–198. doi: 10.1016/0968-0004(90)90161-4. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Liao J, Ruland J, Mak T W, Cohen S N. Proc Natl Acad Sci USA. 2001;98:1619–1624. doi: 10.1073/pnas.98.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 40.Oughtred R, Bedard N, Vrielink A, Wing S S. J Biol Chem. 1998;273:18435–18442. doi: 10.1074/jbc.273.29.18435. [DOI] [PubMed] [Google Scholar]
- 41.Sancho E, Vila M R, Sanchez-Pulido L, Lozano J J, Paciucci R, Nadal M, Fox M, Harvey C, Bercovich B, Loukili N, et al. Mol Cell Biol. 1998;18:576–589. doi: 10.1128/mcb.18.1.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shih S C, Sloper-Mould K E, Hicke L. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kay B K, Williamson M P, Sudol M. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 45.Garnier L, Wills J W, Verderame M F, Sudol M. Nature (London) 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 46.Laney J D, Hochstrasser M. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 47.Nuber U, Scheffner M. J Biol Chem. 1999;274:7576–7582. doi: 10.1074/jbc.274.11.7576. [DOI] [PubMed] [Google Scholar]
- 48.Bishop N, Woodman P. J Biol Chem. 2001;276:11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- 49.Townsley F M, Aristarkhov A, Beck S, Hershko A, Ruderman J V. Proc Natl Acad Sci USA. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hofmann R M, Pickart C M. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 51.Harty R N, Brown M E, Wang G, Huibregtse J, Hayes F P. Proc Natl Acad Sci USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. . (First Published November 28, 2000; 10.1073/pnas.250277297) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.