Abstract
A total of 3941 rodents were captured during a 46-month prospective (mark-recapture) study on the ecology of Catarina virus in southern Texas. Antibody reactive against Catarina virus was found in 73 (11.9%) of 611 southern plains woodrats (Neotoma micropus) and none of 3330 other rodents; strains of Catarina virus were isolated from 6 antibody-negative and 9 antibody-positive southern plains woodrats; and the infections in at least 3 southern plains woodrats were chronic. These results affirm the notion that the southern plains woodrat is the principal host of Catarina virus and suggest that Catarina virus infection is highly specific to N. micropus.
Key Words: Arenavirus, Arenaviridae, Catarina virus, Neotoma micropus, Southern plains woodrat, Tacaribe serocomplex
Introduction
The North American members of the Tacaribe serocomplex (family Arenaviridae, genus Arenavirus) include Bear Canyon virus (BCNV), Catarina virus (CTNV), Tamiami virus (TAMV), and Whitewater Arroyo virus (WWAV) (Cajimat et al. 2011). Specific members of the rodent family Cricetidae (Musser and Carleton 2005) are the principal hosts of the Tacaribe serocomplex viruses for which natural host relationships have been well characterized. For example, the hispid cotton rat (Sigmodon hispidus) in southern Florida is the principal host of TAMV (Bigler et al. 1975, Calisher et al. 1970, Jennings et al. 1970).
The results of a previous study (Fulhorst et al. 2002a) suggested that the southern plains woodrat (Neotoma micropus) in southern Texas is the principal host of CTNV. The objective of this study was to extend our knowledge of the ecology of CTNV.
Materials and Methods
Study area
The Chaparral Wildlife Management Area (CWMA) comprises 6151 hectares (ha) of brush country in southern Texas (Ruthven and Synatzske 2002). The topography is level to gently rolling (average elevation, 152.4 meters). The dominant vegetation includes blackbrush (Acacia rigidula), whitebrush (Aloysia gratissima), guayacan (Guaiacan angustifolium), prickly pear (Opuntia spp.) and other cacti, mesquite (Prosopsis glandulosa), and a number of different forbs and grasses (McLendon 1991, Ruthven and Synatzske 2002). The terrestrial mammalian assemblage comprises coyote (Canis latrans), bobcat (Lynx rufus), common raccoon (Procyon lotor), white-tailed deer (Odocoileus virginianus), collared peccary (Tayassu tajacu), eastern cottontail (Sylvilagus floridana), striped skunk (Mephitis mephitis), northern pygmy mouse (Baiomys taylori), northern grasshopper mouse (Onychomys leucogaster), white-footed mouse (Peromyscus leucopus), fulvous harvest mouse (Reithrodontomys fulvescens), hispid cotton rat, southern plains woodrat, hispid pocket mouse (Chaetodipus hispidus), Ord's kangaroo rat (Dipodomys ordii), Merriam's pocket mouse (Perognathus merriami), Mexican ground squirrel (Spermophilus mexicanus), desert shrew (Notiosorex crawfordi), domestic cow (Bos taurus), and feral pig (Sus scrofa).
Trapping sites
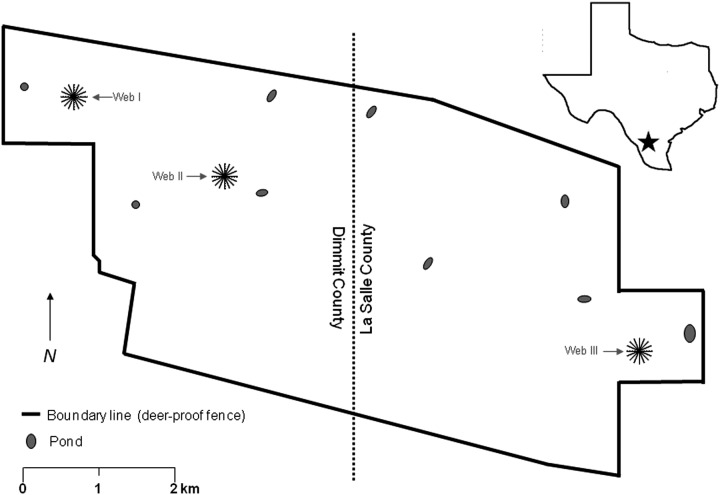
Rodents were captured at 3 sites (Fig. 1) in January 4, 2001 to October 8, 2004 (Table 1). A trapping web (Anderson et al. 1983) was established on site I (Universal Transverse Mercator coordinates, 14-465980E-3131609N) in January, 2001, site II (14-454285N-3136441E) in March, 2001, and site III (14-458569N-3134894E) in January, 2001. Each web (31,400 square meters) consisted of 16 trap lines and 320 trap stations, with 20 stations on each line at 5-meter intervals, beginning 5 meters from the center of the web.
FIG. 1.
Map of the Chaparral Wildlife Management Area showing the locations of Webs I, II, and III. The inset shows the location of the area in Texas.
Table 1.
Prevalence of Catarina Virus Infection in Rodents Captured at 3 Sites on the Chaparral Wildlife Management Area, January 4, 2001–October 8, 2004, by Trapping Sessiona
| Web Ib | Jan 01 | Mar 01 | Jun 01 | Oct 01 | Jan 02 | Mar 02 | Jun 02 | Oct 02 | Jan 03 | Mar 03 | Jun 03 | Oct 03 | Jan 04 | Mar 04 | Jun 04 | Oct 04 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Btay | – | – | – | – | – | – | – | – | – | – | – | 0/3 | 0/2 | 0/10 | 0/28 | 0/4 |
| Nmic | 0/4 | 2/11 | 9/29 | 5/20 | 0/4 | 1/11 | 1/4 | 0/11 | 2/11 | 0/8 | 2/22 | 0/27 | 1/21 | 0/16 | 0/18 | 0/20 |
| Oleu | 0/1 | – | – | – | 0/4 | 0/2 | – | – | 0/4 | 0/3 | 0/5 | 0/5 | 0/10 | 0/9 | 0/13 | 0/4 |
| Pleu | 0/19 | 0/20 | 0/13 | 0/16 | 0/30 | 0/21 | 0/4 | – | 0/53 | 0/34 | 0/25 | 0/36 | 0/32 | 0/21 | 0/11 | 0/22 |
| Rful | 0/7 | 0/1 | 0/3 | 0/3 | 0/12 | 0/3 | – | – | 0/7 | 0/1 | 0/3 | 0/4 | 0/20 | 0/8 | – | 0/1 |
| Shis | 0/3 | 0/5 | 0/30 | 0/27 | 0/6 | 0/6 | 0/1 | – | 0/25 | 0/20 | 0/23 | 0/57 | 0/64 | 0/49 | 0/120 | 0/146 |
| Chis | – | 0/6 | 0/12 | 0/10 | 0/2 | 0/5 | 0/15 | 0/24 | 0/11 | 0/20 | 0/24 | 0/49 | 0/5 | 0/3 | 0/3 | 0/6 |
| Dord | 0/1 | 0/2 | 0/1 | – | 0/4 | 0/4 | 0/3 | – | 0/9 | 0/11 | 0/3 | 0/9 | 0/6 | 0/3 | 0/3 | 0/2 |
| Pmer | 0/1 | 0/4 | 0/22 | 0/10 | – | 0/5 | 0/20 | 0/29 | 0/35 | 0/24 | 0/36 | 0/55 | 0/13 | 0/10 | 0/19 | 0/12 |
| Smex | – | – | – | 0/1 | – | 0/2 | 0/1 | – | – | – | – | – | – | – | – | – |
| Total | 0/36 | 2/49 | 9/110 | 5/87 | 0/62 | 1/59 | 1/48 | 0/64 | 2/155 | 0/121 | 2/141 | 0/245 | 1/173 | 0/129 | 0/215 | 0/217 |
| Web II | Jan 01 | Mar 01 | Jun 01 | Oct 01 | Jan 02 | Mar 02 | Jun 02 | Oct 02 | Jan 03 | Mar 03 | Jun 03 | Oct 03 | Jan 04 | Mar 04 | Jun 04 | Oct 04 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Btay | – | – | – | – | – | – | – | – | – | 0/1 | 0/2 | 0/4 | 0/3 | – | 0/4 | |
| Nmic | 3/11 | 5/15 | 6/19 | 5/21 | 3/11 | 0/4 | 3/22 | 1/16 | 0/8 | 0/39 | 1/36 | 0/17 | 0/9 | 0/21 | 0/36 | |
| Oleu | – | – | 0/1 | – | – | – | – | 0/1 | – | – | 0/1 | – | 0/4 | 0/1 | – | |
| Pleu | 0/17 | 0/17 | 0/18 | 0/24 | 0/11 | 0/6 | 0/2 | 0/32 | 0/27 | 0/43 | 0/37 | 0/23 | 0/18 | 0/3 | 0/2 | |
| Rful | – | – | – | 0/4 | 0/2 | – | – | 0/3 | 0/2 | 0/1 | – | 0/10 | 0/2 | – | – | |
| Shis | 0/2 | 0/2 | 0/8 | 0/3 | 0/1 | 0/1 | 0/1 | 0/14 | 0/17 | 0/46 | 0/104 | 0/68 | 0/97 | 0/128 | 0/110 | |
| Chis | 0/9 | 0/5 | 0/11 | 0/1 | 0/1 | 0/6 | – | 0/2 | 0/4 | 0/12 | 0/18 | 0/1 | – | 0/3 | 0/5 | |
| Dord | 0/2 | 0/2 | – | – | – | – | – | – | – | – | 0/4 | 0/1 | 0/2 | 0/2 | 0/1 | |
| Pmer | 0/10 | 0/14 | 0/14 | 0/1 | – | 0/16 | 0/1 | 0/19 | 0/32 | 0/28 | 0/62 | 0/8 | 0/5 | 0/3 | – | |
| Smex | – | – | – | – | 0/2 | – | – | – | – | – | – | – | – | – | – | |
| Total | 3/51 | 5/55 | 6/71 | 5/54 | 3/28 | 0/33 | 3/26 | 1/87 | 0/90 | 0/170 | 1/264 | 0/132 | 0/140 | 0/161 | 0/158 |
| Web III | Jan 01 | Mar 01 | Jun 01 | Oct 01 | Jan 02 | Mar 02 | Jun 02 | Oct 02 | Jan 03 | Mar 03 | Jun 03 | Oct 03 | Jan 04 | Mar 04 | Jun 04 | Oct 04 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Btay | – | – | – | – | – | – | – | – | – | – | 0/4 | 0/7 | 0/21 | 0/13 | 0/4 | – |
| Nmic | 1/5 | 3/11 | 4/32 | 5/27 | 1/8 | 1/4 | 0/12 | 3/19 | 0/8 | 0/11 | 0/14 | 0/18 | 0/8 | 0/13 | 0/20 | 0/39 |
| Oleu | 0/2 | 0/1 | – | 0/4 | 0/7 | 0/2 | – | – | 0/9 | 0/8 | 0/6 | 0/8 | 0/17 | 0/13 | 0/13 | 0/12 |
| Pleu | 0/25 | 0/27 | 0/28 | 0/20 | 0/47 | 0/34 | 0/16 | 0/2 | 0/86 | 0/45 | 0/52 | 0/45 | 0/31 | 0/32 | 0/18 | 0/10 |
| Rful | 0/8 | 0/3 | – | 0/1 | 0/3 | 0/3 | – | – | 0/12 | 0/6 | 0/8 | 0/2 | 0/18 | 0/10 | 0/7 | 0/3 |
| Shis | 0/8 | 0/9 | 0/11 | 0/17 | 0/5 | 0/1 | 0/2 | 0/1 | 0/11 | 0/12 | 0/87 | 0/103 | 0/93 | 0/92 | 0/132 | 0/135 |
| Chis | – | 0/9 | 0/8 | 0/7 | 0/2 | 0/1 | 0/19 | 0/13 | 0/12 | 0/9 | 0/20 | 0/19 | 0/3 | 0/1 | 0/3 | 0/1 |
| Dord | – | – | – | – | 0/2 | 0/3 | 0/1 | – | – | – | – | 0/1 | – | – | – | 0/1 |
| Pmer | – | 0/3 | 0/6 | 0/10 | 0/1 | 0/4 | 0/13 | 0/14 | 0/25 | 0/13 | 0/14 | 0/25 | 0/6 | – | 0/7 | 0/1 |
| Smex | – | 0/1 | 0/1 | 0/1 | – | 0/2 | 0/3 | – | – | 0/1 | 0/1 | – | – | – | – | – |
| Total | 1/48 | 3/64 | 4/86 | 5/87 | 1/75 | 1/54 | 0/66 | 3/49 | 0/163 | 0/105 | 0/206 | 0/228 | 0/197 | 0/174 | 0/204 | 0/202 |
Trapping on Webs I and III began in January, 2001; trapping on Web II began in March, 2001. A total of 1049 (26.6%) of the 3941 rodents captured during the 46-month study period were captured in 2 or more trapping sessions.
Btay, Baiomys taylori; Nmic, Neotoma micropus; Oleu, Onychomys leucogaster; Pleu, Peromyscus leucopus; Rful, Reithrodontomys fulvescens; Shis, Sigmodon hispidus; Chis, Chaetodipus hispidus; Dord, Dipodomys ordii; Pmer, Perognathus merriami; Smex, Spermophilus mexicanus.
Collection and processing of rodents
A Sherman live trap (H.B. Sherman Traps, Inc., Tallahassee, FL, model LFATDG) was set at each trap station on Web I (W-I) and Web III (W-III) on 3 consecutive nights in January, March, June, and October in January, 2001–October, 2004; a Sherman live trap was set at each trap station on Web II (W-II) on 3 consecutive nights in March, June, and October in 2001, and 3 consecutive nights in January, March, June, and October in January, 2002–October, 2004; a Tomahawk trap (Tomahawk Live Traps, Mfg., Hazelhurst, WI, model 201) was set at a trap station on the second and third nights of a trapping session if the Sherman trap was moved from the station, presumably by a small mammal, on the first night of the same trapping session; and a Tomahawk trap was set at a trap station on the third night of a trapping session if the Sherman trap was moved from the station on the second night of the same trapping session. Traps were set 1 h prior to sunset; baited with a mixture of rolled oats, cracked corn, and birdseed; and checked at dawn the following day. Animals captured in the traps or found dead on the webs were processed in a laboratory on the CWMA.
Individual animals were processed only once each trapping session. The live animals were restrained with ketamine hydrocholoride (1 μg/g body weight, administered by intramuscular injection) to facilitate accurate measurement of the animals and the collection of samples. At first capture, each animal was assigned a unique identification (TK) number and “marked” with a unique toe-clip pattern (Gannon et al. 2007). Information collected on a standardized form included date of capture, web and trap station, species identity (based on external morphological features), gender, body weight, total length (nose to tip of tail), length of tail, and age class (juvenile, subadult, adult based on body size, and condition and coloration of the pelage). Blood samples, throat (oropharyngeal) swabs, and urine samples were collected from the woodrats. Only blood samples were collected from the other mammals. Blood from a bleeding (amputated) toe was collected on a Nobuto strip (Advantec MFS, Inc., Pleasanton, CA); blood from a retro-orbital venous plexus was collected with a sterile microhematocrit tube or sterile Pasteur pipette; cardiac blood from rodents that were euthanized or found dead was collected in an aseptic manner by trans-thoracic puncture with a sterile 25-gauge×5/8” needle on a tuberculin syringe. Oropharyngeal secretions (OPsec) and other materials in the oropharynx were collected on a sterile cotton-tipped swab wetted with 0.01 M phosphate-buffered saline (PBS; pH 7.4) and then mixed with 0.3 mL of PBS containing 10% v/v heat-inactivated fetal bovine serum (FBS). Urine was collected by midstream catch or by antepubic cystocentesis, using a 25-gauge×5/8” needle on a tuberculin syringe. The animals captured before the third day in the October, 2004, trapping session were processed, then provided small pieces of apple for hydration, and, when fully recovered from the effects of the ketamine HCl and stress of processing, were released at the trap stations at which they were captured the previous night. The animals captured on the third day in the October, 2004, trapping session, animals found moribund, and animals with incapacitating injuries were euthanized by intraperitoneal injection of a lethal dose of a mixture of ketamine hydrochloride and xylazine hydrochloride (0.8 mg and 0.16 mg/10 grams body weight, respectively).
The samples of blood, OPsec, and urine were transported on dry ice to a biosafety level 3 (BSL-3) laboratory on the campus of The University of Texas Medical Branch, Galveston. Skins, skulls, and other voucher materials from the animals that died during processing and animals that were euthanized or found dead were deposited into the Museum of Texas Tech University.
Antibody assay
The blood samples were tested for anti-arenavirus immunoglobulin G (IgG), using an enzyme-linked immunosorbent assay (ELISA) (Bennett et al. 2000). The test antigen was a detergent lysate of Vero E6 cells infected with CTNV strain AV A0400135 (Cajimat et al. 2007), the negative control (comparison) antigen was a detergent lysate of uninfected Vero E6 cells. Serial fourfold dilutions (from 1:80 through 1:5,120) of each blood sample were tested against both antigens. Woodrat, grasshopper mouse, pygmy mouse, white-footed mouse, cotton rat, and harvest mouse IgG bound to antigen were detected by using a mixture of a goat anti-rat IgG peroxidase conjugate and goat anti-Peromyscus leucopus IgG peroxidase conjugate in conjunction with the ABTS Microwell Peroxidase Substrate System (Kirkegaard and Perry Laboratories, Gaithersburg, MD). Kangaroo rat, hispid pocket mouse, Merriam's pocket mouse, squirrel, and shrew IgG bound to antigen were detected by using Pierce ImmunoPure® Protein A/G, Peroxidase Conjugated (Pierce, Rockford, IL) in conjunction with the ABTS Microwell Peroxidase Substrate System. Optical densities (OD) were measured at 410 nm (reference, 490 nm). The adjusted OD (AOD) of a blood–antigen reaction was the OD of the well coated with the test antigen less the OD of the well coated with the negative control antigen. A blood sample was considered positive for anti-CTNV antibody if the AOD at 1:80 was ≥0.250, the AOD at 1:320 was ≥0.250, and the sum of the AOD for the series of fourfold dilutions (from 1:80 through 1:5,120) was ≥0.750 (Fulhorst et al. 2001). End point titers were measured in all samples from the antibody-positive rodents, using serial fourfold dilutions from 1:80 through 1:1,310,720. The antibody titer in an antibody-positive sample was the reciprocal of the highest dilution for which the AOD was ≥0.250.
Virus assay
The samples of cardiac blood, OPsec, and urine from the woodrats, and samples of kidney from the woodrats captured on the third day of the October, 2004, trapping session were tested for arenavirus by cultivation in Vero E6 cells (Fulhorst et al. 1996). Briefly, 0.2 mL of a 10% v/v suspension of blood or urine in sterile PBS, 0.2 mL of OPsec stored in 0.3 mL of PBS-FBS, or 0.2 mL of a crude 10% w/v homogenate of kidney in sterile PBS was inoculated onto a confluent monolayer of Vero E6 cells in a 12.5-cm2 plastic culture flask. Cell spots were prepared from the monolayer on the 13th or 14th day after inoculation, and arenaviral antigen in the cell spots was detected by using an indirect fluorescent antibody test (IFAT) in which the primary antibody was a hyperimmune mouse ascitic fluid raised against WWAV strain AV 9310135.
Genetic characterization of viruses
The nucleotide sequences of a 587-nucleotide fragment of the nucleocapsid (N) protein genes of isolates AV C0410166, AV C0410175, AV C0410194, AV D0660002, AV D1030087, and AV D1030150 (Table 2) were determined from RNA isolated from monolayers of infected Vero E6 cells. Reverse transcription of N protein gene RNA and amplification of first-strand cDNA were done by using the ABgene Reverse iT™ One-Step RT-PCR Kit (ABgene House, Surrey, UK) in conjunction with oligonucleotides AVNP119 (5′-ACAGCCAATGATTCCACACTCTTC-3′) and AVNP121 (5′-GTCAGGTCAAAGATGCATCACTCATGATG-3′). Both strands of each gel-purified PCR product were sequenced directly, using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Inc., Foster City, CA). The nucleotide sequences of the 587-nucleotide fragment of the N protein genes of the isolates were deposited into the GenBank nucleotide sequence database under accession nos. JQ063083 through JQ063088.
Table 2.
Arenaviruses Isolated from 6 Antibody-Negative Woodrats and 9 Antibody-Positive Woodrats Captured on the CWMA, January, 2001–January, 2003a
| |
|
|
|
|
Arenavirus (strain) isolated from |
||
|---|---|---|---|---|---|---|---|
| Animal identifier | Web | Sample date | Age class | Antibody titerb | Blood | OPsecc | Urine |
| TK100150d | W-I | Mar 2001 | Subadult | 0 | AV C0410175 | – | – |
| TK100313d | W-I | Jun 2001 | Juvenile | 0 | AV C0690111 | – | – |
| TK100206d | W-II | Mar 2001 | Subadult | 0 | AV C0410194 | AV C0410273 | – |
| TK100126d | W-III | Mar 2001 | Adult | 0 | AV C0410166 | AV C0410246 | – |
| TK100453d | W-III | Oct 2001 | Juvenile | 0 | AV C1170025 | AV C1170026 | – |
| TK100529d | W-III | Oct 2001 | Adult | 0 | – | AV C1170108 | – |
| TK100312 | W-I | Jun 2001 | Adult | 7 | – | AV C0690110 | – |
| TK100312 | W-I | Oct 2001 | Adult | 7 | – | – | AV C1170179 |
| TK100316d | W-1 | Jun 2001 | Adult | 6 | – | – | AV C0690122 |
| TK100380 | W-I | Jun 2001 | Adult | 7 | AV C0690203 | AV C0690204 | – |
| TK100380 | W-I | Jan 2003 | Adult | 7 | – | – | AV E0020168 |
| TK100381d | W-I | Jun 2001 | Adult | 3 | – | AV C0690206 | – |
| TK100385d | W-I | Jun 2001 | Subadult | 4 | – | – | AV C0690218 |
| TK100445 | W-I | Oct 2001 | Adult | 7 | – | – | AV C1170006 |
| TK100445 | W-I | Jun 2002 | Adult | 6 | – | – | AV D0660002 |
| TK100190 | W-II | Mar 2001 | Adult | 7 | AV C0410190 | – | – |
| TK100190 | W-II | Jun 2001 | Adult | 6 | – | – | – |
| TK102276d | W-II | Oct 2002 | Adult | 7 | – | – | AV D1030087 |
| TK102318d | W-III | Oct 2002 | Adult | 7 | – | – | AV D1030150 |
None of the woodrats captured in March, 2003–October, 2004 were positive for arenavirus.
0, <320; 1=320; 2=1280; 3=5120; 4=20,480; 5=81,920; 6=327,680; 7, ≥1,310,720.
Oropharyngeal secretions.
Captured in only 1 trapping session.
The analyses of the N protein gene sequences included CTNV strain AV A0400135 (GenBank accession no. DQ865244), CTNV strain AV A0400212 (DQ865245), BCNV strain AV A0070039 (AY924390), WWAV strain AV 9310135 (AF228063), and TAMV strain W 10777 (AF512828). Strains AV A0400135 and AV A0400212 were originally isolated from southern plains woodrats captured on the CWMA in 1999 (Fulhorst et al. 2002a); AV A0070039 from a California mouse (Peromyscus californicus) captured in California in 1998 (Fulhorst et al. 2002b); W 10777 from a hispid cotton rat captured in Florida in 1965 (Calisher et al. 1970); and AV 9310135 from a white-throated woodrat (N. albigula) captured in New Mexico in 1993 (Fulhorst et al. 1996). The 11-taxa nucleotide sequence alignment was generated by using the computer program CLUSTAL W1.7 (Thompson et al. 1994); the neighbor-joining analysis of genetic (p) distances was done with MEGA, version 4.0 (Tamura et al. 2007); and bootstrap support (Felsenstein 1985) for the results of the neighbor-joining analysis was based on 1000 pseudoreplicate datasets generated from the original multiple nucleotide sequence alignment.
Results
A total of 3941 rodents, representing 10 species, were captured in 16 trapping sessions in January 4, 2001–October 8, 2004 (Table 3). A desert shrew was the only other mammal captured on the webs during the 46-month study period.
Table 3.
Prevalence of Arenavirus Infection in 611 Southern Plains Woodrats and Antibody to Catarina Virus in 3330 Other Rodents Captured at 3 Sites on the Chaparral Wildlife Management Area, January 4, 2001–October 8, 2004a
| Species | Web I | Web II | Web III | Total |
|---|---|---|---|---|
| Baiomys taylori | 0/8 | 0/11 | 0/35 | 0/54 |
| Neotoma micropus | 17/183 | 24/232 | 16/196 | 57/611 |
| Onychomys leucogaster | 0/40 | 0/8 | 0/65 | 0/113 |
| Peromyscus leucopus | 0/212 | 0/177 | 0/335 | 0/724 |
| Reithrodontomys fulvescens | 0/57 | 0/21 | 0/69 | 0/147 |
| Sigmodon hispidus | 0/460 | 0/441 | 0/525 | 0/1426 |
| Chaetodipus hispidus | 0/174 | 0/65 | 0/97 | 0/336 |
| Dipodomys ordii | 0/32 | 0/9 | 0/5 | 0/46 |
| Perognathus merriami | 0/192 | 0/180 | 0/96 | 0/468 |
| Spermophilus mexicanus | 0/4 | 0/2 | 0/10 | 0/16 |
| Total | 17/1362 | 24/1146 | 16/1433 | 57/3941 |
Trapping on Webs I and III began in January, 2001; trapping on Web II began in March, 2001.
In all, 220 (5.6%) of the 3941 rodents were found dead in traps (n=128), died during processing (n=91), or were killed for humane reasons (n=1). The dead rodents included 6 northern grasshopper mice, 95 white-footed mice, 23 fulvous harvest mice, 40 hispid cotton rats, 16 southern plains woodrats, 11 hispid pocket mice, 2 Ord's kangaroo rats, and 27 Merriam's pocket mice. The deaths of a majority of these rodents were attributed to hypothermia and/or severe fatigue associated with multiple captures in a single trapping session.
Counting multiple captures of individual rodents in the same trapping session, the 3941 rodents were captured 8559 times in 45,120 trap station-nights, with an overall trap success rate of 19.0%. Individual rodents were captured on as many as 18 nights (median, 2 nights); 1049 rodents each were captured in 2 or more trapping sessions; and 232 rodents each were captured in 2 or more trapping sessions separated by 12 months or longer (Table 4).
Table 4.
Summary of 3941 Rodents Captured in 45,120 Trap Station-Nights on the Chaparral Wildlife Management Area, January 1, 2001–October 8, 2004
| Species | Number captured | Total number of capturesa | Number captured in ≥2 trapping sessionsb | Number recaptured ≥1 yr after initial capture |
|---|---|---|---|---|
| Baiomys taylori | 54 | 87 (6, 1) | 12 (22.2%) | 2 (3.7%) |
| Neotoma micropus | 611 | 950 (7, 1) | 123 (20.1%) | 47 (7.6%) |
| Onychomys leucogaster | 113 | 257 (11, 2) | 33 (29.2%) | 8 (7.1%) |
| Peromyscus leucopus | 724 | 2081 (18, 2) | 261 (36.0%) | 55 (7.6%) |
| Reithrodontomys fulvescens | 147 | 240 (8, 1) | 27 (18.4%) | 4 (2.7%) |
| Sigmodon hispidus | 1426 | 3248 (11, 2) | 370 (25.9%) | 33 (2.3%) |
| Chaetodipus hispidus | 336 | 713 (9, 2) | 75 (22.3%) | 28 (8.3%) |
| Dipodomys ordii | 46 | 114 (8, 2) | 19 (41.3%) | 11 (23.9%) |
| Perognathus merriami | 468 | 852 (12, 1) | 129 (27.6%) | 44 (9.4%) |
| Spermophilus mexicanus | 16 | 17 (2, 1) | 0 (0.0%) | 0 (0.0%) |
| Total | 3941 | 8559 | 1049 (26.6%) | 232 (5.9%) |
Total number of captures (maximum number of times an individual rodent was captured, median of the number of times that individual rodents were captured).
Number (%) captured in 2 or more trapping sessions.
Number (%) captured ≥12 months after initial capture.
Antibody (IgG) reactive with CTNV strain AV A0400135 was found in 73 (11.9%) of 611 woodrats and none of the 3330 other rodents. The antibody titers in the antibody-positive blood samples ranged from 320 to ≥1,310,720. One woodrat seroconverted between first capture (January, 2002) and second capture (January, 2003). The 72 other antibody-positive woodrats were positive for antibody to CTNV at first capture.
Arenavirus was isolated from samples of blood, OPsec, and/or urine from 6 antibody-negative woodrats and 9 antibody-positive woodrats (Table 2). The assays for arenavirus in the kidneys of the 51 woodrats captured on the last day in the October, 2004, trapping session were negative.
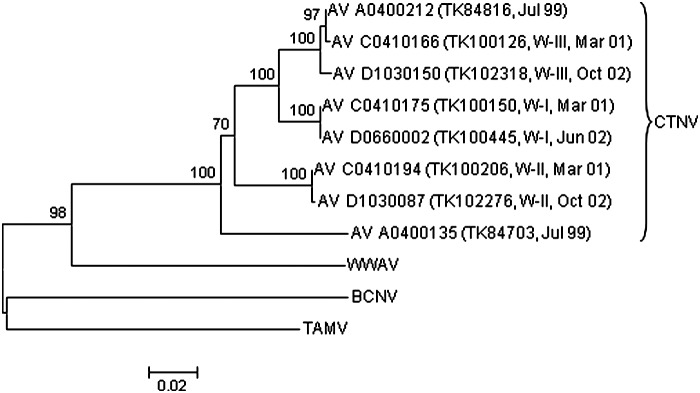
The results of the neighbor-joining analysis of genetic (p) distances (Fig. 2) indicated that arenaviruses AV C0410166, AV C0410175, AV C0410194, AV D0660002, AV D1030087, and AV D1030150 are strains of CTNV. Non-identities among the nucleotide sequences of these 6 strains, CTNV strain AV A0400135, and CTNV strain AV A0400212 ranged from 0/587 (AV C0410175 and AV D0660002) to 60/587 (AV A0400135 and AV D1030150).
FIG. 2.
Phylogenetic relationships among 8 strains of Catarina virus (CTNV) isolated from southern plains woodrats (Neotoma micropus) captured on the Chaparral Wildlife Management Area (CWMA), based on a neighbor-joining analysis of nucleocapsid protein gene sequence data. BCNV, Bear Canyon virus strain AV A0070039; TAMV, Tamiami virus strain W 10777; WWAV, Whitewater Arroyo virus strain AV 9310135. Branch lengths are proportional to genetic (p) distances; the numbers at the nodes indicate the percentage of 1000 bootstrap replicates that supported the interior branches; bootstrap support values less than 70% are not listed; and BCNV was the designated outgroup. The branch labels for the CTNV strains include (in the following order) virus strain, woodrat (TK) number, and month and year in which the virus-positive sample was collected.
The antibody titers in the first blood samples from 16 of the 19 antibody-positive, culture-negative juvenile woodrats and 5 of the 7 antibody-positive, culture-negative subadult woodrats were low (320 or 1280) relative to the antibody titers in the first blood samples from 44 of the 46 antibody-positive adult woodrats (Table 5). Hereafter, it was presumed that the anti-arenavirus antibody in the 21 young woodrats with antibody titers ≤1280 was maternal rather than a consequence of infection.
Table 5.
Antibody Titers to Catarina Virus in the First Blood Samples from 73 Antibody-Positive Woodrats Captured on the CWMA, January, 2001–October, 2004, by Age Class
| |
Antibody titera,b |
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Age class | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Total |
| Juvenile | – | 9 | 7 | 2 | 1 | – | – | – | 19 |
| Subadult | – | 2 | 3 | – | 1c | 1 | 1 | – | 8 |
| Adult | 1b | – | 1 | 7c | 11 | 13 | 5c | 8c | 46 |
| Total | 1 | 11 | 11 | 9 | 13 | 14 | 6 | 8 | 73 |
0, <320; 1=320; 2=1280; 3=5120; 4=20,480; 5=81,920; 6=327,680; 7, ≥1,310,720.
One adult woodrat seroconverted between first capture (January, 2002) and recapture (January, 2003).
Arenavirus was isolated from samples collected at first capture from the subadult woodrat with a titer of 20,480; 1 of 7 adult woodrats with titers of 5120; 1 of 5 adult woodrats with titers of 327,680; and 6 of 8 adult woodrats with titers ≥1,310,720.
Overall, 57 (9.3%) of the 611 woodrats were infected; that is, positive for CTNV and/or positive for CTNV-reactive antibody in response to infection (Table 6). By study site, the prevalence of infection among woodrats ranged from 8.2% (W-III) to 10.3% (W-II) (Table 3).
Table 6.
Prevalence of Catarina Virus Infections among Southern Plains Woodrats Captured on the Chaparral Wildlife Management Area, 2001–2004, by Study Site
| Web I | Web II | Web III | Total | |
|---|---|---|---|---|
| Prevalence of infection among all woodrats captured in | ||||
| January, 2001–October 2004 | 17/183 (9.3%) | 24/232 (10.3%) | 16/196 (8.2%) | 57/611 (9.3%) |
| March–October, 2001–2004 | 17/162 (10.5%) | 20/202 (9.9%) | 14/182 (7.7%) | 51/546 (9.3%) |
| March–October, 2001 | 15/55 (27.3%) | 13/41 (31.7%) | 11/65 (16.9%) | 39/161 (24.2%) |
| March–October, 2002 | 2/25 (8.0%) | 6/35 (17.1%) | 4/33 (12.1%) | 12/93 (12.9%) |
| March–October, 2003 | 3/54 (5.6%) | 1/74 (1.4%) | 0/37 (0.0%) | 4/165 (2.4%) |
| March–October, 2004 | 0/48 (0.0%) | 0/62 (0.0%) | 0/64 (0.0%) | 0/174 (0.0%) |
| Prevalence of infection among adult male woodrats captured in | ||||
| January 2001–October, 2004 | 5/49 (10.2%) | 10/56 (17.9%) | 5/49 (10.2%) | 20/154 (13.0%) |
| March–October, 2001–2004 | 5/40 (12.5%) | 7/41 (17.1%) | 5/45 (11.1%) | 17/126 (13.5%) |
| March–October, 2001 | 4/11 (36.4%) | 4/6 (66.7%) | 4/16 (25.0%) | 12/33 (36.4%) |
| March–October, 2002 | 0/7 (0.0%) | 3/7 (42.9%) | 1/5 (20.0%) | 4/19 (21.1%) |
| March–October, 2003 | 1/6 (16.7%) | 1/16 (6.3%) | 0/9 (0.0%) | 2/31 (6.5%) |
| March–October, 2004 | 0/16 (0.0%) | 0/12 (0.0%) | 0/15 (0.0%) | 0/43 (0.0%) |
| Prevalence of infection among adult female woodrats captured in | ||||
| January, 2001–October, 2004 | 8/61 (13.1%) | 11/75 (14.7%) | 9/55 (16.4%) | 28/191 (14.7%) |
| March–October, 2001–2004 | 8/53 (15.1%) | 10/68 (14.7%) | 8/52 (15.4%) | 26/173 (15.0%) |
| March–October, 2001 | 7/15 (46.7%) | 6/16 (37.5%) | 7/16 (43.8%) | 20/47 (42.6%) |
| March–October, 2002 | 2/9 (22.2%) | 4/10 (40.0%) | 2/10 (20.0%) | 8/29 (27.6%) |
| March–October, 2003 | 2/19 (10.5%) | 0/18 (0.0%) | 0/11 (0.0%) | 2/48 (4.2%) |
| March–October, 2004 | 0/20 (0.0%) | 0/28 (0.0%) | 0/22 (0.0%) | 0/70 (0.0%) |
Fifty-four (21.3%) of the 254 woodrats captured in January, 2001–October, 2002, and 3 (1.2%) of the 339 woodrats captured in January, 2003–October, 2004, were infected (Fig. 3). Only 1 of the 3 infected woodrats captured in January, 2003–October, 2004 (i.e., TK100380) was positive for arenavirus (Table 2).
FIG. 3.
Prevalence of Catarina virus infections among 611 southern plains woodrats captured on the Chaparral Wildlife Management Area, January, 2001–October, 2004, by trapping session.
Forty-eight (89.5%) of the 54 infected woodrats captured in January, 2001–October, 2002, and the 3 infected woodrats captured in January, 2003–October, 2004, were adult and infected at first capture. The prevalence of infection among male woodrats was 26/289 (9.0%), the prevalence of infection among female woodrats was 31/322 (9.6%), and the difference between 26/289 and 31/322 was not statistically significant (Fisher's exact test, two-tailed, p>0.50).
Counting multiple captures of individual rodents in the same trapping session, infected woodrats were captured 70 times at 55 trap stations in January, 2001–October, 2002; 801 rodents other than woodrats were captured 1606 times at 585 trap stations in January, 2001–October, 2002; and 52 rodents (35 white-footed mice, 1 fulvous harvest mouse, 5 hispid cotton rats, 3 hispid pocket mice, 2 northern grasshopper mice, 1 Ord's kangaroo rat, 3 Merriam's pocket mice, and 2 Mexican ground squirrels) were captured 63 times at 26 of the 55 stations at which infected woodrats were captured in January, 2001–October, 2002. Collectively, these observations suggest that close physical interactions between rodents other than woodrats and infected woodrats in January, 2001–October, 2002, were rare.
A total of 297 woodrats in this study were juvenile or subadult at first capture (Table 7). In all, 123 (99.2%) of the 124 juvenile woodrats were captured in March, June, or October, indicating that breeding in N. micropus on the CWMA in 2001–2004 usually began in early spring and ended in fall.
Table 7.
Numbers of Juvenile Woodrats and Subadult Woodrats Captured on the Chaparral Wildlife Management Area, 2000–2004, by Month of Trapping Session and Yeara
| |
Juvenile woodratsb |
Subadult woodratsb |
||||||
|---|---|---|---|---|---|---|---|---|
| Month | 2001 | 2002 | 2003 | 2004 | 2001 | 2002 | 2003 | 2004 |
| January | – | – | – | 0/1 | 0/3 | 0/7 | 0/11 | 0/7 |
| March | 0/14 | – | 0/4 | 0/11 | 2/6 | 0/4 | 0/6 | 0/4 |
| June | 1/17 | 0/1 | 0/16 | 0/4 | 2/19 | 0/7 | 0/25 | 0/16 |
| October | 2/12 | 2/23 | 0/13 | 0/8 | 1/15 | 0/12 | 0/16 | 0/15 |
| Total | 3/43 | 2/24 | 0/33 | 0/24 | 5/43 | 0/30 | 0/58 | 0/42 |
Traps were set on Webs I and III but not Web II in January, 2001.
Number infected/number captured.
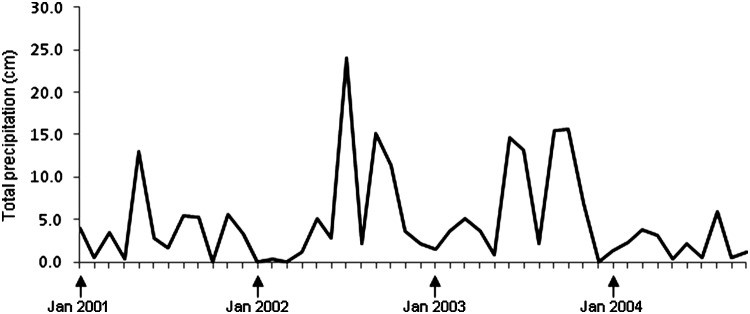
By year, the proportion of juvenile woodrats captured in March or June ranged from 1/24 (4.2%) in 2002 to 31/43 (72.1%) in 2001 (Table 7), and the amount of precipitation in January–April ranged from 1.5 cm in 2002 to 14.1 cm in 2003 (Fig. 4). There was a positive association between the number of pups captured in March or June and total precipitation in January–April (r=0.81).
FIG. 4.
Precipitation on the Chaparral Wildlife Management Area, January, 2001–October, 2004, by month and year. Precipitation was measured with a Vantage Pro2™ weather station (Davis Instruments Corp., Hayward, CA) located on the Area. Total precipitation in January–April in 2001, 2002, 2003, and 2004 was 8.2 cm, 1.5 cm, 14.1 cm, and 10.6 cm, respectively. The precipitation data were kindly provided by D. Synatzske and D. Walker (Texas Parks & Wildlife Department).
Discussion
In a previous study (Fulhorst et al. 2002a), strains of CTNV were isolated from 5 southern plains woodrats, anti-arenavirus was found in 4 (14.3%) of 28 other southern plains woodrats, and neither arenavirus nor anti-arenavirus antibody was found in 60 hispid cotton rats, 13 white-footed mice, 3 northern grasshopper mice, 1 northern pygmy mouse, 1 fulvous harvest mouse, 3 Ord's kangaroo rats, 62 Merriam's pocket mice, or 32 hispid pocket mice captured on the CWMA in a 1-week period in July 1999. Collectively, these findings suggested that the southern plains woodrat is the principal host of CTNV on the CWMA and that CTNV infection is highly specific to N. micropus. The infections in 57 (9.3%) of 611 woodrats and none of the 3330 other rodents in this study affirm the notion that CTNV infection is highly specific to N. micropus.
The analysis of capture data suggested that close physical interactions between infected woodrats and rodents other than woodrats in January, 2001–October, 2002, were rare. A low frequency of intimate physical interactions with infected woodrats may explain the failure to find evidence of arenavirus infections in rodents other than woodrats in this study. Alternatively, rodents other than woodrats are refractory to CTNV infection or CTNV-infected woodrats are rarely infectious to rodents other than woodrats.
A characteristic of arenaviruses is their ability to establish chronic infections in their respective principal hosts. The isolation of CTNV from samples collected from TK100312 in June, 2001, and October, 2001, TK100380 in June, 2001, and January, 2003, and TK100445 in October, 2001, and June, 2002 (Table 2) indicate that CTNV infections in some southern plains woodrats are chronic. Whether virus shedding in chronically infected woodrats is persistent remains to be determined.
The results of a previous study (Suchecki et al. 2004) suggested that breeding in N. micropus on the CWMA begins in early spring and ends some time in fall. As such, chronic infections in individual woodrats likely are critical to long-term maintenance, in particular—overwintering, of CTNV on the CWMA.
The results of a laboratory study (Milazzo and Fulhorst 2012) suggested that the duration of CTNV infection in southern plains woodrats is dependent upon age of woodrat at the onset of infection; for example, 6 (85.7%) of 7 southern plains woodrats inoculated at age 1 day with strain AV A0400135 were viruric through 5 months of age, whereas all 4 adult southern plains woodrats inoculated with the same strain, same dose, and by the same route sterilized their infections within 30 days of inoculation. Hypothetically, the majority of chronic infections in southern plains woodrats on the CWMA result from exposure to CTNV early in life; and, as such, vertical (dam-to-progeny or sire-to-progeny) virus transmission in N. micropus plays an important role in the long-term maintenance of CTNV on the CWMA.
The prevalence of infection among woodrats captured in March–October in this study decreased from 39/161 (24.2%) in 2001 to 0/174 (0.0%) in 2004 (Table 6). The majority (67.0%) of the 100 juvenile woodrats captured in 2001, 2003, and 2004 were captured in January, March, or June (Table 7). In contrast, only 1 (4.2%) of the 24 juvenile woodrats captured in 2002 was captured before the October trapping session (Table 7). The onset of breeding in N. micropus in 2002 may have been delayed by conditions related to the drought in January–April in 2002; alternatively, the majority of pups whelped in spring of 2002 did not survive through early summer. Regardless, a high level of reproductive success in N. micropus early in spring may be essential to long-term maintenance of CTNV on the CWMA.
The analysis of N protein gene sequence data in this study revealed a significant level of genetic diversity among CTNV strains isolated from woodrats captured on the CWMA in July, 1999–October, 2002. Previous studies (Méndez-Harclerode et al. 2005, 2007) revealed a high level of genetic diversity among southern plains woodrats captured on the CWMA in January, 2001–October, 2004. Hypothetically, virus strain and host genetics, as well as age of woodrat at onset of infection, affect the duration of CTNV infection in the southern plains woodrat (Childs and Peters 1993).
The results of a previous study (Milazzo et al. 2011) suggested that arenaviruses naturally associated with woodrats (Neotoma spp.) are etiological agents of acute central nervous system disease or undifferentiated febrile illnesses in humans in the United States. It is generally accepted that humans usually become infected with arenaviruses by inhalation of virus in aerosolized droplets of saliva, respiratory secretions, urine, or blood from infected rodents. The isolation of CTNV from samples of OPsec or urine from woodrats in this study suggests that secretions and excretions from naturally infected southern plains woodrats may be infectious to humans. Neotoma micropus in Texas is principally associated with habitats dominated by cactus or thorny desert shrubs (Braun and Mares 1989). As such, persons who work, live, or enjoy outdoor activities in rural areas in southern Texas may be at risk of CTNV infection.
Acknowledgments
Joel Brant, Nevin D. Durish, Heath Garner, Kara Graham, Mariko Kageyama, Michelle Knapp, Peter Larson, Lisa K. Longhofer, L. Rex McAliley, Lisa Mitchell, Vicki Swier, and Amy Vestal (Texas Tech University); Jason E. Comer, Eleanor Deardorff, and Sammie Hartmann (The University of Texas Medical Branch, Galveston); Emmanuel Rollin (University of Georgia, Athens, GA); and Stephen G. Bennett (Orange County Vector Control District, Santa Ana, CA) assisted with the capture and processing of rodents. Bal M. Mishra (The University of Texas Medical Branch, Galveston) assisted with the genetic characterization of the CTNV strains.
This study was supported by National Institutes of Health (NIH) grant AI-41435, “Ecology of emerging arenaviruses in the southwestern United States.” James Meegan and Patricia Repik (National Institute of Allergy and Infectious Diseases) facilitated the grant support for this study.
Author Disclosure Statement
No competing financial interests exist.
References
- Anderson DR. Burnham KP. White GC. Otis DL. Density estimation of small-mammal populations using a trapping web and distance sampling methods. Ecology. 1983;64:674–680. [Google Scholar]
- Bennett SG. Milazzo ML. Webb JP., Jr Fulhorst CF. Arenavirus antibody in rodents indigenous to coastal southern California. Am J Trop Med Hyg. 2000;62:626–630. doi: 10.4269/ajtmh.2000.62.626. [DOI] [PubMed] [Google Scholar]
- Bigler WJ. Lassing E. Buff E. Lewis AL, et al. Arbovirus surveillance in Florida: Wild vertebrate studies 1965–1974. J Wildl Dis. 1975;11:348–356. doi: 10.7589/0090-3558-11.3.348. [DOI] [PubMed] [Google Scholar]
- Braun JK. Mares MA. Mammalian Species. The American Society of Mammalogists; 1989. Neotoma micropus; pp. 1–9. no. 330. [Google Scholar]
- Cajimat MNB. Milazzo ML. Bradley RD. Fulhorst CF. Catarina virus, an arenaviral species principally associated with Neotoma micropus (southern plains woodrat) in Texas. Am J Trop Med Hyg. 2007;77:732–726. [PubMed] [Google Scholar]
- Cajimat MNB. Milazzo ML. Haynie ML. Hanson JD, et al. Diversity and phylogenetic relationships among the North American Tacaribe serocomplex viruses (family Arenaviridae) Virology. 2011;421:87–95. doi: 10.1016/j.virol.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher CH. Tzianabos T. Lord RD. Coleman PH. Tamiami virus, a new member of the Tacaribe group. Am J Trop Med Hyg. 1970;19:520–526. doi: 10.4269/ajtmh.1970.19.520. [DOI] [PubMed] [Google Scholar]
- Childs JE. Peters CJ. Ecology and epidemiology of arenaviruses and their hosts. In: Salvato MS, editor. The Arenaviridae. New York: Plenum Press; 1993. pp. 331–384. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF. Bowen MD. Ksiazek TG. Rollin PE, et al. Isolation and characterization of Whitewater Arroyo virus, a novel North American arenavirus. Virology. 1996;224:114–120. doi: 10.1006/viro.1996.0512. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF. Charrel RN. Weaver SC. Ksiazek TG, et al. Geographic distribution and genetic diversity of Whitewater Arroyo virus in the southwestern United States. Emerg Infect Dis. 2001;7:403–407. doi: 10.3201/eid0703.010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulhorst CF. Milazzo ML. Carroll DS. Charrel RN, et al. Natural host relationships and genetic diversity of Whitewater Arroyo virus in southern Texas. Am J Trop Med Hyg. 2002a;67:114–118. doi: 10.4269/ajtmh.2002.67.114. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF. Bennett SG. Milazzo ML. Murray HL, et al. Bear Canyon virus: An arenavirus naturally associated with Peromyscus californicus (California mouse) Emerg Infect Dis. 2002b;8:717–721. doi: 10.3201/eid0807.010281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon WL. Sikes RS, et al. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mammal. 2007;83:809–823. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings WL. Lewis AL. Sather GE. Pierce LV, et al. Tamiami virus in the Tampa Bay area. Am J Trop Med Hyg. 1970;19:527–536. doi: 10.4269/ajtmh.1970.19.527. [DOI] [PubMed] [Google Scholar]
- McLendon T. Preliminary description of the vegetation of south Texas exclusive of coastal saline zones. Texas J Sci. 1991;43:13–32. [Google Scholar]
- Méndez-Harclerode FM. Hanson JD. Fulhorst CF. Milazzo ML. Genetic diversity within the southern plains woodrat (Neotoma micropus) in southern Texas. J Mammal. 2005;86:180–190. doi: 10.1644/1545-1542(2005)086<0180:gdwtsp>2.0.co;2. e al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Harclerode FM. Strauss RE. Fulhorst CF. Milazzo ML, et al. Molecular evidence for high levels of intrapopulation genetic diversity in woodrats (Neotoma micropus) J Mammal. 2007;88:360–370. doi: 10.1644/05-MAMM-A-377R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo ML. Campbell GL. Fulhorst CF. Novel arenavirus infection in humans, United States. Emerg Infect Dis. 2011;17:1417–1420. doi: 10.3201/eid1708.110285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo ML. Fulhorst CF. Duration of Catarina virus infection in the southern plains woodrat (Neotoma micropus) Vector Borne Zoonotic Dis. 2012;12:321–324. doi: 10.1089/vbz.2011.0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser GG. Carleton MD. Family Cricetidae. In: Wilson DE, editor; Reeder DM, editor. Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd. Baltimore: The Johns Hopkins University Press; 2005. pp. 955–1189. [Google Scholar]
- Ruthven DC., III Synatzske DR. Response of herbaceous vegetation to summer fire in the western south Texas plains. Texas J Sci. 2002;54:195–210. [Google Scholar]
- Suchecki JR. Ruthven DC., III Fulhorst Bradley RD. Natural history of the southern plains woodrat Neotoma micropus (Rodentia: Muridae) from southern Texas. Texas J Sci. 2004;56:131–140. [Google Scholar]
- Tamura K. Dudley J. Nei M. Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software, version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD. Higgins DG. Gibson TJ. CLUSTAL W (1.7): Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choices. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]