Abstract
The residential regions of Yunnan province, canton of Jing Hong, in China were surveyed for Japanese encephalitis virus (JEV) infection in mosquito and swine vectors to determine the frequency of JEV-carrying zoonotic vectors in 2009–2010. A total of 21,500 mosquitoes were collected and divided by species, and brain tissue was collected from 108 stillborn piglets. The infection rates for the different JEV species were 13.2% for Culex tritaeniorhynchus, 2.7% for Anopheles sinensis, 0.7% for Armigeres subalbatus, and 18.5% for stillborn piglets. The complete genomes of two JEV samples that were collected in different seasons and different regions, Yunnan 0901 and Yunnan 0902, were sequenced from a pool of Culex mosquitoes and stillborn piglets that had been collected randomly from several piggeries. Multiple sequence alignment with 24 fully-sequenced genes and 93 complete sequences of the JEV-encoded E gene revealed nucleotide homologies ranging from 97.2–99.6% and 94.5–99.7% in mosquitoes and piglets, respectively, and deduced amino acid homologies ranging from 97.4–98.1% and 96.0–98.2%, respectively. Phylogenetic analyses of the Yunnan 0901 and Yunnan 0902 strains' full-length genomes and E gene sequences indicated that these strains are most closely related to six Chinese SA14-derived viruses, and distantly related to the Australian FU, vellore P20778, and Japanese Ishikawa strains, and the previously isolated YN86-B8639 strains. The phylogenetic relationships based on the full-length genome were similar to those found for the E gene, indicating that phylogenetic analysis of the E gene will be a useful approach for genotyping of JEV, but not to better understand the potential changes in the biological characteristics and genetic relationship of JEV isolates.
Key Words: Genotype, Japanese encephalitis virus, Molecular epidemiology, Mosquito vector, RT-PCR
Introduction
Japanese encephalitis virus (JEV) belongs to the genus Flavivirus within the family Flaviviridae. JEV is the most important cause of viral encephalitis in Asia (Gubler et al. 2007). The virus is maintained in a natural cycle that primarily involves mosquito and swine vectors (Vaughn and Hoke 1992; Endy and Nisalak 2002). Infection with JEV has serious consequences for sow reproduction due to the high death rate of piglets (World Health Organization 1998).
JEV (Nakayama strain) was first isolated from the human brain in 1935 in Japan. Since then, a number of geographically-diverse JEV strains have been isolated at different times from several sources. Numerous JEV isolates have been found in different geographical areas and at different times, from humans, mosquitoes, and pigs (Wang et al. 2007). JEV was first isolated in China in 1940 (Yin et al. 1997).
The Flavivirus genome contains a single open reading frame (ORF) that encodes a polyprotein approximately 11 kb in length (Burke and Monath 2001). This polyprotein encodes three structural proteins which are encoded in the 5′ third of the ORF sequences: the capsid (C), pre-membrane/membrane (prM/M), and envelope (E) proteins. Seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) are encoded in the remaining 3′ two-thirds sequences. The 5′ and 3′ non-coding regions (NCRs) are about 95 and 582 nucleotides in length, respectively (Sumiyoshi et al. 1987; Hashimoto et al. 1988). Recently several authors have classified JEV into four genotypes based on the analysis of highly variable nucleotide sequences in the prM and E gene regions, and in the 3′ NCR (Chen et al. 1990; Ali et al. 1995; Nam et al. 2001; Uchil and Satchidanandam 2001).
In this study, we have determined the complete nucleotide and deduced amino acid sequences of the Yunnan 0901 and Yunnan 0902 strains (GenBank accession numbers JQ086762 and JQ086763) isolated in the Yunnan province in the Jing Hong region during 2009 and 2010, and compared the full-length genome sequence with that of the 24 fully-sequenced JEV strains currently available. We have fully characterized the sequence at the molecular level, and established its relationship to the other fully-sequenced JEV strains. We compared the genetic relationship of the Yunnan 0901 and Yunnan 0902 strains to a large and heterogeneous selection of 93 JEV E genes from various strains that were isolated from different geographic regions at different time periods. The findings of this study add to the overall JEV data, and may help in future studies to predict the virus's evolutionary trends.
Materials and Methods
Virus collection
We collected 21,500 mosquitoes and divided them by species, before selecting 50 insects of each species and collecting them in single tubes. In total, 430 tubes were used for analysis. A total of 108 brain tissue samples from stillborn piglets were also used.
Virus proliferation
The two JEV strains used in the present study were isolated from pools of Culex tritaeniorhynchus and brain tissue from stillborn piglets, respectively, collected from villages in Yunnan Province, in the canton of Jing Hong (Fig. 1). The two strains were designated as Yunnan 0901 and Yunnan 0902, respectively.
FIG. 1.
Map showing the canton of Jing Hong in Yunnan Province, where the study was carried out.
Emulsions [10% (w/v)] of mosquitoes or piglet brain tissue suspensions were prepared in Eagle's minimum essential medium (EMEM) that contained 2% heat-inactivated fetal bovine serum (FBS). The virus isolates were propagated in Aedes albopictus C6/36 cells. The cytopathogenic effects (CPEs) were observed by inoculating the C6/36 cell cultures with the emulsions made from the mosquito and piglet brain tissue suspensions. A litter of suckling mice (n=15) was divided into three groups (n=5 each). Five mice in one group were inoculated intracerebrally with 50 μL of each of the emulsions made from the mosquito or piglet brain tissue suspensions. The other five mice from the same litter were inoculated intracerebrally with EMEM that contained 2% FBS as controls. The mice inoculated with the emulsion made from the mosquito and piglet brain tissue suspension showed apparent paralysis with a lack of appetite on day 3 post-inoculation when all animals, including the healthy control mice, were euthanized. Brain tissues were collected from the paralyzed and control mice, and pooled for each group.
Experimental infection of animals
The suckling mice were inoculated intracranially with 0.02 mL JEV isolate of Yunnan 0901 and Yunnan 0902 and observed for 15 days. As soon as the suckling mice demonstrated illness, the mice were collected. The LD50 was determined by the Reed-Muench method (Yin et al. 1997).
Isolation of viral RNA
Total RNA was extracted from the supernatants of the cell cultures that showed CPE, and from 10% (w/v) brain emulsion samples prepared from the brain tissues of the paralyzed mice and control mice, using a QIAamp viral RNA extraction kit (Qiagen China, Shanghai, P.R. China), according to the manufacturer's instructions. The denatured RNA was incubated at 42°C for 50 min to perform first-strand cDNA synthesis using reverse-transcriptase polymerase chain reaction (PCR) with random primers, as previously described (Zhang et al. 2009).
Nucleotide sequencing of the JEV Yunnan 0901 and Yunnan 0902 genomes
Sequences of oligonucleotide primers (Table 1) were designed according to the JEV of SA14-14-2 sequence to amplify segments that coded for the capsid, pre-membrane, envelope, NS1, NS2, NS3, NS4, and NS5 protein regions. The 3′ and 5′ termini of these viral genes were amplified by 3′ and 5′ RACE (Krishnamurthy and Samal, 1998; Kumar et al. 2008). PCR was carried out using 30 cycles of amplification. The presence of the correct PCR products was confirmed by electrophoresing 10 μL through 1.0% agarose gels. To achieve high-quality consensus sequences and to avoid laboratory PCR artifacts, each entire genome was sequenced at least three times. The amplified fragments were cloned into the pMD18-T vector and confirmed by sequencing (Shanghai Biological Engineering Co., Ltd., Shanghai, P.R. China). The full-length genome was compiled using the DNASTAR software program (Yang et al. 2004a).
Table 1.
Oligonucleotide Primers for PCR Amplification
| Gene name | Sequence 5′–3′ | Amplified length |
|---|---|---|
| C | JEVC1: 5-ATGACTAAAAAACCAGGAGGG-3 | 381 bp |
| JEVC2: 5-TGGAAATTCGACAACTTCATG-3 | ||
| Prm | JEVP/M1: 5-ATGAAGTTGTCGAATTTCCAG-3 | 501 bp |
| JEVP/M2: 5-CCCATTCCCAGACAATTAAAA-3 | ||
| E | JEVE1: 5-TTTAATTGTCTGGGAATGGGC-3 | 1500 bp |
| JEVE2: 5-TCAATGGCACATCCAGTGTCA | ||
| NS1 | JEVNS12: 5-CTGTTGGCCTCTGCGAAAGCA-3 | 1265 bp |
| JEV NS2A1: 5-GCTTTCGCAGAGGCCAACAGT-3 | ||
| NS2A | JEV NS2A1: 5-GCTTTCGCAGAGGCCAACAGT-3 | 512 bp |
| JEV NS2A2: 5-AACTCAGTAGCTGGCCACCCT-3 | ||
| NS2B | JEV NS2B1: 5-GGGTGGCCAGCTACTGAGTTT-3 | 413 bp |
| JEV NS2B2: 5-GTGTCCCAAAACACGCCCCCT-3 | ||
| NS3 | JEV NS31: 5-GGGGGCGTGTTTTGGGACACG-3 | 1877 bp |
| JEV NS32: 5-TCTATGAAGCTAACGGCTGAT-3 | ||
| NS4A | JEV NS4A1: 5-TCAGCCGTTAGCTTCATAGAG-3 | 821 bp |
| JEV NS4A2: 5-ATGTACCCATAGTGAAGTGTC-3 | ||
| NS4B | JEV NS4B1: 5-ACACTTCACTATGGGTACATG-3 | 431 bp |
| JEV NS4B2: 5-GTCCTGCCCCCAGGCCTTCCC-3 | ||
| NS5 | JEV NS51: 5-GGAAGGCCTGGGGGCAGGACG-3 | 2735 bp |
| JEV NS52: 5-TTCTACCTTAAATCACACTAG-3 |
Public datasets
Full-length genes from the Yunnan 0901 and Yunnan 0902 strains were deposited in the GenBank database under accession numbers JQ086762 and JQ086763 (http://www.ncbi.nlm.nih.gov/GenBank/).
Multiple alignments and phylogenetic analyses
The JEV strains used for multiple sequence alignments and phylogenetic analyses, with a description of the history of these strains and their GenBank accession numbers, are shown in Table 2. Multiple sequence alignments and sequence similarity calculations between aligned nucleotide and amino acid sequences were performed using DNASTAR software (Madison, WI). Multiple sequence alignments and phylogenetic trees were produced using MEGA 4.1 software and constructed from aligned nucleotide sequences using the neighbor-joining method. The stability of the tree obtained was established by bootstrapping analysis with 1000 replications (Kumar et al. 2004).
Table 2.
Japanese Encephalitis Virus (JEV) Strains Used in this Study
| Nation | Gene type | Strain | Year isolated | Source | GenBank accession number |
|---|---|---|---|---|---|
| Australia | II | Fu | 1995 | Human serum | AF217620 |
| Cambodia | I | M859 | 1967 | Mosquito | U70410 |
| China | III | SA14 | 1954 | Mosquito | U14163 |
| III | SA14-14-2 | 1954 | SA-14 derivative | AF315119 | |
| III | Beijing-1 | 1949 | Human brain | L48916 | |
| III | P3 | 1949 | Mosquito | U47032 | |
| III | SA14-2-8 | 1954 | SA-14 derivative | U02367 | |
| III | SA(V) | 1954 | SA14 derivative | D90194 | |
| III | SA(A) | 1954 | SA14-14-2 derivative | D90195 | |
| I | SH-53 | 1987 | Human brain | AY555757 | |
| I | SH-96 | 2001 | Culex tritaeniorhynchus | AY555760 | |
| I | SH-101 | 2001 | Culex tritaeniorhynchus | AY555761 | |
| III | TLA | 1971 | Human brain | AY243832 | |
| III | G35 | 1954 | Mosquito pool | AY243831 | |
| III | 02-29 | 2002 | Human cerebrospinal fluid | AY243834 | |
| III | SH04-5 | 2004 | Culex tritaeniorhynchus | DQ404106 | |
| III | GZ04-4 | 2004 | Armigeres | DQ404110 | |
| III | FJ03-31 | 2003 | Human blood | DQ404117 | |
| III | YNDL04-29 | 2004 | Culex theiler | DQ404139 | |
| III | HLJ02-134 | 2002 | Genus culicoids | DQ404081 | |
| III | HLJ02-170 | 2002 | Aedes vexans | DQ404084 | |
| III | GZ04-71 | 2004 | Armigeres | DQ404114 | |
| III | FJ03-97 | 2003 | Human blood | DQ404127 | |
| III | YNDL04-19 | 2004 | Culex theileri | DQ404147 | |
| I | HN04-11 | 2004 | Culex | DQ404087 | |
| I | HN04-40 | 2004 | Culex | DQ404089 | |
| I | SC04-25 | 2004 | Culex | DQ404094 | |
| I | SH03-103 | 2003 | Culex tritaeniorhynchus | DQ404096 | |
| III | SA14-12-1-7 | 1954 | SA14 derivative | AF416457 | |
| III | GP78 | 1978 | Human brain | AF075723 | |
| India | III | P20778 | 1958 | Human brain | AF080251 |
| III | 782219 | 1978 | Human | U70402 | |
| III | 733913 | 1973 | Human brain | Z34095 | |
| III | Vellore P20778 | 1958 | Human brain | AF080251 | |
| III | G8924 | 1958 | Mosquito | U70394 | |
| IV | JKT7003 | 1981 | Mosquito | U70408 | |
| Indonesia | II | JKT5441 | 1981 | Mosquito | U70406 |
| II | JKT6468 | 1968 | Mosquito | U70407 | |
| II | JKT1749 | 1979 | Mosquito | U70405 | |
| IV | JKT9092 | 1981 | Mosquito | U70409 | |
| III | JaOAr8982 | 1982 | Mosquito | M18370 | |
| Japan | I | Ishikawa | 1998 | Mosquito | AB051292 |
| III | JaGAr01 | 1959 | Mosquito | AF069076 | |
| III | Kamiyama | 1966 | Human brain | S49265 | |
| III | Nakayama-NIH | 1935 | Human brain | U70413 | |
| III | JaNAr516 | 1999 | NA | AB028270 | |
| III | Oita100 | 1999 | NA | AB028269 | |
| III | JaOH0566 | 1997 | NA | AY029207 | |
| III | JaOArK5789 | 1989 | IU | AB028285 | |
| III | Sagayama | 1988 | cDNA clone | E02136 | |
| III | Osaka | 1979 | Mosquito | U70414 | |
| III | B18A | 1978 | Mosquito | U70390 | |
| III | Mis44-1 | 1969 | Mosquito | U70411 | |
| III | JaOH3767 | 1967 | Human brain | U70400 | |
| III | Sagiyama | 1957 | Mosquito | U70419 | |
| I | K94P05 | 1994 | Mosquito | AF045651 | |
| Korea | I | KV1899 | 1999 | Pig | AY316157 |
| III | K87P39 | 1987 | Mosquito | U34927 | |
| III | K82P01 | 1982 | Mosquito | U34926 | |
| III | Anyang | 1969 | Pig | Unpublished | |
| I | K91P55 | 1991 | Mosquito | U34928 | |
| II | WTP7022 | 1970 | Mosquito | U70421 | |
| Malaysia | III | T1P1 | 1997 | Mosquito | AF254453 |
| Taiwan | III | CH2195LA | 1994 | NA | AF221499 |
| III | CH1392 | 1990 | Mosquito | AF254452 | |
| III | RP-Zms | 1985 | Mosquito | AF014160 | |
| III | RP-9 | 1985 | Mosquito | AF014161 | |
| III | YL | NA | NA | AF486638 | |
| III | TC | NA | Mosquito | AF098736 | |
| III | TL | NA | Mosquito | AF098737 | |
| III | HVI | NA | Mosquito | AF098735 | |
| III | CH2195SA | 1994 | CH2195 derivative | AF221500 | |
| III | cc27 | 1994 | Mosquito | U44957 | |
| III | CH1302 | 1990 | Mosquito | AF254452 | |
| III | CH392 | 1987 | Mosquito | U44961 | |
| III | CH109 | 1986 | Mosquito | U44959 | |
| III | NT113 | 1985 | Mosquito | U44968 | |
| III | ML117 | 1985 | Pig blood | U44965 | |
| III | NT109 | 1984 | Mosquito | U44967 | |
| III | HK 8256 | 1972 | Mosquito | U70396 | |
| I | ThCMAr4492 | 1992 | Mosquito | D45360 | |
| I | ThCMAr6793 | 1963 | Mosquito | D45363 | |
| Thailand | I | B2239 | 1984 | Pig | U70391 |
| I | B1065 | 1983 | Pig | U70388 | |
| I | P19Br | 1982 | Human | U70416 | |
| I | 2372 | 1979 | Human | U70401 | |
| III | Chiang Mai | 1964 | Human | U70393 | |
| Vietnam | III | VN118 | 1979 | Mosquito | U70420 |
Fully sequenced JEV strains are indicated in bold type.
NA, not available.
Results
The JEV infection rate for C. tritaeniorhynchus was 13.2% (57/430), A. sinensis was 2.7% (3/430), A. subalbatus was 0.7% (3/430), and for stillborn piglets was 18.5% (20/108). Studies have shown that pig infection rates are closely related to annual mosquito seasonal fluctuations (Table 3).
Table 3.
Mosquitos and Swine with Japanese Encephalitis Virus Infection in the Canton of Jing Hong in Yunnan Province, China, 2009–2010
| |
|
No. of mosquitoes and swine tested |
No. (%) of mosquitoes and swine testing positive |
||||
|---|---|---|---|---|---|---|---|
| Year | Month | Mosquitoes (50/tube) | Swine brain | C. tritaeniorhynchus | A. sinensis | A. subalbatus | Swine brain |
| 2009 | 8 | 75 | 15 | 9(12) | 3(4.0) | 0 | 1(6.7) |
| 9 | 66 | 20 | 12(18) | 4(6.0) | 2(3.0) | 5(25) | |
| 10 | 65 | 10 | 10(15) | 2(3.0) | 1(1.5) | 1(10) | |
| 2010 | 8 | 82 | 21 | 8(9.7) | 1(1.2) | 0 | 2(9.5) |
| 9 | 74 | 25 | 10(13.5) | 2(2.7) | 0 | 7(28) | |
| 10 | 68 | 17 | 8(11.7) | 0 | 0 | 4(24) | |
| Total | 430 | 108 | 57(13.2) | 12(2.7) | 3(0.7) | 20(18.5) | |
The maximum number of mosquitoes tested per pool was 50.
Other mosquito species tested included C. tritaeniorhynchus, A. sinensis, and A. subalbatus.
Brain tissue from stillborn piglets was collected randomly from several piggeries.
In the experimental infection of animals, suckling mice inoculated with isolates Yunnan 0901 and Yunnan 0902 showed 103.33 LD50/0.02 mL and 103.5 LD50/0.02 mL, respectively.
Nucleotide sequences of individual cDNA fragments and their junctions in the viral genome were obtained. These sequences were assembled and the genome of JEV isolates Yunnan 0901 and Yunnan 0902 was found to be 10,976 nucleotides in length, and to contain one ORF that coded for proteins of 3432 amino acids. The lengths of the 5′ NCR and the 3′ NCR were 96 and 565 nucleotides, respectively. Nucleotide sequences of individual cDNA fragments and their junctions on the viral genome were obtained. We compared the complete Yunnan 0901 and Yunnan 0902 genomic sequences with sequences of SA14-14-2 to characterize the molecular structure of the Yunnan 0901 and Yunnan 0902 genes, and to determine how they were related to other fully-sequenced genomes and E gene sequences of previously-reported JEV strains. The sequence comparison showed that Yunnan 0901 and Yunnan 0902 had several nucleotide substitutions scattered throughout the genome, except for at the 3′ NCR, where the sequence was totally conserved. A total of 36 and 68 nucleotide substitutions were found, respectively, which represented a 0.33% and 0.62% nucleotide difference. Also, 18 and 46 amino acid substitutions were found, which represented a 0.52% and 1.34% amino acid difference, respectively. The structural proteins had 4 and 10 amino acid substitutions, 14 and 36 of which of which were non-conservative changes (Tables 4 and 5).
Table 4.
Genome Sequence Analysis of the Yunnan 0901 Strain of Japanese Encephalitis Virus
| |
Size |
Nucleotide substitution |
Amino acid substitution |
||||
|---|---|---|---|---|---|---|---|
| Genome segment | Nucleotides | Amino acids | No. of substitutions | % substitutions | No. of substitutions | % substitutions | % NSRAAC |
| 5′-NCR | 95 | 0 | 1 | 1.05 | 0 | 0.0 | 0.0 |
| Capsid | 381 | 127 | 1 | 0.26 | 1 | 0.79 | 100 |
| Membrane | 501 | 167 | 1 | 0.20 | 0 | 0.0 | 0.0 |
| Envelope | 1500 | 500 | 4 | 0.27 | 3 | 0.60 | 75.0 |
| NS1 | 1245 | 415 | 3 | 0.24 | 2 | 0.48 | 66.7 |
| NS2A | 492 | 164 | 2 | 0.41 | 1 | 0.61 | 50.0 |
| NS2B | 393 | 131 | 3 | 0.76 | 2 | 1.52 | 66.7 |
| NS3 | 1857 | 619 | 1 | 0.05 | 0 | 0.0 | 0.0 |
| NS4A | 801 | 267 | 1 | 0.13 | 0 | 0.0 | 0.0 |
| NS4B | 411 | 137 | 3 | 0.73 | 1 | 0.73 | 33.3 |
| NS5 | 2735 | 911 | 16 | 0.59 | 9 | 0.99 | 56.2 |
| 3′-NCR | 565 | 0 | 0 | 0 | 0 | 0.0 | 0.0 |
| Complete | 10976 | 3432 | 36 | 0.33 | 18 | 0.52 | 50.0 |
The nucleotide sequence of the Yunnan 0901 genome was compared with that of the SA14-14-2 strain.
NCR, non-coding region; NS, non-structural; NSRAAC, nucleotide substitution resulting in an amino acid change.
Table 5.
Genome Sequence Analysis of the Yunnan 0902 Strain of Japanese Encephalitis Virus
| |
Size |
Nucleotide substitution |
Amino acid substitution |
||||
|---|---|---|---|---|---|---|---|
| Genome segment | Nucleotides | Amino acids | No. of substitutions | % substitutions | No. of substitutions | % substitutions | % NSRAAC |
| 5′-NCR | 95 | 0 | 1 | 1.05 | 0 | 0.0 | 0.0 |
| Capsid | 381 | 127 | 2 | 0.52 | 0 | 0.0 | 0.0 |
| Membrane | 501 | 167 | 3 | 0.59 | 1 | 0.60 | 33.3 |
| Envelope | 1500 | 500 | 10 | 0. 67 | 9 | 1.80 | 90.0 |
| NS1 | 1245 | 415 | 6 | 0.48 | 5 | 1.20 | 83.3 |
| NS2A | 492 | 164 | 2 | 0.41 | 1 | 0.61 | 50.0 |
| NS2B | 393 | 131 | 3 | 0.76 | 2 | 1.52 | 66.7 |
| NS3 | 1857 | 619 | 7 | 0.38 | 4 | 0.65 | 57.1 |
| NS4A | 801 | 267 | 2 | 0.25 | 1 | 0.37 | 50.0 |
| NS4B | 411 | 137 | 7 | 1.70 | 5 | 3.65 | 71.4 |
| NS5 | 2735 | 911 | 25 | 0.91 | 18 | 1.98 | 72.0 |
| 3′-NCR | 565 | 0 | 0 | 0 | 0 | 0.0 | 0.0 |
| Complete | 10976 | 3432 | 68 | 0.62 | 46 | 1.34 | 67.6 |
The nucleotide sequence of the Yunnan 0902 genome was compared with that of the SA14-14-2 strain.
NCR, non-coding region; NS, non-structural; NSRAAC, nucleotide substitution resulting in an amino acid change.
We used 95 representative JEV strains to perform multiple sequence alignments and phylogenetic analyses. This number included 24 strains with complete genomic sequences, including several Chinese strains (such as Beijng-1, P3, SA14 and its derivative SA14-14-2, HW, and SH0601), and 71 isolates with distinctive E gene sequences.
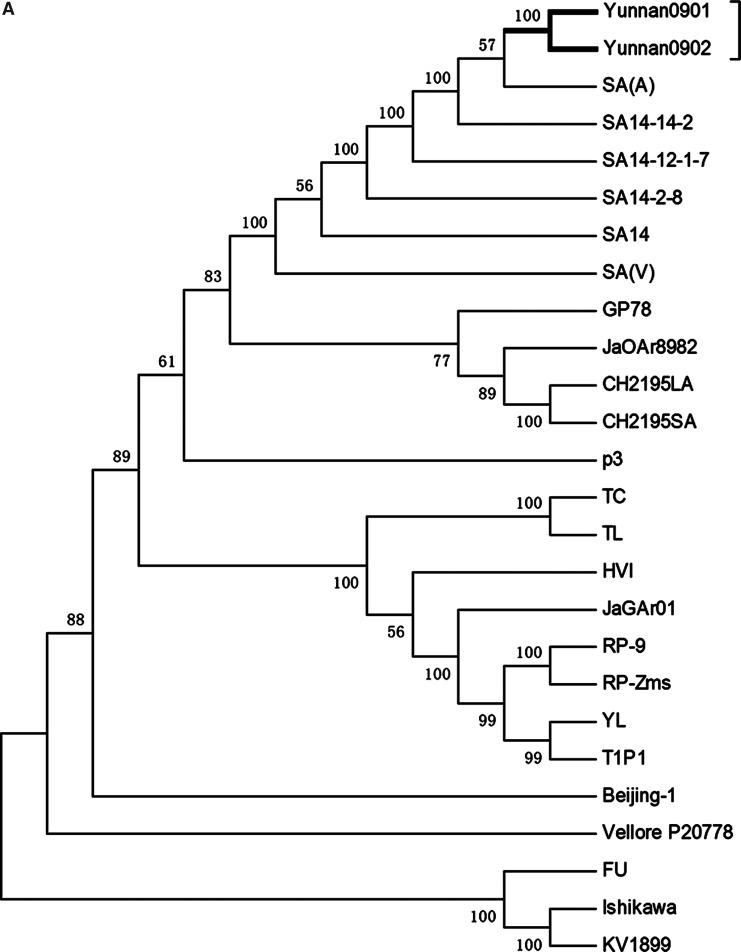
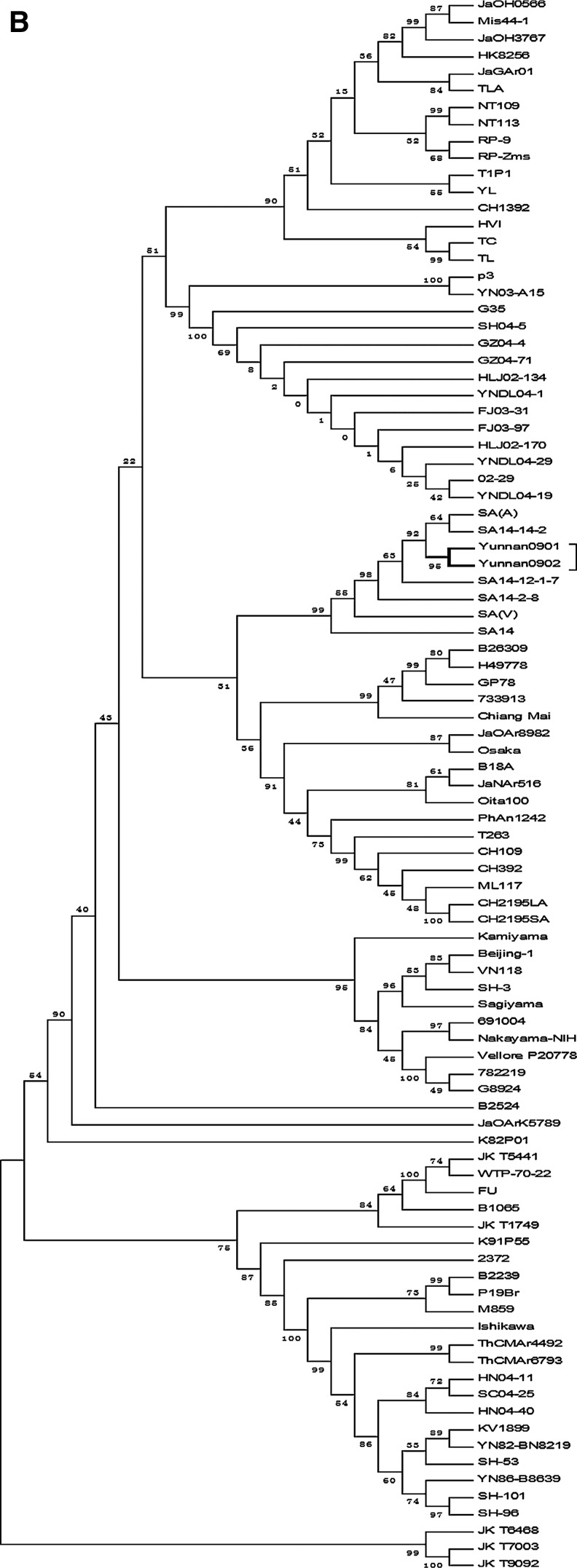
We compared complete sequence and nucleotide sequences of the E gene. The homology of the Yunnan 0901 and Yunnan 0902 isolates, compared with the genotype JEV III strain, was 97.2–99.6% and 94.5–99.7%, respectively, which was higher than that for the other genotypes (88.5–88.8% and 83.1–87.4%). At the deduced amino acid level, the homologies were 97.4–98.1% and 96.0–98.2%, respectively. The analyses showed that phylogenetic profiles based on the E genome are less similar to the basic phylogenetic profile of gene evolution (Fig. 2A and B). From these results, it was decided that the Yunnan 0901 and Yunnan 0902 isolates should be classified as genotype III. Our results indicated that genotype III was the major JEV subtype circulating in China.
FIG. 2.
The complete genome of JEV and the E gene for the aligned sequence and phylogenetic tree. (A) Multiple sequence alignment of the complete JEV genome was carried out with MEGA software, and (B) the phylogenetic tree was constructed by the neighbor-joining method using 1000 bootstrapping replicates.
Our results confirmed that high levels of nucleotide and amino acid sequence identity exist among JEV strains. It is possible that JEV strains isolated from the canton of Jing Hong in villages in Yunnan may be representative of the strains currently circulating in the poultry population in that region. The JQ086762 and JQ086763 strains differed in sequence from the previously isolated YN86-B8639 strains. This report contains the first description of a JEV complete genome sequence from the isolates from Jing Hong villages of Yunnan. These results suggest that one JEV genotype is currently circulating and undergoing evolution in Yunnan.
The close genetic relatedness of JEV strains isolated in 2009 and 2010 in the canton of Jing Hong villages in Yunnan suggests that these strains are endemic, and that mosquitoes and pigs are effective amplifying host populations in this province.
Furthermore, these results suggest that the JEV vaccine used in this province may not be effective in stopping virus shedding. This situation would allow the circulation of a virulent virus to go unnoticed in the vaccinated swine population until the development of an outbreak. Therefore it may be necessary to evaluate the effectiveness of the current vaccine used in the Yunnan province of China against circulating JEV strains.
Discussion
Japanese encephalitis viruses are RNA viruses that have a high potential for evolution due to their lack of repair mechanisms that would otherwise act during the replication of their genome (Holland et al. 1982). To our knowledge, only limited data are available on the selection pressures acting on JEV (Yang et al. 2000).
Understanding the epidemiological situation and genetic changes in the envelope gene is an important step in the study of JEV evolution. Extensive surveillance helps us to understand geographical movement of and genotype shift in JEV. This study is a localized example of JEV molecular evolution occurring in nature. Recent data have shown that many people in the Yunnan region of China are infected with JEV (Fang et al. 2010). This area is the first Yunnan province of the Jing Hong region in China from which the complete sequences of isolates of JEV from mosquito and swine (Yunnan 0901 and Yunnan 0902 strains) have become available, providing insights into the prevention of JEV infection in humans.
Experimental infections prove that suckling mice infected with the Yunnan 0901 and Yunnan 0902 strains had high morbidity, similarly to previously reported results (Yang et al. 2004b).
Complete genome sequences of other JEV isolates were obtained from the GenBank database (Table 2). Here we have carried out pair-wise alignment of the complete nucleotide and E gene sequences of these viruses to establish phylogenetic relatedness (Fig. 2A and B). The sequence comparison showed that the Yunnan 0901 and Yunnan 0902 strains had a number of nucleotide substitutions that were scattered throughout the genome, except in the 3′-NCR, which was totally conserved (Tables 4 and 5). Nucleotide substitution rates were 0.27% and 0.67% in the part of the genome that codes for the envelope protein (nucleotides 978–2477). This finding indicates that there is less variation between these two strains in the E gene coding region than in the rest of the genome. Similar variation was found when amino acid divergence, based on partial sequence analysis, was compared with that which had been calculated on the basis of the complete sequence of the polyprotein. However, we noted that even though there were differences in the extent of sequence variation as discussed above, this difference did not result in changes in genotype among the strains. Thus the E gene sequence can be used as the basis for genotyping, but not as a basis for monitoring gene evolution.
The 3′ NCR showed no changes in sequence, but the 5′ NCR nucleotide substitution rate was 1.05%. Both the 5′ and 3′ NCRs are involved in virus replication. In addition, the NS5 protein is an important non-structural protein that functions as the viral RNA replicase (Chen et al. 1997). Regions in the 5′ NCR are also involved in the control of viral RNA translation, which is necessary for producing viral proteins that are subsequently required for genome replication. This fact could explain why RNA viruses have a high potential for evolution. The nucleotide substitution rate was 0.59% and 0.91%, and amino acid substitution rate was 0.99% and 1.98%, for the Yunnan 0901 and Yunnan 0902 strains, respectively (Tables 3 and 4). This situation also explains JEV virus evolution and how it constantly adapts to environmental and host pressures.
Using criteria established by Chen and associates and others, phylogenetic analysis based on the complete genome and the E region demonstrated that the two newly-isolated JEV strains from Yunnan belonged to genotype III (Fig. 2A and B). The complete genomic analysis showed that they were closely related to SA-14. However, E gene region analysis indicated that they were closely related to both SA-14 and SA-14-14-2. The E protein of flaviviruses plays an important role in immunogenicity, tissue tropism, cell fusion, infectivity, and virus maturation (McMinn 1997; Yun et al. 2003). Sequencing of this gene, however, did not provide any insights into the biological characteristics of the Yunnan 0901 and Yunnan 0902 strains, or their genetic relationship with other JEV isolates.
One of the isolates was obtained from stillborn piglets from swine in the canton of Jing Hong villages in Yunnan Province. These pigs had been vaccinated using the live attenuated vaccine strain SA14-14-2 to provide immunity against JEV-related viruses, and with the secondary aim of reducing their circulation in vaccinated regions. Thus the Yunnan 0901 and Yunnan 0902 strains may have originated from pigs immunized with the SA-14-14-2 virus. The live vaccine virus was derived from the parental SA-14. It is unlikely that the SA-14 virus genome is so naturally stable that it is capable of such long-term survival. Thus we speculate that the virus has recently re-emerged from the natural environment.
We propose that phylogenetic analysis of the full JEV genome sequences for all gene sets should be employed in future studies to help determine the relationships among JEV strains. Development of a new vaccine that includes genotype III strains may be necessary. Since Yunnan is located on the border between China, Myanmar, and Vietnam, this epidemiological survey not only provides a basis for the prevalence of JEV in this region, but also for neighboring countries. Thus the data from this study may aid in the design of effective control strategies for each of these regions. Further investigation of the distribution and seasonality of JEV in China needs to be continued as this virus continues to evolve.
Acknowledgments
We wish to express our sincere gratitude to the Yunnan Institute of Parasitic Diseases for assistance in mosquito collection, and especially to Hong-Ning Zhou and Zhong-Hua Yang for their excellent technical assistance. This work was supported by a grant from the Special Fund for Agro-Scientific Research in the Public Interest (no. 201203082).
Author Disclosure Statement
No competing financial interests exist.
References
- Ali A. Igarashi A. Paneru LR, et al. Characterization of two Japanese encephalitis virus strains isolated in Thailand. Arch Virol. 1995;140:1557–1575. doi: 10.1007/BF01322530. [DOI] [PubMed] [Google Scholar]
- Burke DS. Monath TP. Fields Virology. 4th. Philadelphia: Lippincott; 2001. Flavivirus; pp. 991–1024. [Google Scholar]
- Chen C-J. Kuo M-D. Chien L-J, et al. RNA-protein interactions: involvement of NS3, NS5,and 39 noncoding region of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WR. Tesh RB. Rico-Hesse R. Genetic variation of Japanese encephalitis virus in nature. J Gen Virol. 1990;71:2915–2922. doi: 10.1099/0022-1317-71-12-2915. [DOI] [PubMed] [Google Scholar]
- Endy TP. Nisalak A. Japanese encephalitis virus: ecology and epidemiology. Curr Top Microbiol Immunol. 2002;267:11–48. doi: 10.1007/978-3-642-59403-8_2. [DOI] [PubMed] [Google Scholar]
- Fang GD. Fang GX. Ning ZH. Investigation of the prevalence of Japanese encephalitis in Jingdong County, Yunnan. J Pathogen Biol. 2010;5:57–59. [in Chinese]. [Google Scholar]
- Gubler DJ. Kuno G. Markoff L. Flaviviruses. In: Knipe DM, editor; Howley PM, editor. Fields Virology. 4th. Philadelphia: Lippincott & Wilkins; 2007. pp. 1153–1252. [Google Scholar]
- Hashimoto H. Nomoto A. Watanabe K, et al. Molecular cloning and complete nucleotide sequence of the genome of Japanese encephalitis virus Beijing-1 strain. Virus Genes. 1988;1:305–317. doi: 10.1007/BF00572709. [DOI] [PubMed] [Google Scholar]
- Holland J. Spindler K. Horodyski F, et al. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S. Samal SK. Nucleotide sequences of the trailer, nucleocapsid protein gene and intergenic regions of Newcastle disease virus strain Beaudette C and completion of the entire genomesequence. J Gen Virol. 1998;79:2419–2424. doi: 10.1099/0022-1317-79-10-2419. [DOI] [PubMed] [Google Scholar]
- Kumar S. Nayak B. Collins PL, et al. Complete genome sequence of avian paramyxovirus type 3 reveals an unusually long trailer region. Virus Res. 2008;137:189–197. doi: 10.1016/j.virusres.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Tamura K. Nei M. Mega 3: Integrated software for molecular evolutionary. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- McMinn PC. The molecular basis of virulence of the encephalitogenic flaviviruses. J Gen Virol. 1997;78:2711–2722. doi: 10.1099/0022-1317-78-11-2711. [DOI] [PubMed] [Google Scholar]
- Nam JH. Chae SL. Won SY, et al. Genetic heterogeneity of Japanese encephalitis virus assessed via analysis of the full-length genome sequence of Korean isolate. Am J Trop Med Hyg. 2001;65:388–392. doi: 10.4269/ajtmh.2001.65.388. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi H. Mori C. Fuke I, et al. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987;161:497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- Uchil PD. Satchidanandam V. Phylogenetic analysis of Japanese encephalitis virus: envelope gene based analysis reveals a fifth genotype, geographic clustering, and multiple introductions of the virus into the Indian subcontinent. Am J Trop Med Hyg. 2001;65:242–251. doi: 10.4269/ajtmh.2001.65.242. [DOI] [PubMed] [Google Scholar]
- Vaughn DW. Hoke CH. The epidemiology of Japanese encephalitis: prospects for prevention. Am J Epidemiol. 1992;14:197–221. doi: 10.1093/oxfordjournals.epirev.a036087. [DOI] [PubMed] [Google Scholar]
- Wang HY. Takasaki T. Fu SH, et al. Molecular epidemiological analysis of Japanese encephalitis virus in China. J Gen Virol. 2007;88:885–894. doi: 10.1099/vir.0.82185-0. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Japanese encephalitis vaccines. Wkly Epidemiol Rec. 1998;73:334–344. [Google Scholar]
- Yang DK. Kim BH. Kweon CH, et al. Biophysical characterization of Japanese encephalitis virus (KV1899) isolated from pigs in Korea. J Vet Sci. 2004a;5:125–130. [PubMed] [Google Scholar]
- Yang DK. Kim BH. Kweon CH, et al. Molecular characterization of full-length genome of Japanese encephalitis virus (KV1899) is isolated from pigs in Korea. J Vet Sci. 2004b;5:197–205. [PubMed] [Google Scholar]
- Yang Z. Nielsen R. Goldman N, et al. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z. Liu JH. Animal Virology. 2nd. Marrickville, Australia: Science Press; 1997. pp. 329–331. 633–641. [Google Scholar]
- Yun SI. Kim SY. Choi WY, et al. Molecular characterization of the full-length genome of the Japanese encephalitis viral strain K87P39. Virus Res. 2003;96:129–140. doi: 10.1016/s0168-1702(03)00181-3. [DOI] [PubMed] [Google Scholar]
- Zhang J-S. Zhao Q-M. Zhang P-H, et al. Genomic sequence of a Japanese encephalitis virus isolate from southern China. Arch Virol. 2009;154:1177–1180. doi: 10.1007/s00705-009-0421-x. [DOI] [PubMed] [Google Scholar]