Abstract
c-Myc is a transcriptional factor that functions as a central regulator of cell growth, proliferation, and apoptosis. Overexpression of c-Myc also enhances DNA double-strand breaks (DSBs), genetic instability, and tumorigenesis. However, the mechanism(s) involved remains elusive. Here, we discovered that γ-ray ionizing radiation-induced DSBs promote c-Myc to form foci and to co-localize with γ-H2AX. Conditional expression of c-Myc in HO15.19 c-Myc null cells using the Tet-Off/Tet-On inducible system results in down-regulation of Ku DNA binding and suppressed activities of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and DNA end-joining, leading to inhibition of DSB repair and enhanced chromosomal and chromatid breaks. Expression of c-Myc reduces both signal and coding joins with decreased fidelity during V(D)J recombination. Mechanistically, c-Myc directly interacts with Ku70 protein through its Myc box II (MBII) domain. Removal of the MBII domain from c-Myc abrogates its inhibitory effects on Ku DNA binding, DNA-PKcs, and DNA end-joining activities, which results in loss of c-Myc's ability to block DSB repair and V(D)J recombination. Interestingly, c-Myc directly disrupts the Ku/DNA-PKcs complex in vitro and in vivo. Thus, c-Myc suppression of DSB repair and V(D)J recombination may occur through inhibition of the nonhomologous end-joining pathway, which provides insight into the mechanism of c-Myc in the development of tumors through promotion of genomic instability.
Introduction
DNA double-strand breaks (DSBs) result not only from by-products of endogenous metabolism, such as oxidative metabolism, DNA replication, or V(D)J recombination for immune system development, but also from exogenous sources, including ionizing radiation (IR), chemical agents, UV light, and cancer chemotherapeutics. DSBs are extremely dangerous, toxic lesions with severe consequences for cellular survival and genomic integrity [1]. The failure to repair or inappropriate repair of DSBs can cause cell death or large-scale chromosomal changes including deletions, translocations, and chromosome fusions that may lead to neoplastic transformation and ultimately cancer development [2]. In mammalian cells, DSBs are repaired mainly by two kinds of mechanisms: nonhomologous end-joining (NHEJ) and homologous recombination (HR), and deficiencies in either pathway can lead to genomic instability and promote tumorigenesis [3,4].
DSBs trigger a series of signaling cascades that lead to the recruitment of DNA repair proteins to the sites of genomic damage and formation of repair complex foci at the site of the break. One of the earliest events in the DNA damage response is the phosphorylation of histone H2A variant at the serine 139 position leading to the generation of γ-H2AX, a key regulator of IR-induced nuclear foci formation [5–7]. Ataxia telangiectasia mutated and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) play essential but partially redundant roles in phosphorylating H2AX in response to DSBs [8,9]. The phosphorylated H2AX functions as a scaffold for the accumulation and retention of the central components of the signaling cascade initiated by DNA damage [1]. DNA ligase IV, along with its binding partners, XRCC4 and XLF (also called Cernunnos) [10], seals the break in the final step of the DSB repair. The nuclease Artemis helps repair a subset of IR-induced DSBs by NHEJ and is important for opening hairpins.
It is known that DSBs induced by exogenous agents are potentially harmful lesions for cells. However, physiological DSBs are also generated by recombination activating genes (RAG1/2) during V(D)J recombination [11], which are essential for immune system development. This reaction produces recombination signal (RS) ends containing blunt 5′ phosphorylated DSBs and coding ends in the form of covalently sealed hairpins [12]. Importantly, joining of RS and coding ends during V(D)J recombination also requires the NHEJ DNA repair pathway [13,14]. Because V(D)J recombination is a physiological process in B and t cell development [11], suppression of the NHEJ DNA repair pathway not only negatively affects general DSB repair but may also inhibit V(D)J recombination and subsequently impair immune system development.
c-Myc is a transcription factor and functions as a central signaling switch for a variety of cellular processes including cellular growth and proliferation, differentiation, angiogenesis, cell adhesion, and apoptosis [15]. c-Myc has been implicated as an important factor in the onset or progression of many types of cancer and is frequently overexpressed in up to 70% of all human malignancies [16,17]. c-Myc dysregulation directly promotes genomic instability and tumorigenesis [15,18–21]. Additionally, c-Myc can induce DNA breakage, alter DNA break repair [22,23], and cause chromosomal breaks and translocations in normal human cells [24]. Such Myc-mediated activities are undoubtedly central to its role in tumorigenesis. However, the mechanisms by which c-Myc dysregulation leads to DNA breakage and genomic instability are poorly understood. It was previously reported that expression of c-Myc upregulates reactive oxygen species (ROS) leading to increased DNA damage [25]. In contrast, a more recent study demonstrated that c-Myc activation in vivo or in vitro under a physiologically relevant oxygen environment does not induce ROS production but can still result in the formation of DNA breaks [26]. These results support the concept that c-Myc overexpression can induce widespread DNA damage independent of ROS. Although a link between c-Myc dysregulation and DSBs has been known for many years, the direct mechanism(s) by which c-Myc enhances DSBs remains elusive. Because the NHEJ factors (i.e., Ku70 and Ku86) have recently been identified as c-Myc-associated proteins by a combined TAP/MudPIT approach [27], this provides possibility that c-Myc could directly regulate the repair of DSBs. Here, we provide direct evidence demonstrating that c-Myc inhibits DSB repair and V(D)J recombination through direct suppression of the NHEJ pathway. These findings have important implications for understanding the relationship between pleiotropic oncogenes, DNA repair pathways, and tumorigenesis.
Materials and Methods
Materials
Ku70, Ku86, DNA-PKcs, and c-Myc antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Purified recombinant c-Myc protein was purchased from Active Motif (Carlsbad, CA). Purified recombinant Ku70 protein was obtained from GenWay Biotech, Inc (San Diego, CA). The γ-H2AX antibody was purchased from Upstate Biotech (Charlotteville, VA). Ku70/Ku86 DNA Repair Kit was obtained from Active Motif. The SignaTECT DNA-PK Assay Kit was purchased from Promega (Madison, WI). c-Myc small interfering RNA (siRNA) and control siRNA were purchased from Ambion (Austin, TX). DNase I was obtained from Sigma-Aldrich (St Louis, MO). The pJH290, pJH200, RAG1, and RAG2 constructs as well as Escherichia coli MC1061 were obtained from Dr Frederick W. Alt (Harvard Medical School). HO15.19 c-Myc null cells were provided by Dr John M. Sedivy (Brown University). Other reagents used were obtained from commercial sources unless otherwise stated.
Generation of Various c-Myc Deletion Mutants
A series of c-Myc deletion mutants, including ΔMBI, ΔMBII, and ΔHLH, were created using Transformer Site-Directed Mutagenesis Kit (Clontech, Mountain View, CA) according to the manufacturer's instructions. The 5′-phosphorylated mutagenic primers for various precise deletion mutants were synthesized as follows: ΔMBI [amino acids (aa) 45–63], 5′-GCA GAG CGA GCT GCA GCC CCC GGC GAG CCG CCG CTC CGG GCT-3′; ΔMBII (aa 129–143), 5′-GAC CCG GAC GAC GAG ACC TTC ATC AAA AAC ATC CTC GTC TCA GAG AAG CTG GCC TCC TAC CAG GCT G-3′; ΔHLH (aa 368–410), 5′-GTC TTG GAG CGC CAG AGG AGG CAA AAG CTC ATT TCT G-3′. The wild-type (WT) c-Myc/pUC19 construct was used as the target plasmid, which contains a unique NdeI restriction site for selection against the unmutated plasmid. The NdeI selection primer is 5′-GAG TGC ACC ATGGGCGGT GTG AAA-3′. Each single mutant was confirmed by sequencing of the cDNA. WT and each of the c-Myc deletion mutants were then cloned into the response plasmid pTRE2hyg (Clontech) or the pcDNA3.1 (Invitrogen, Grand Island, NY) mammalian expression vector, respectively.
Cell Lines, Plasmids, and Conditional Expression of c-Myc
H460, H157, and H1299 cells were maintained in RPMI 1640 medium with 10% FBS. HO15.19 c-Myc null cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% calf serum as described [28]. For conditional expression of c-Myc, the Tet-On inducible gene expression system (Clontech) was established in HO15.19 cells according to the manufacturer's instructions. First, pTet-On plasmids containing E. coli lacZ under control of the Tet response element (TRE) were transfected into HO15.19 cells. Stable clones were selected by G418 (0.8 mg/ml). Second, WT c-Myc cDNA was cloned into the pTRE2hyg vector between the BamHI and EcoRV restriction sites. Finally, the stable Tet-On cells were transfected with the response plasmid pTRE2hyg containing c-Myc and selected using hygromycin. c-Myc expression under control of the TRE in double-stable cells was turned on by the addition of doxycycline (DOX, 1 µg/ml).
Immunofluorescence
The cells were washed with 1x phosphate-buffered saline (PBS), fixed with cold methanol and acetone (1:1) for 5 minutes, and then blocked with 10% normal mouse or rabbit serum for 60 minutes at room temperature. Cells were incubated with a rabbit (i.e., c-Myc) or mouse (i.e., γ-H2AX) primary antibody for 90 minutes. After washing, samples were incubated with Alexa 488 (green)-conjugated anti-rabbit or Alexa 594 (red)-conjugated anti-mouse secondary antibodies or 4′,6-diamidino-2-phenylindole (DAPI) for 60 minutes. Cells were washed with 1x PBS and observed under a fluorescent microscope (Zeiss). Pictures were taken and colored with the same exposure setting for each experiment. Individual green- and red-stained images derived from the same field were merged using Openlab 3.1.5 software from Improvision, Inc (Lexington, MA).
Measurement of Ku70/86 Activity
Ku70/Ku86 DNA binding activity was analyzed using a Ku70/86 DNA Repair Kit (Active Motif). Briefly, cells were washed and resuspended in hypotonic buffer. The nuclear extract was isolated for Ku activity analysis. Five micrograms of nuclear protein was loaded into the oligonucleotide-coated 96-well plate and incubated for 60 minutes at room temperature. Ku70 or Ku86 antibody was added and incubated for another 60 minutes. After washing, HRP-conjugated secondary antibody was added and incubated for 60 minutes. After washing, developing solution and stop solution were added. Optical density was read on a spectrophotometer at 450 nm. Each experiment was repeated three times and data represent the mean µ SD of three separate determinations.
Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis (PFGE) was performed as described previously [29]. Briefly, cells were harvested, resuspended in ice-cold buffer L [0.1 M Na2-EDTA, 0.01 M Tris, 0.02 M Na Cl (pH 8.0)] at a concentration of 5 x 106 cells/ml, and mixed with an equal volume of 1% low melting point agarose (Beckman) at 42°C. The mixture was pipetted into a small length of Tygon tubing, clamped tight at both ends, and chilled to 0°C. The solidified agarose “snake” was extruded from the tubing, added to 10x volume of buffer L containing 1 mg/ml proteinase K and 1% sarkosyl, and incubated for 16 hours at 50°C. Following lysis, the agarose snake was washed four times with TE buffer and then cut into 0.5-cm plugs. The plugs were inserted into the wells of a pre-cooled 1% low melting point agarose gel (4°C). PFGE (200-s pulse time, 150 V, 15 hours at 14°C) was performed using Clamped Homogenous Electric Fields Mapper (Bio-Rad, Hercules, CA). After electrophoresis, the gel was stained with ethidium bromide for photography.
Telomere-Fluorescence In Situ Hybridization Analysis
Telomere-Fluorescence In Situ Hybridization (T-FISH) was performed using Telomere peptide nucleic acid (PNA) FISH Kit/Cy3 (DakoCytomation, Copenhagen, Denmark) as described [30]. Cells were incubated with colcemid (KaryoMAX; Gibco, Grand Island, NY) at 100 ng/ml for 1.5 hours and then harvested by trypsinization. Cells were swollen in pre-warmed 30 mM sodium citrate for 30 minutes at 37°C, fixed in methanol/acetic acid (3:1), and air dried on slides overnight. After pepsin digestion, slides were denatured at 80°C for 5 minutes, hybridized with Cy3-labeled PNA telomeric probe (Cy3-[TTAGGG]3) in 70% formamide at room temperature for 3 to 4 hours, washed, dehydrated, and mounted in Vectashield with DAPI (Vector Laboratories, Burlingame, CA). Metaphase images were captured using a fluorescent microscope (Zeiss) and x63 objective lens. At least 30 metaphases of each cell line were scored for chromosomal aberrations.
Transient V(D)J Recombination Assay
Recombination substrates pJH200 and pJH290 were used in the transient V(D)J recombination assay to test for RS and coding joins as described [31]. Briefly, pJH200 or pJH290 was co-transfected into cells with the full-length RAG1 and RAG2 expression constructs under the control of the Rous sarcoma virus long terminal repeat promoter using Lipofectamine 2000. Extrachromosomal DNA was recovered after 48 hours by alkaline lysis as previously described [32] and electroporated into E. coli MC1061. The V(D)J recombination activity was measured by comparing the ratio of ampicillin resistant (Ampr) and chloramphenicol-resistant (Camr) colonies versus Ampr colonies in cells. Because a precise signal join generates an ApaLI site, ApaLI digestion of polymerase chain reaction products from the recombinant substrate pJH 200 was used to measure the fidelity of the RS joins as described [31]. The coding join sequence was determined by comparison of the nucleotide sequence isolated from individual recombined pJH290 clones.
Results
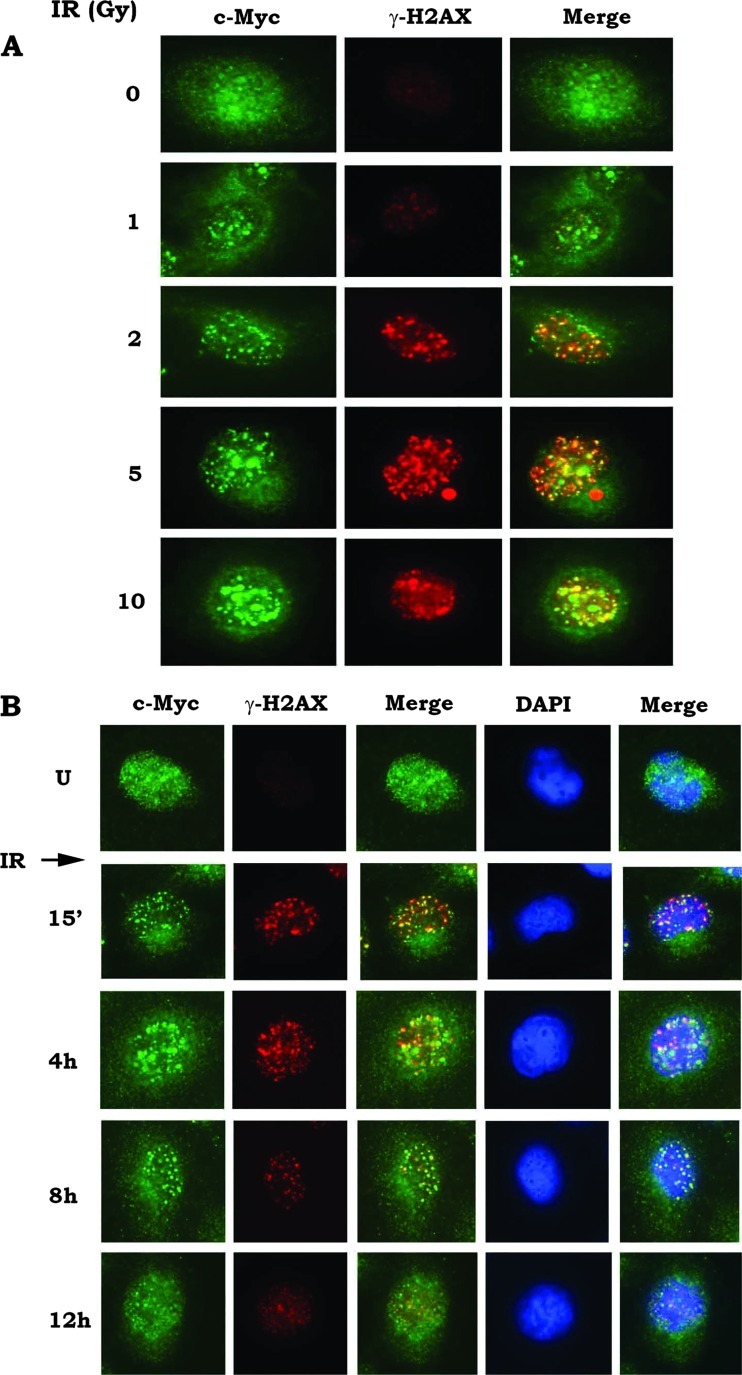
IR Induces Formation of c-Myc Foci and DSBs
It has been reported that c-Myc overexpression is associated with abnormalities in chromosomal number, chromosomal breaks, and translocations [33]. It is possible that c-Myc-induced genetic instability may result from increased DNA damage and/or reduced DNA repair. Formation of γ-H2AX foci is considered to be a sensitive and selective signal for the presence of DSBs and an early event following DSB induction [34,35]. Here, we found that, in addition to γ-H2AX foci, IR can also induce formation of c-Myc foci in a dose-dependent manner in H1299 cells (Figure 1A). Both c-Myc and γ-H2AX foci are gradually reduced over a 12-hour time period following IR exposure (Figure 1B). These findings suggest that IR-induced DNA damage signals may facilitate the recruitment of c-Myc molecules to the vicinity of broken ends of double-strand DNA. Importantly, majority of IR-induced c-Myc foci are co-localized with γ-H2AX because the strong yellow foci could be observed in the merged images (Figures 1 and W1), indicating that c-Myc may play a role in regulating the repair of IR-induced DSBs. Similar results were observed in H460 and H157 cells after IR exposure (Figure W1A), suggesting that the IR-induced formation of c-Myc foci is not confined to a specific cell type. Importantly, after silence of c-Myc using c-Myc siRNA, only γ-H2AX (red) without green signal of c-Myc could be observed following DNA damage (Figure W1B), indicating that the antibody used is truly staining c-Myc.
Figure 1.
IR induces formation of c-Myc foci and DSBs. (A) H1299 cells were treated with increasing doses of IR (0–10 Gy). After radiation, c-Myc and γ-H2AX were analyzed immediately by immunofluorescent staining. (B) H1299 cells were treated with IR (5 Gy). Cells were harvested at various time points as indicated. c-Myc and γ-H2AX were analyzed by immunofluorescent staining.
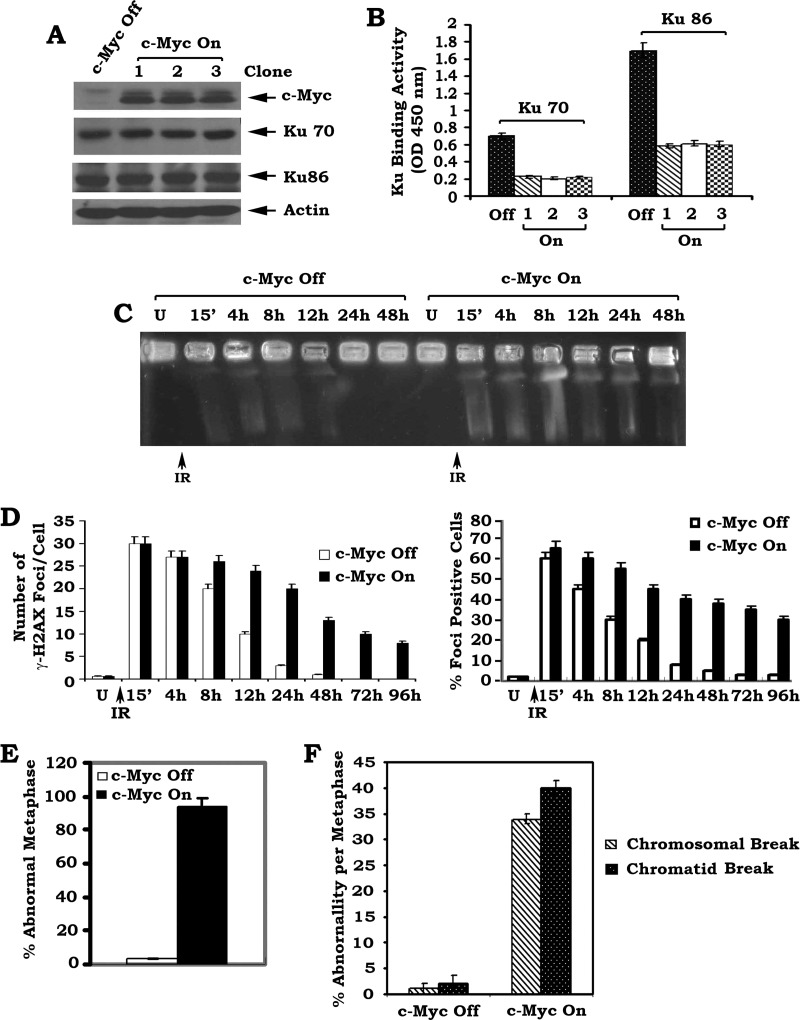
Conditional Expression of c-Myc Results in Suppression of DSB Repair and Increased Genetic Instability in Association with Decreased Ku, DNA-PKcs, and DNA End-joining Activities
DSBs can be repaired by two pathways: HR and NHEJ [36,37].However, NHEJ is the major pathway in mammalian cells for repairing DSBs [36,38]. Ku70, Ku86, and DNA-PKcs are the major components of the NHEJ machinery required for DSB repair [36–38]. We observed that c-Myc molecules may be recruited to the broken ends of double-strand DNA following DNA damage (Figure 1). To further address the role of c-Myc in DSB repair, c-Myc was conditionally expressed in the HO15.19 c-Myc null cells using the Tet-Off/Tet-On inducible gene expression system. In this system, c-Myc expression can only be turned on by the addition of DOX (Figure 2A). DOX itself has no effect on expression of c-Myc and Ku in HO15.19 parental cells without the Tet-Off/Tet-On inducible gene expression system (Figure W2). This helps to rule out potential effect of DOX treatment. Intriguingly, conditional expression of c-Myc significantly inhibits the DNA binding activity of both Ku70 and Ku86 by more than 50% compared to the c-Myc-Off control (Figure 2, A and B). Similar results were obtained from three independent clones expressing similar levels of exogenous c-Myc, indicating that these findings are reliable. Electrophoretic mobility shift assays (EMSAs) were employed to assess the extent of Ku DNA binding and provided further confirmation that Ku DNA binding is dramatically decreased in the c-Myc-On cells compared to c-Myc-Off cells (Figure W3A). Collectively, these results suggest that conditional overexpression of c-Myc may interfere with the binding of Ku to dsDNA broken ends.
Figure 2.
Conditional expression of c-Myc downregulates Ku DNA binding activity and suppresses DSB repair leading to increased genetic instability. (A) HO15.19 cells bearing inducible Tet-Off/Tet-On c-Myc gene were established as described in Materials and Methods section. c-Myc was turned on in three independent stable clones by the addition of DOX (1 µg/ml) for 24 hours. Expression levels of c-Myc, Ku70, and Ku86 were analyzed by Western blot. (B) DNA binding activity of Ku70 or Ku86 in c-Myc-Off or c-Myc-On cells was measured using a Ku70/86 DNA Repair Kit. Error bars represent ± SD. (C and D) The c-Myc-Off and c-Myc-On HO15.19 cells were exposed to 5 Gy of IR. Cells were harvested at various time points. DSBs were determined by PFGE or analysis of γ-H2AX by immunostaining. The number of γ-H2AX foci per cell was determined on a cell-to-cell basis by the quantitative analysis of at least 30 randomly chosen cells as described [58]. The percentage of γ-H2AX foci-positive cells was determined by analyzing 100 randomly chosen cells as described [34]. Error bars represent ± SD. (E and F) c-Myc was turned on by addition of DOX for 4 weeks. Then, percentage of abnormal metaphases in the c-Myc-Off or c-Myc-On cells was quantified by T-FISH analysis. At least 30 metaphases per culture were analyzed. Error bars represent ± SD. Frequency of cytogenetic abnormality per metaphase in c-Myc-Off or c-Myc-On cells. Each experiment was repeated three times and error bars represent ± SD.
To test the effect of c-Myc on DNA-PKcs activity in HO15.19 cells, a substrate phosphorylation assay was employed and revealed that conditional expression of c-Myc also results in down-regulation of DNA-PKcs activity (Figure W3B). In addition, c-Myc also inhibits DNA end-joining activity (Figure W3C). Because NHEJ is thought to be the major DSB repair pathway in mammalian cells [39–41], c-Myc-mediated inhibition of Ku, DNA-PKcs, and DNA end-joining activities may lead to suppression of DSB repair. To test this possibility, PFGE of genomic DNA under neutral conditions was employed to directly measure the level of DSBs [29]. Following IR exposure, a significant amount of genomic DNA from all cells migrated into the gel, indicating DNA fragmentation (i.e., DSBs). In c-Myc-Off cells, DSB repair was nearly completed in 24 hours following IR exposure as evidenced by little or no shortened DNA fragments migrating into the gel (Figure 2C). In contrast, DNA fragmentation (presence of DSBs) could still be observed 48 hours following IR exposure in the c-Myc-On cells (Figure 2C). DSBs were also evaluated by analysis of γ-H2AX foci [7]. IR is a potent inducer for formation of γ-H2AX foci in both c-Myc-Off and c-Myc-On cells (Figure W3D). The intensity of foci staining, the number of γ-H2AX foci per cell, as well as the overall percentage of foci-positive cells are significantly reduced by 12 to 24 hours post-IR exposure in c-Myc-Off cells. An increased number of γ-H2AX foci with greater staining intensities per cell and a higher percentage of foci-positive cells were observed in c-Myc-On cells compared to c-Myc-Off cells 12 to 24 hours post-IR exposure (Figures 2D and W3D). Western blot analysis of γ-H2AX levels confirmed that expression of c-Myc significantly prolongs the presence of γ-H2AX in cells following IR exposure (Figure W3E). Collectively, these findings directly indicate that c-Myc inhibits the repair of IR-induced DSBs through suppression of the NHEJ pathway.
c-Myc can cause chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus [24]. To directly test the effect of c-Myc on chromosomal aberrations, a FISH assay that combines DAPI staining with a telomere-specific PNA probe (T-FISH) was employed to evaluate metaphase chromosomes for instability [30]. Increased levels of cytogenetic abnormalities, including chromosomal and chromatid breaks, were observed in c-Myc-On cells compared to c-Myc-Off control cells (Figures 2, E and F, and W4). These findings demonstrate that c-Myc suppression of DSB repair may lead to increased genetic instability.
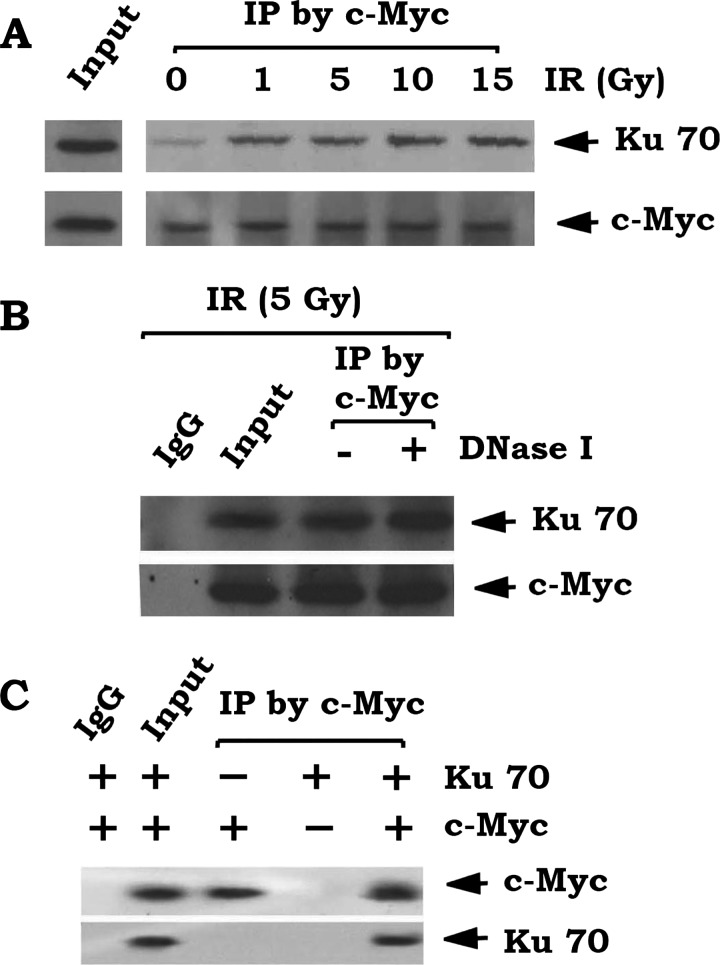
c-Myc Directly Interacts with Ku
To test the effect of IR-induced DSBs on c-Myc and Ku interaction, a co-immunoprecipitation (co-IP) experiment using a c-Myc antibody was performed following exposure of H1299 cells with increasing doses of IR. IR-induced DSBs promote c-Myc to directly associate with Ku70 (Figure 3A). To rule out the possible involvement of DNA in this interaction, cell lysates were first treated with 150 KU DNase I for 5 minutes. A co-IP was performed with c-Myc antibody as described [42]. Results reveal that c-Myc still interacts with Ku70 after digestion of DNA (Figure 3B). Additionally, a direct interaction between c-Myc and Ku was also confirmed by co-IP using purified recombinant c-Myc and Ku70 proteins in vitro (Figure 3C).
Figure 3.
c-Myc interacts with Ku70. (A) H1299 cells were treated with increasing doses of IR (0–15 Gy). co-IP was carried out using c-Myc antibody. Ku70 and c-Myc were analyzed by Western blot. (B) H1299 cells were treated with IR (5 Gy). co-IP was performed with anti-Myc antibody or normal IgG in the presence or absence of 150 KU of DNase I. Ku70 and c-Myc were analyzed by Western blot. (C) Purified recombinant c-Myc was incubated with purified Ku70 in lysis buffer at 4°C for 2 hours. A co-IP was performed using a c-Myc antibody or normal IgG control. Ku70 and c-Myc were analyzed by Western blot.
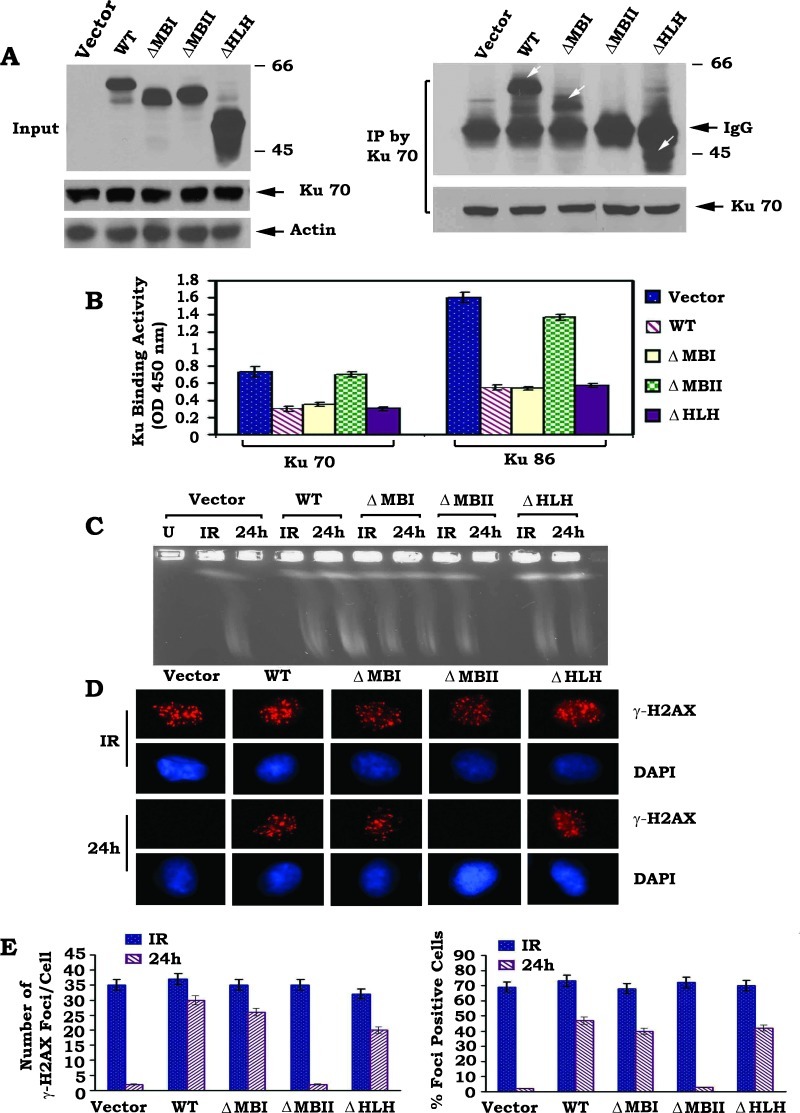
c-Myc Interacts with Ku70 through Its MBII Domain that Is Required for c-Myc Suppression of NHEJ-mediated DSB Repair
The c-Myc protein comprised several functional domains. The N-terminal structure contains the highly conserved Myc box I (MBI, aa 45–63) and Myc box II (MBII, aa 129–143) domains that determine the transcription-regulatory activity of c-Myc [43]. The C-terminal structure is composed of helix-loop-helix (HLH, aa 368–410) and leucine zipper (aa 410–439) regions, which are essential for Myc-Max heterodimerization [43,44]. To identify the Ku binding site in c-Myc protein, a series of c-Myc deletion mutants, including ΔMBI (aa 45–63), ΔMBII (aa 129–143), and ΔHLH (aa 368–410), were generated (Materials and Methods section). WT c-Myc or each of the c-Myc deletion mutants were expressed in HO15.19 c-Myc null cells (Figure 4A). Interestingly, expression of WT c-Myc or any of the c-Myc deletion mutants does not affect the expression level of Ku70 (Figure 4A, left panel). This result is consistent with the recent report that c-Myc knockdown inhibits Rad51 but does not affect Ku70 expression [45]. A co-IP experiment revealed that Ku70 is able to associate with WT c-Myc, ΔMBI, and ΔHLH c-Myc mutants but not with the ΔMBII c-Myc mutants (Figure 4A, right panel), indicating that the MBII domain is the Ku binding site on c-Myc.
Figure 4.
Ku70 directly interacts with c-Myc through its MBII domain, which is essential for c-Myc suppression of DSB repair. (A) WT or each of the c-Myc deletion mutants was transfected into HO15.19 cells. Expression levels of c-Myc were analyzed by Western Blot. Additionally, a co-IP was performed using Ku70 antibody. The Ku-associated c-Myc was analyzed by Western blot. White arrow(s) points c-Myc or c-Myc deletion mutant protein band(s). (B) DNA binding activity of Ku70 or Ku86 in HO15.19 cells expressing WT or each of the c-Myc deletion mutants was measured using a Ku70/86 DNA Repair Kit. Error bars represent ± SD. (C–E) HO15.19 cells expressing WT or each of the c-Myc deletion mutants were exposed to 5 Gy of IR. After 24 hours, DSBs were measured by PFGE or analysis of γ-H2AX by immunostaining with quantitative evaluation as described in the legend of Figure 2.
To test the functional effects of c-Myc/Ku binding on NHEJ and DSB repair, Ku DNA binding, DNA-PKcs, and DNA end-joining activities, as well as DSB repair, were compared in HO15.19 cells expressing similar levels of WT c-Myc or each of the c-Myc deletion mutants. Expression of WT c-Myc, ΔMBI, and ΔHLH c-Myc mutants but not the ΔMBII c-Myc mutant significantly suppresses Ku DNA binding, DNA-PKcs, and DNA end-joining activities (Figures 4B and W5, A and B). Functionally, deletion of the MBII domain from c-Myc results in failure of c-Myc to inhibit the repair of IR-induced DSBs, as determined by PFGE or analysis of γ-H2AX by immunostaining (Figure 4, C–E). In contrast, removal of the MBI or HLH domain does not affect the ability of c-Myc to inhibit DSB repair. This supports the notion that the binding of c-Myc to Ku through the MBII domain may be required for the inhibitory effects of c-Myc on Ku DNA binding, DNA-PKcs, and DNA end-joining activities resulting in inhibition of DSB repair.
A T-FISH assay revealed that more than 80% of cells were engaged in abnormal metaphase with increased chromosomal and chromatid breaks observed in HO15.19 cells expressing WT c-Myc, ΔMBI, or ΔHLH c-Myc mutants (Figure W5, C and D). Intriguingly, deletion of MBII from c-Myc results in loss of c-Myc's effect on genetic instability, as demonstrated by the significant reduction in chromosomal and chromatid breaks in HO15.19 cells expressing ΔMBII c-Myc mutant compared to WT c-Myc (Figure W5, C and D). These findings indicate that the MBII domain may also be required for c-Myc-mediated genomic instability.
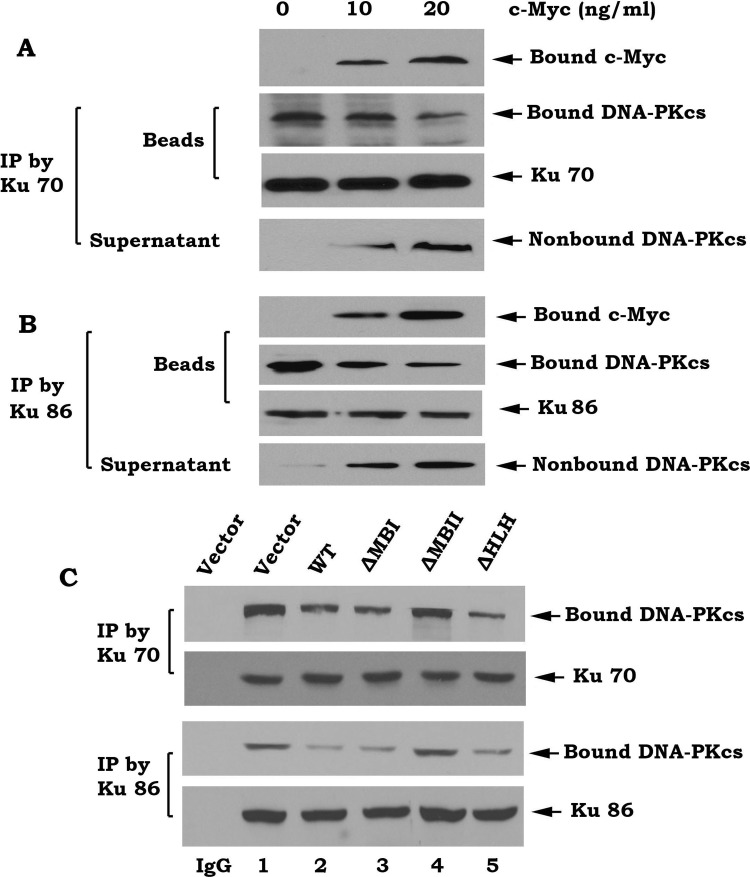
c-Myc Directly Disrupts the Ku/DNA-PKcs Complex
DNA-PK is composed of the Ku70/86 heterodimer and the DNA-PK catalytic subunit (DNA-PKcs) that exists as a large functional complex [46]. To directly test whether c-Myc affects Ku70/DNA-PKcs association, the Ku70/DNA-PKcs complex was co-immunoprecipitated from H1299 cells using a Ku70 or Ku86 antibody. The immune complex was incubated with purified recombinant c-Myc and proteins released from the complex were identified in the supernatant following centrifugation. Because the addition of purified c-Myc results in decreased levels of bound DNA-PKcs and increased levels of nonbound DNA-PKcs (Figure 5, A and B), this suggests that c-Myc can directly disrupt the Ku70/DNA-PKcs complex under cell-free conditions in vitro. To further test whether c-Myc affects Ku/DNA-PKcs complex in intact cells, WT c-Myc or each of the c-Myc deletion mutants was overexpressed in HO15.19 c-Myc null cells. Co-IP experiments revealed that overexpression of WT c-Myc, ΔMBI, or ΔHLH c-Myc mutants but not the ΔMBII c-Myc mutant potently suppresses the Ku/DNA-PKcs association (Figure 5C). This result indicates that deletion of the MBII domain from c-Myc results in loss of c-Myc's ability to disrupt the Ku/DNA-PKcs binding. Thus, the inhibitory effect of c-Myc on Ku/DNA-PKcs binding requires the MBII domain (i.e., Ku binding site).
Figure 5.
c-Myc directly disrupts the Ku/DNA-PKcs complex. (A and B) The Ku70/DNA-PKcs or Ku86/DNA-PKcs complex was coimmunoprecipitated from HO15.19 cells and incubated with purified c-Myc (10–20 ng/ml) in EBC lysis buffer at 4°C for 2 hours. After centrifugation, the resulting supernatant and immunocomplex beads were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Ku, c-Myc, and DNA-PKcs that bound to Ku or unbound DNA-PKcs present in the supernatant were then analyzed by Western blot. (C) HO15.19 cells expressing WT or each of the c-Myc deletion mutants were disrupted in EBC lysis buffer. co-IP was performed using an agarose-conjugated Ku70 or Ku86 antibody, respectively. The Ku-associated DNA-PKcs was then analyzed by Western blot. Normal rabbit IgG was used as a control.
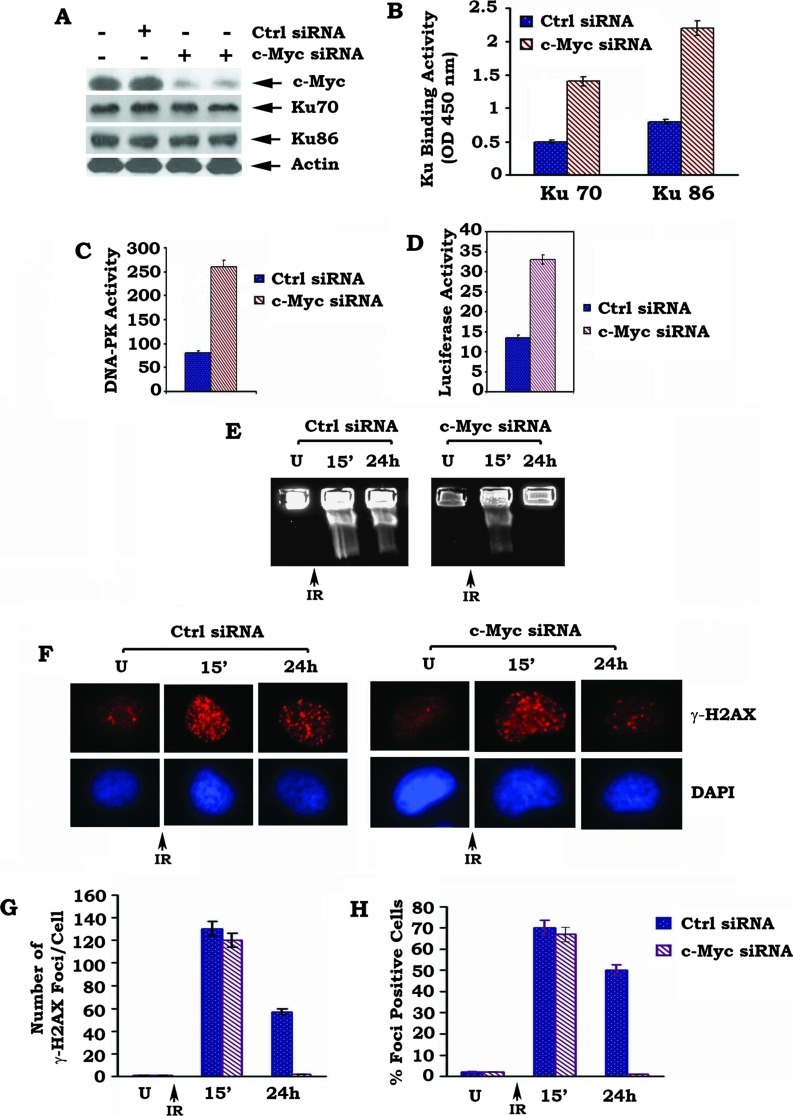
Knockdown of c-Myc by RNA Interference Results in Up-regulation of Ku DNA Binding, DNA-PKcs, and DNA End-joining Activities and Accelerates DSB Repair
To test the effect of c-Myc at physiological level on NHEJ and DSB repair, endogenous c-Myc was depleted from H460 cells by RNA interference (RNAi). Transfection of c-Myc siRNA significantly reduces the expression level of endogenous c-Myc by more than 80% but does not affects Ku expression in H460 cells (Figure 6A). This effect of siRNA on c-Myc expression is highly specific because the control siRNA has no effect. Functionally, knockdown of endogenous c-Myc by RNAi upregulates Ku DNA binding, DNA-PKcs, and DNA end-joining activities in association with accelerated repair of IR-induced DSBs, as determined by both PFGE and analysis of γ-H2AX by immunostaining (Figure 6). These findings strongly suggest that physiologically expressed c-Myc in cells is able to suppress DSB repair by a mechanism involving the NHEJ pathway.
Figure 6.
Depletion of endogenous c-Myc by RNAi enhances Ku DNA binding, DNA-PKcs, and DNA end-joining activities in association with accelerated DSB repair. (A) H460 cells expressing high levels of endogenous c-Myc were transfected with c-Myc siRNA or control siRNA. Expression of c-Myc was analyzed by Western blot. (B–D) Ku DNA binding, DNA-PKcs, and DNA end-joining activities were measured in H460 cells expressing c-Myc siRNA and control siRNA. (E–H) H460 cells expressing c-Myc siRNA or control siRNA were exposed to 5 Gy of IR. After 24 hours, DSBs were measured by PFGE or analysis of γ-H2AX by immunostaining with quantitative evaluation as described in the legend of Figure 2.
Conditional Expression of c-Myc Inhibits Formation of RS and Coding Joins in V(D)J Recombination
Ku and DNA-PKcs are required for joining of both RS and coding ends during V(D)J recombination [14,31]. Because our results showed that c-Myc suppresses both Ku and DNA-PKcs activities (Figures 2 and W3), it is also possible that c-Myc may negatively affect V(D)J recombination. To test this possibility, we assessed the ability of RAG-transfected HO15.19 cells expressing exogenous WT c-Myc or each of the c-Myc deletion mutants (i.e., ΔMBI, ΔMBII or ΔHLH) to undergo V(D)J recombination within the context of transiently transfected substrates designed to test the formation of either RS joins (pJH200) or coding joins (pJH290), as described previously [31]. At least four independent transfection experiments were carried out with each cell line. Expression of WT c-Myc or ΔMBI or ΔHLH c-Myc mutants results in significant impairment in the ability to form both RS and coding joins compared to vector-only control (Table 1). The Ku binding capacity of c-Myc may be essential for this effect because overexpression of the Ku binding-deficient ΔMBII c-Myc mutant has no significant effect on either RS or coding joins formation (Table 1). It is known that a precise RS join generates an ApaLI site and that the loss of nucleotides from the RS ends will prevent the formation of the ApaLI site. Digestion of polymerase chain reaction products generated from the recombination substrate pJH 200 with the ApaLI restriction enzyme was carried out for the analysis of fidelity of the RS joins [31]. Intriguingly, only 28% to 71% of the RS joins recovered from HO15.19 cells expressing WT c-Myc, ΔMBI, or ΔHLH c-Myc mutants could be cut by ApaLI (Table 1), indicating that expression of c-Myc also reduces the fidelity of RS joins. This is dependent on the Ku binding capacity of c-Myc, because expression of the Ku binding-deficient ΔMBII c-Myc mutant does not affect the fidelity of RS joins (Table 1). Thus, in addition to the general DSB repair process, c-Myc may also negatively regulate the repair of DSBs generated by RAG1/2 during the V(D)J recombination reaction.
Table 1.
Effect of c-Myc on RS and Coding Joining.
| Cell Line | pJH200 (RS Joining) | pJH290 (Coding Joining) | ||||||
| (Ampr + Camr)/Ampr | % | Relative Level | Fidelity % | (Ampr + Camr)/Ampr | % | Relative Level | ||
| Experiment 1 | ||||||||
| HO15.19 | ||||||||
| Vector | 95/12111 | 0.78 | 1 | 100 (14/14) | 103/6250 | 1.64 | 1 | |
| WT | 24/40101 | 0.06 | 0.08 | 28 (4/14) | 10/15430 | 0.06 | 0.04 | |
| ΔMBI | 30/38400 | 0.08 | 0.1 | 42 (6/14) | 12/22800 | 0.05 | 0.03 | |
| ΔMBII | 105/14101 | 0.74 | 0.95 | 100 (14/14) | 172/10867 | 1.58 | 0.95 | |
| ΔHLH | 24/12577 | 0.19 | 0.24 | 35 (5/14) | 17/16110 | 0.1 | 0.06 | |
| Experiment 2 | ||||||||
| HO15.19 | ||||||||
| Vector | 119/14500 | 0.82 | 1 | 100 (14/14) | 161/17980 | 0.89 | 1 | |
| WT | 33/40110 | 0.08 | 0.09 | 42 (6/14) | 52/44560 | 0.12 | 0.12 | |
| ΔMBI | 51/45890 | 0.11 | 0.13 | 28 (4/14) | 111/33420 | 0.33 | 0.37 | |
| ΔMBII | 77/10900 | 0.7 | 0.85 | 100 (14/14) | 255/29840 | 0.85 | 0.95 | |
| ΔHLH | 22/25990 | 0.08 | 0.09 | 35 (7/14) | 67/33220 | 0.2 | 0.22 | |
| Experiment 3 | ||||||||
| HO15.19 | ||||||||
| Vector | 129/19500 | 0.66 | 1 | 100 (14/14) | 322/36745 | 0.88 | 1 | |
| WT | 32/32300 | 0.09 | 0.13 | 57 (8/14) | 59/32412 | 0.18 | 0.2 | |
| ΔMBI | 21/20310 | 0.1 | 0.15 | 64 (9/14) | 119/38567 | 0.31 | 0.35 | |
| ΔMBII | 48/8430 | 0.57 | 0.86 | 100 (14/14) | 234/31010 | 0.75 | 0.85 | |
| ΔHLH | 45/35400 | 0.13 | 0.2 | 57 (8/14) | 131/45991 | 0.28 | 0.32 | |
| Experiment 4 | ||||||||
| HO15.19 | ||||||||
| Vector | 145/23500 | 0.62 | 1 | 100 (14/14) | 245/31300 | 0.8 | 1 | |
| WT | 25/30800 | 0.08 | 0.12 | 57 (8/14) | 78/43200 | 0.18 | 0.23 | |
| ΔMBI | 21/20100 | 0.1 | 0.16 | 71 (10/14) | 87/28400 | 0.31 | 0.38 | |
| ΔMBII | 50/10300 | 0.49 | 0.79 | 100 (14/14) | 153/23200 | 0.66 | 0.82 | |
| ΔHLH | 43/32300 | 0.13 | 0.21 | 71 (10/14) | 178/63800 | 0.27 | 0.34 | |
RS and coding joins were measured in HO15.19 cells expressing WT or each of the c-Myc deletion mutants. Four independent transfection experiments were carried out with each cell line.
To test whether c-Myc induces mutation or deletion during the process forming coding joins, the nucleotide sequences of the products of coding joins were compared in HO15.19 cells expressing WT c-Myc and each of the c-Myc deletion mutants [14,47]. Comparison of coding junctions revealed that extensive nucleotide deletions (i.e., >10 bp) were observed in the clones from HO 15.19 cells expressing WT c-Myc and ΔMBI or ΔHLH c-Myc mutants compared to cells expressing the Ku binding-deficient ΔMBII c-Myc mutant or vector-only control (Table 2). These findings indicate that, in addition to RS joins, c-Myc also reduces the fidelity of coding joins formation, which requires the MBII domain (Tables 1 and 2).
Table 2.
Effect of c-Myc on Coding Joining Sequence.
| Left Coding Flank Right Coding Flank | No. Clones | ||
| TTGGGCTGCAGGTCGAC | GGATCCCCGGGGATCAGC | ||
| Vector | TTGGGCTGCAGGT | ATCCCCGGGGATCAGC | 1 |
| TTGGGCTGCAGGTCG | GGATCCCCGGGGATCAGC | 3 | |
| TTGGGCTGCAGG | GGATCCCCGGGGATCAGC | 2 | |
| TTGGGCTGCAGGTCGA | ATCCCCGGGGATCAGC | 3 | |
| TTGGGCTGCAGGTCGA | CCCCGGGGATCAGC | 1 | |
| WT | TTGGGCTGCAGGTCG | GGATCCCCGGGGATCAGC | 1 |
| TTGG | TCAGC | 2 | |
| TTGGGCTG | GGATCAGC | 2 | |
| TT | GC | 3 | |
| TTGGGCTG | GGGATCAGC | 2 | |
| ΔMBI | TTGGGCTGCAGG | 2 | |
| TTGGGCTGCA | AGC | 2 | |
| TTGGG | GGGGATCAGC | 2 | |
| TTGGGC | CGGGGATCAGC | 1 | |
| TTGGGCTGCA | AGC | 1 | |
| TTGGG | 2 | ||
| ΔMBII | TTGGGCTGCAGGT | CCCCGGGGATCAGC | 2 |
| TTGGGCTGCAGGTC | ATCCCCGGGGATCAGC | 2 | |
| TTGGGCTGCAGGTCGAC | GGATCCCCGGGGATCAGC | 2 | |
| TTGGGCTGCAG | GATCCCCGGGGATCAGC | 2 | |
| ΔHLH | TTGGGCTGCAG | CCGGGGATCAGC | 1 |
| GATCCCCGGGGATCAGC | 3 | ||
| TTGGGCTG | 3 | ||
| TTGGGCTGCAGGT | ATCCCCGGGGATCAGC | 3 |
Aberrant coding join sequences from HO15.19 cells expressing WT or each of the c-Myc deletion mutants. Individual clones from the double selection plates were isolated and sequenced.
Discussion
c-Myc is a potent oncogenic molecule and its overexpression contributes to the development of human cancers [48]. The genomic damage induced by c-Myc can be broadly grouped into two classes of abnormalities. First, overexpression of c-Myc induces loss of chromosomal integrity associated with chromosomal aberrations such as gene amplifications, double minutes, and fusions. c-Myc may also contribute to tumorigenesis by destabilizing the cellular genome. Second, c-Myc overexpression can cause inappropriate DNA replication, resulting in endoreduplication. c-Myc uncouples DNA replication from mitosis with implications for genomic instability [49].The former type of genomic abnormality can be caused by defects in the repair of DSBs because overexpression of c-Myc not only causes DSBs but also disrupts their repair [23,48,50]. However, the direct molecular mechanism(s) by which c-Myc enhances occurrence of DSBs and genetic instability are not fully understood. In the present study, we discovered at first time that conditional expression of c-Myc in HO15.19 cells downregulates Ku DNA binding, DNA-PKcs, and DNA end-joining activities resulting in suppression of DSB repair and increased genomic instability (Figures 2, W3, and W4), indicating that the inhibitory effect of c-Myc on DSB repair is likely to occur through a mechanism involving the NHEJ pathway. IR-induced DSB signals not only promote the formation of c-Myc foci that co-localize with γ-H2AX but also facilitate c-Myc interaction with Ku70 (Figures 1 and 3). This suggests that c-Myc may locate at DSB sites and bind to Ku proteins following IR exposure, which could prevent NHEJ factors from binding broken DNA ends, leading to suppression of DNA end-joining. Additionally, the inhibitory effect of DSB repair by c-Myc can help explain why c-Myc could enhance apoptosis seen in the context of DNA-damaging agents [51,52]. Previous reports indicate that c-Myc can also inhibit HR [23,45], suggesting that c-Myc may also regulate DSB repair through the HR pathway. HR appears to be the predominant mechanism of DSB repair in yeast, which is operative only in the S/G2 phases of the cell cycle when a sister chromatid is available. By contrast, NHEJ, which simply pieces together the broken DNA ends, can function in all phases of the cell cycle. Therefore, NHEJ is the major pathway in mammalian cells for repairing DSBs [36,38]. Here, we discovered that, in addition to HR, c-Myc may play a more important role in regulating DSB repair by suppressing NHEJ in mammalian cells. This uncovers a novel mechanism by which c-Myc negatively regulates DSB repair.
In addition to the well-known functions as transcription factor, c-Myc also has nontranscriptional functions [53,54]. Our results reveal that expression of WT c-Myc or the c-Myc deletion mutant(s) did not alter the Ku expression level (Figures 2A and 4A). On the basis of these findings, the inhibitory effect of c-Myc on DSB repair may not be a consequence of c-Myc regulation of Ku DNA repair gene transcription. This may occur through a nontranscriptional function of c-Myc (e.g., a direct interaction with Ku protein). It has been recently reported that c-Myc can interact with Ku70, but the Ku binding domain in c-Myc is not clear [27]. It is known that the highly conserved MBII domain of c-Myc is critical for the transactivation of target genes [43,55]. Here, we found that c-Myc is able to directly interact with Ku70 in vitro and in cells (Figure 3). Furthermore, structure-function studies identified that the MBII domain of c-Myc is the Ku binding site (Figure 4A). Importantly, removal of the Ku binding site (i.e., ΔMBII domain) from c-Myc results in loss of its ability to suppress Ku DNA binding, DNA-PKcs, DNA end-joining, and DSB repair and failure to enhance chromosomal and chromatid breaks (Figures 4 and W5). These findings not only indicate a role of c-Myc in suppression of NHEJ but also demonstrate that a direct interaction between c-Myc and Ku through MBII domain is required for its inhibitory effects on NHEJ and DSB repair, as well as for its role in chromosomal instability.
Recruitment of DNA-PKcs by the Ku complex to the broken DNA ends and formation of the functional Ku/DNA-PKcs complexes are essential for DSB repair [36]. It has been proposed that c-Myc may disrupt formation of DNA repair complexes and/or disrupt chromatin structure, preventing efficient DNA repair [26]. However, there is to date no published report that substantiates this hypothesis. Here, we discovered that c-Myc not only interacts with Ku through its MBII domain (Figures 3 and 4) but also directly dissociates DNA-PKcs from Ku/DNA-PKcs complex (Figure 5, A and B). Overexpression of WT, ΔMBI, or ΔHLH but not ΔMBII c-Myc mutant in HO15.19 cells significantly suppresses Ku/DNA-PKcs association (Figure 5C), indicating that the MBII domain of c-Myc is required for the c-Myc/Ku binding as well as for disruption of Ku/DNA-PKcs complex. These findings support the notion that c-Myc/Ku binding may functionally interfere with Ku-mediated recruitment of DNA-PKcs to the broken DNA ends, which will eventually lead to suppression of DSB repair.
V(D)J recombination has been considered a physiological DSB repair process in which the NHEJ proteins are recruited to specifically repair broken ends such as the signal and hairpin coding ends generated by RAG1/2 [31,56]. Here, we discovered that overexpression of c-Myc results in decreased formation of RS and coding joins with reduced fidelity (i.e., nucleotide mutations or deletions; Tables 1 and 2), as assayed by co-transfection of RAG1 and RAG2 expression vectors along with plasmid V(D)J recombination substrates (pJH200 or pJH290), as described [31]. Because the Ku binding-deficient c-Myc mutant (i.e., ΔMBII) has no such effect, this indicates that the inhibitory effect of c-Myc on V(D)J recombination may require c-Myc/Ku binding to suppress the NHEJ pathway. Because both general DSB repair and V(D)J recombination require Ku and DNA-PKcs, this explains why c-Myc plays a role in general DSB repair similar to that observed in V(D)J recombination. Translocations occur spontaneously but more often in cells deficient in DSB repair. Misrepair of DSBs introduced during V(D)J recombination can promote oncogenic translocations [11]. Therefore, c-Myc-induced oncogenic translocation may result, at least in part, from its inhibitory effect on DSB repair and V(D)J recombination. Because V(D)J recombination is a required process for immune system development [11], c-Myc-mediated inhibition of V(D)J recombination may also impair the immune system. Thus, the role of c-Myc in DSB repair and genetic instability, together with its negative effects on V(D)J recombination and immune system development, may cooperatively promote tumor formation. Although this is a reconstituted in vitro system, the data from this system discovered at first time that c-Myc has direct role in regulating V(D)J recombination. On the basis of these findings, future studies will be required to further characterize the role of c-Myc on V(D)J recombination in B or T cells or transgenic mice that selectively target B or T cells.
In summary, our results reveal that c-Myc inhibition of DSB repair and V(D)J recombination most likely occurs through direct suppression of the NHEJ pathway. The DNA DSB signal promotes c-Myc foci formation and interaction with Ku through its MBII domain, which is required for c-Myc to inhibit Ku DNA binding, formation of Ku/DNA-PKcs complexes, DNA-PK activity, DNA end-joining, DSB repair, and V(D)J recombination. It is known that V(D)J recombination is crucial to the development of the adaptive immune system, providing B and T cells the capacity to generate a virtually unlimited repertoire of antigen receptors [57]. On the basis of our findings, we propose that the inhibitory effects of c-Myc on DSB repair and V(D)J recombination may lead to accumulation of DSBs in living cells, increased genetic instability, and impairment of the immune system, which eventually contribute to the development of various types of cancer.
Supplementary Material
Supplementary Methods
Preparation of Cell Lysates
Cells were washed with 1x PBS and resuspended in ice-cold 0.5% NP-40 EBC lysis buffer [0.5% NP-40, 50 mM Tris (pH 7.6), 120 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 50 mM NaF, and 1 mM β-mercaptoethanol] with a cocktail of protease inhibitors (Calbiochem, Billerica, MA). Cells were lysed by sonication and centrifuged at 14,000g for 10 minutes at 4°C. The resulting supernatant was collected as the total cell lysate and used for Western blot or co-IP as described [1].
DNA-PK Activity Assay
The SignaTECT DNA-PK Assay Kit was used to measure DNA-PK activity (Promega). First, the following reaction mixtures in the presence or absence of activator was prepared (i.e., 2.5 µl of DNA-PK activation buffer or control buffer, 5.0 µl of DNA-PK in 5x reaction buffer, 2.5 µl of DNA-PK biotinylated peptide substrate, 0.2 µl of BSA (10 mg/ml), and 5 µl of [γ-32P]ATP). The mixture was pre-incubated at 30°C for 4 minutes. The whole-cell extract sample was diluted in the enzyme buffer and the reaction was initiated by adding the appropriate amount of enzyme sample and incubated at 30°C for 5 minutes. The reaction was stopped by addition of termination buffer. Sample was spotted onto the SAM2R Biotin Capture membrane. The membrane was then washed and dried under a heat lamp for 10 minutes. DNA-PK enzyme activity was determined by scintillation counting. Each experiment was repeated three times and data represent the mean ± SD of three separate determinations.
In Vivo DNA End-joining Assay
The pGL3 plasmid (Promega), in which expression of the luciferase gene is controlled by the CMV promoter, was used to evaluate correct NHEJ activity that precisely rejoins broken DNA ends in vivo as described [2]. The pGL3 plasmid was completely linearized by the restriction endonuclease NarI, which cleaves within the luciferase coding region as confirmed by agarose gel electrophoresis. The linearized DNA was purified and then dissolved in sterilized water. A 20:1 mixture of the linearized pGL3 plasmid and pTK Renilla control luciferase reporter vector (an internal control) was transfected into cells. After 48 hours, luciferase activity was measured using a dual luciferase assay system following the manufacturer's instruction (Promega). Because the pGL3 reporter plasmid was digested to completion with NarI within the luciferase coding region, only precise DNA end-joining can restore the luciferase activity. Each experiment was repeated three times and data represent the mean ± SD of three separate determinations.
Electrophoretic Mobility Shift Assay
Ku DNA binding was analyzed by EMSA as described [3]. Briefly, two complementary 79-nucleotide oligonucleotides were denatured and then annealed to generate a 71-bp fragment with BamHI overhangs at each end. These two oligonucleotides are given as follows: Oligonucleotide 1, GAT CCT CTG AGG ACA CAG CCT TGT ATT ACT GTG CAA GAC ACA CAA TGA GCA AAA GTT ACT GTG AGC TCA AAC TAA AAC C and Oligonucleotide 2, GAT CGG TTT TAG TTT GAG CTC ACA GTA ACT TTT GCT CAT TGT GTG TCT TGC ACA GTA ATA CAA GGC TGT GTA CTC AGA G. The fragments were labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP. Ku protein binding activity was measured by a nondenatural gel EMSA. Each reaction mixture of 20 µl contained 0.5 ng of radiolabeled probe, 1.5 µl of nuclear extract, and 1 µg of supercoiled competitor DNA (pJG4-5) in binding buffer [15 mM Hepes (pH 7.9), 50 mM NaCl, 5 mM MgCl, 1 mM DTT, 0.5 mM EDTA, 1 mM Tris-HCl (pH 7.9), and 5% glycerol]. For all binding assays, the protein was added last and the reaction mixture was incubated at room temperature for 20 minutes. The reaction mixture was resolved by nondenaturing electrophoresis through a 6% polyacrylamide gel in 0.53 Tris-borate-EDTA buffer. The gel was dried on Whatman 3M paper and exposed to Kodak film.
RNA Interference
H460 cells expressing endogenous c-Myc were transfected with c-Myc siRNA using Lipofectamine 2000 (Invitrogen). A control siRNA (nonhomologous to any known gene sequence) was used as a negative control. The levels of c-Myc expression were analyzed by Western blot analysis. Specific silencing of the targeted c-Myc gene was confirmed by at least three independent experiments.
Acknowledgments
We are grateful to Frederick W. Alt (Harvard Medical School) for kindly providing the pJH200, pJH290, RAG1, and RAG2 constructs, Shigemi Matsuyama (Case Western Reserve University) for Flag-tagged Ku70 mutants, and John M. Sedivy (Brown University) for HO15.19 c-Myc null cells. We also thank Anthea Hammond for editing the manuscript.
Abbreviations
- HR
homologous recombination
- NHEJ
nonhomologous end-joining
- DSB
DNA double-strand break
- PFGE
pulsed-field gel electrophoresis
- siRNA
small interfering RNA
- RNAi
RNA interference
- DOX
doxycycline
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
Footnotes
This work was supported by National Cancer Institute, National Institutes of Health grants R01CA112183 and R01CA136534, by Flight Attendant Medical Research Institute Clinical Innovator Awards, and by National Aeronautics and Space Administration grant NNX12AC30G. The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Figures W1 to W5 and are available online at www.neoplasia.com.
References
- 1.Kinner A, Wu W, Staudt C, Iliakis G. γ-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 3.Rothkamm K, Kuhne M, Jeggo PA, Lobrich M. Radiation-induced genomic rearrangements formed by nonhomologous end-joining of DNA double-strand breaks. Cancer Res. 2001;61:3886–3893. [PubMed] [Google Scholar]
- 4.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 6.Fillingham J, Keogh MC, Krogan NJ. γH2AX and its role in DNA double-strand break repair. Biochem Cell Biol. 2006;84:568–577. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- 7.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 9.Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 10.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 11.Gostissa M, Alt FW, Chiarle R. echanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- 12.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 13.Blunt T, Finnie NJ, Taccioli GE, Smith GC, Demengeot J, Gottlieb TM, Mizuta R, Varghese AJ, Alt FW, Jeggo PA, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 14.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 15.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in β cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 16.Klapproth K, Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. Br J Haematol. 2010;149:484–497. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- 17.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 18.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 20.Potter M, Wiener F. Plasmacytomagenesis in mice: model of neoplastic development dependent upon chromosomal translocations. Carcinogenesis. 1992;13:1681–1697. doi: 10.1093/carcin/13.10.1681. [DOI] [PubMed] [Google Scholar]
- 21.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 22.Hironaka K, Factor VM, Calvisi DF, Conner EA, Thorgeirsson SS. Dysregulation of DNA repair pathways in a transforming growth factor alpha/c-myc transgenic mouse model of accelerated hepatocarcinogenesis. Lab Invest. 2003;83:643–654. doi: 10.1097/01.lab.0000067483.89649.11. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson A, Deb-Basu D, Cherry A, Turner S, Ford J, Felsher DW. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc Natl Acad Sci USA. 2003;100:9974–9979. doi: 10.1073/pnas.1732638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louis SF, Vermolen BJ, Garini Y, Young IT, Guffei A, Lichtensztejn Z, Kuttler F, Chuang TC, Moshir S, Mougey V, et al. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc Natl Acad Sci USA. 2005;102:9613–9618. doi: 10.1073/pnas.0407512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 26.Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA, Felsher DW. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res. 2006;66:6598–6605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- 27.Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, Yates JR, III, Menssen A, Hermeking H. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle. 2007;6:205–217. doi: 10.4161/cc.6.2.3742. [DOI] [PubMed] [Google Scholar]
- 28.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 29.Ager DD, Dewey WC, Gardiner K, Harvey W, Johnson RT, Waldren CA. Measurement of radiation-induced DNA double-strand breaks by pulsed-field gel electrophoresis. Radiat Res. 1990;122:181–187. [PubMed] [Google Scholar]
- 30.Zha S, Alt FW, Cheng HL, Brush JW, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc Natl Acad Sci USA. 2007;104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Gao F, May WS, Zhang Y, Flagg T, Deng X. Bcl2 negatively regulates DNA double-strand-break repair through a nonhomologous end-joining pathway. Mol Cell. 2008;29:488–498. doi: 10.1016/j.molcel.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guffei A, Lichtensztejn Z, Goncalves Dos Santos Silva A, Louis SF, Caporali A, Mai S. c-Myc-dependent formation of Robertsonian translocation chromosomes in mouse cells. Neoplasia. 2007;9:578–588. doi: 10.1593/neo.07355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Modesti M, Kanaar R. DNA repair: spot(light)s on chromatin. Curr Biol. 2001;11:R229–R232. doi: 10.1016/s0960-9822(01)00112-9. [DOI] [PubMed] [Google Scholar]
- 36.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Pastwa E, Blasiak J. Non-homologous DNA end joining. Acta Biochim Pol. 2003;50:891–908. [PubMed] [Google Scholar]
- 38.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 39.Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 40.Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat Res. 2001;480–481:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 41.Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11:S52–S59. doi: 10.1016/s0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- 42.Ricke RM, Bielinsky AK. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 43.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 44.Vervoorts J, Lüscher-Firzlaff J, Luscher B. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem. 2006;281:34725–34729. doi: 10.1074/jbc.R600017200. [DOI] [PubMed] [Google Scholar]
- 45.Luoto KR, Meng AX, Wasylishen AR, Zhao H, Coackley CL, Penn LZ, Bristow RG. Tumor cell kill by c-MYC depletion: role of MYC-regulated genes that control DNA double-strand break repair. Cancer Res. 2010;70:8748–8759. doi: 10.1158/0008-5472.CAN-10-0944. [DOI] [PubMed] [Google Scholar]
- 46.Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yarnell Schultz H, Landree MA, Qiu JX, Kale SB, Roth DB. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol Cell. 2001;7:65–75. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 48.Pusapati RV, Rounbehler RJ, Hong S, Powers JT, Yan M, Kiguchi K, McArthur MJ, Wong PK, Johnson DG. ATM promotes apoptosis and suppresses tumorigenesis in response to Myc. Proc Natl Acad Sci USA. 2006;103:1446–1451. doi: 10.1073/pnas.0507367103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q, Dang CV. c-Myc overexpression uncouples DNA replication from mitosis. Mol Cell Biol. 1999;19:5339–5351. doi: 10.1128/mcb.19.8.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mai S, Hanley-Hyde J, Fluri M. c-Myc overexpression associated DHFR gene amplification in hamster, rat, mouse and human cell lines. Oncogene. 1996;12:277–288. [PubMed] [Google Scholar]
- 51.El-Kady A, Sun Y, Li YX, Liao DJ. Cyclin D1 inhibits whereas c-Myc enhances the cytotoxicity of cisplatin in mouse pancreatic cancer cells via regulation of several members of the NF-κB and Bcl-2 families. J Carcinog. 2011;10:24. doi: 10.4103/1477-3163.90437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Bueren AO, Oehler C, Shalaby T, von Hoff K, Pruschy M, Seifert B, Gerber NU, Warmuth-Metz M, Stearns D, Eberhart CG, et al. c-MYC expression sensitizes medulloblastoma cells to radio- and chemotherapy and has no impact on response in medulloblastoma patients. BMC Cancer. 2011;11:74. doi: 10.1186/1471-2407-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 54.Cole MD, Cowling VH. Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat Rev Mol Cell Biol. 2008;9:810–815. doi: 10.1038/nrm2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mu ZM, Yin XY, Prochownik EV. Pag, a putative tumor suppressor, interacts with the Myc Box II domain of c-Myc and selectively alters its biological function and target gene expression. J Biol Chem. 2002;277:43175–43184. doi: 10.1074/jbc.M206066200. [DOI] [PubMed] [Google Scholar]
- 56.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 57.Johnson K, Chaumeil J, Skok JA. Epigenetic regulation of V(D)J recombination. Essays Biochem. 2010;48:221–243. doi: 10.1042/bse0480221. [DOI] [PubMed] [Google Scholar]
- 58.Balajee AS, Geard CR. Replication protein A and γ-H2AX foci assembly is triggered by cellular response to DNA double-strand breaks. Exp Cell Res. 2004;300:320–334. doi: 10.1016/j.yexcr.2004.07.022. [DOI] [PubMed] [Google Scholar]
Supplemental References
- 1.Deng X, Gao F, Flagg T, Anderson J, May WS. Bcl2's flexible loop domain regulates p53 binding and survival. Mol Cell Biol. 2006;26:4421–4434. doi: 10.1128/MCB.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin KH, Kang MK, Dicterow E, Kameta A, Baluda MA, Park NH. Introduction of human telomerase reverse transcriptase to normal human fibroblasts enhances DNA repair capacity. Clin Cancer Res. 2004;10:2551–2560. doi: 10.1158/1078-0432.ccr-0669-3. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Lieber MR. Protein-protein and protein-DNA interaction regions within the DNA end-binding protein Ku70-Ku86. Mol Cell Biol. 1996;16:5186–5193. doi: 10.1128/mcb.16.9.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.