Abstract
The expression of the angiogenic phenotype is regulated by a balance of pro-angiogenic and anti-angiogenic factors released into the tumor microenvironment. Nuclear protein 7 (NOL7), a novel tumor suppressor, acts as a master regulator of angiogenesis by downregulating pro-angiogenic factors and upregulating anti-angiogenic factors. Using cervical cancer as a model of investigation, we have previously shown that loss of NOL7 mRNA and protein expression is observed as early as the premalignant phase. Analysis of the gene failed to identify tumor-specific promoter methylation or coding region mutations, suggesting that NOL7 loss may be mediated by aberrant expression of its upstream regulators. In this study, we show that the RB tumor suppressor gene (RB) positively regulates NOL7 at the transcriptional level by recruiting transcription factors and transcription machinery proteins to its promoter region. Conversely, the loss of RB represses NOL7 transcription by inhibiting assembly of these proteins. This loss of NOL7 expression is also observed in RB-deficient human malignancies. Together, this work further characterizes the transcriptional activator function of RB and defines a potential role for RB in regulating angiogenesis through activation of NOL7. Current anti-angiogenic therapies lack long-term efficacy, as they are unable to target the diverse angiogenic signals generated by tumors. Our data provide evidence to support the hypothesis that reactivation of pRB can potentially modulate the expression of the angiogenic phenotype through regulation of NOL7. Therefore, this knowledge may be employed to design more comprehensive and effective therapies.
Introduction
Angiogenesis is a hallmark of cancer and is considered an essential step for tumor growth and metastasis [1,2]. The expression of the angiogenic phenotype is regulated by a balance of pro-angiogenic and anti-angiogenic factors released in the tumor microenvironment [1]. Anti-angiogenic therapies often lack long-term efficacy because they are generally designed to target specific pathways or molecules, and blocking one such step may activate alternative mechanisms or promote resistance through alternative mechanisms [3–5]. To better understand the complex interplay between angiogenesis regulation and other important processes in the pathogenesis of cancer, it is therefore important to characterize novel angiogenesis regulators and their control by key tumor suppressor pathways.
Using cervical cancer (CC) as model of investigation, we have previously identified a novel tumor suppressor gene, NOL7 [6]. Nuclear protein 7 (NOL7) inhibits angiogenesis by downregulating pro-angiogenic factors, including vascular endothelial growth factor, and upregulating anti-angiogenic factors, such as thrombospondin-1 [6,7]. This ability to differentially modulate angiogenesis-related proteins suggests that NOL7 may function as a master regulator of angiogenesis [7]. NOL7 expression is significantly reduced in CC cell lines and tumors [6,8], and recent reports have demonstrated that reduced NOL7 expression is significantly associated with higher relapse risk in CC [9]. However, the mechanisms explaining NOL7 loss of expression are not known.
Previously, we evaluated NOL7 expression in normal, premalignant, and CC tissues and determined that the initial loss of protein expression was observed at the early premalignant phases [8]. While NOL7 allelic loss and genomic alterations have been reported, these changes are typically not observed until after the development of malignant disease, suggesting that alternative mechanisms are responsible for the loss of NOL7 expression at the premalignant stage [10,11]. We identified the NOL7 promoter region and assessed its methylation and mutation status [8,12]. This analysis failed to identify any directly deleterious methylation or mutation patterns specific in either CC tissue or cell lines [8,12]. In addition, analysis of the NOL7 3′untranslated region (UTR) failed to identify conserved cis-elements or microRNA binding sites, suggesting that loss of NOL7 expression does not occur by way of post-transcriptional or translational mechanisms. These data suggest that down-regulation of NOL7 at the premalignant phases of CC results from aberrant expression of its upstream regulators.
CC development is driven in part by the HPV oncoproteins E6 and E7 that contribute to malignant transformation by inducing genomic instability, inactivating the p53 and RB tumor suppressors, and promoting angiogenesis [13,14]. In CC, the RB, p107, and p130 pocket proteins are inactivated by the HPV E7 gene product that both targets the pocket proteins for degradation and inhibits their interactions with E2Fs [15,16]. Previous studies have detected a significant loss of RB protein (pRB) expression between the premalignant cervical intraepithelial neoplasia II and III phases [17]. Interestingly, our analysis of premalignant cervical tissue depicted a similar loss of NOL7 expression at the same interval [8]. Analysis of pRB expression in CC cell lines demonstrated a strong correlation between pRB and NOL7 expression levels in 81% of cell lines (data not shown) [8]. In addition, reactivation of RB in CC cell lines results in the induction of NOL7 transcript levels [18]. This suggested that pRB positively regulates NOL7 expression and that RB loss and reduced NOL7 expression observed in CC are mechanistically linked. Although known primarily as a transcriptional repressor, recent data have suggested that pRB can activate gene transcription by interacting with site-specific transcription factors (TFs) such as E2F1 and SP1 [19,20]. The NOL7 promoter region has predicted binding sites for these TFs, and we have previously demonstrated that SP1 associates with the NOL7 promoter [12]. The objective of this study was to determine if pRB regulates NOL7 transcription and characterize the mechanism behind this regulation.
In this report, we describe a novel role for the RB tumor suppressor in transcriptionally activating the anti-angiogenic protein, NOL7. We demonstrate that pRB stimulates NOL7 transcription by interacting with and recruiting TFs, coactivators, and chromatin remodeling enzymes to the NOL7 promoter region. We also show that RB loss significantly downregulates NOL7 promoter activity and protein levels. This correlation between RB inactivation and decreased NOL7 expression is also observed in different types of human malignancies. This suggests that RB-mediated NOL7 regulation is not restricted to a single cell type and that RB loss results in subsequent NOL7 down-regulation during malignant transformation. Taken together, our data identify a potential role for pRB in regulating angiogenesis through modulation of NOL7 expression.
Materials and Methods
Cell Lines
HeLa and Hec-1A (ATCC, Manassas, VA) were cultured as described previously [8]. GP2-293 (Clontech, Mountain View, CA) and HaCaT (ATCC) cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA). HPV E7 expression was induced by treating cells with 1 µM 4-hydroxytamoxifen (Sigma, St. Louis, MO) containing media for 20 hours. For generation of stable cell lines, media were supplemented with puromycin (1 µg/ml; Sigma) or geneticin (300 µg/ml; Invitrogen).
Polymerase Chain Reaction Amplification, Plasmids, and Cloning
Polymerase chain reaction (PCR) was carried out using Phusion High-Fidelity Master Mix (New England Biolabs, Ipswich, MA) [12]. The full-length NOL7 promoter (FL) was subcloned into the pGL4.20 vector (Promega, Madison, WI) [12]. The promoter subregions R1, R2, R3, ΔR1, and ΔR3 were PCR amplified using primers (Table W1) and cloned into the pGL3-basic vector (Promega). Biotinylated fragments of the NOL7 promoter, subregions, p21 promoter, and a random nongenic region (R02; Switchgear Genomics, Menlo Park, CA) were obtained by PCR amplification using biotin-labeled primers (Table W2). The pCMV-VSV-G, E7, and E1A (wild-type and LXCXE mutant) vectors were obtained from the following laboratories: DiMaio, Munger, Schlegel, and Weinberg [18,21–23]. The pIRES-GFP and pBabe-puro-ER-E7 plasmids were generously provided by Dr Kay Macleod. The wild-type and mutant E7 and E1A genes were subcloned into the pIRES-green fluorescent protein (GFP) vector.
Transfections
Transfections were performed using X-fect (Clontech) as per the manufacturer's guidelines. For stable RB knockdown, cells were transfected with pGFP-V-RS plasmid (Origene Technologies, Rockville, MD) containing either scrambled or RB shRNA (5′-AGCAGTTCGATATCTACTGAAA-3′) followed by selection with puromycin. HaCaT and Hec-1A cell lines stably expressing the NOL7 promoter-luciferase were obtained by transfecting cells with pGL4.20-NOL7 promoter plasmid followed by selection with puromycin. These HaCaT-luc or Hec-1A-luc cells were transiently transfected with pIRES-GFP vector containing either wild-type or LXCXE mutant E7 or E1A genes. HaCaT cells were transiently transfected with FL, R1, R2, R3, ΔR1, or ΔR3 vectors along with pIRES-GFP.
Retrovirus Production and Transduction
GP2-293 cells were co-transfected with pBabe-puro-ER-E7 and pCMV-VSV-G plasmids. Forty-eight hours after transfection, the culture medium containing the retrovirus particles was harvested, concentrated using the Retro-X Concentrator (Clontech), and reconstituted in media containing polybrene (8 µg/ml; Millipore, Billerica, MA). Cells were incubated in this media for 8 hours followed by a media change. This infection was repeated one more time, followed by selection in media containing puromycin to obtain cells stably expressing a tamoxifen-inducible HPV E7.
Luciferase and GFP Assays
Luciferase assays were performed as described previously [12]. GFP expression was measured using the Synergy HT reader (BioTek, Winooski, VT). The GFP values were used to normalize the luciferase activity. Experiments were performed in triplicates.
Chromatin Immunoprecipitation and Reverse Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) assays were performed for the TFs pRB, E2F1 to E2F4, or SP1 using NOL7 promoter-specific primers as described previously [12]. One microgram of pRB, phospho-RB (Ser 780-790-807-811; Cell Signaling Technology, Danvers, MA), E2F1 to E2F4 (Santa Cruz Biotechnology, Santa Cruz, CA), SP1, and mouse or rabbit IgG (Millipore) antibodies were used. For the reverse ChIP (ReChIP) experiments, the pRB-bound chromatin-protein complexes were precipitated by incubation with 1 µg of E2F3, BRG-1, BRM, p300 (Santa Cruz Biotechnology), PCAF (Cell Signaling Technology), SP1, RNA Polymerase II (RNA Pol II), and mouse IgG or rabbit IgG (Millipore) antibodies, followed by elution and PCR analysis as described previously [12]. Each ChIP or ReChIP experiment was repeated twice. ChIP-grade antibodies were used for these assays, which have also been previously validated in other studies [12,20,24–27].
Western Blot Analysis
Western blot analysis was performed as described previously [12]. The following primary antibodies were used: β-actin (Abcam, Cambridge, MA), E2F4 (Bethyl Laboratories, Montgomery, TX), pRB, BRM, PCAF (Cell Signaling Technology), E2F1, RNA Pol II, SP1 (Millipore), E2F2, BRG-1, p300 (Santa Cruz Biotechnology), E2F3, NOL7, p107, and p130 (Sigma). The blots were incubated with the appropriate secondary antibodies, developed as described before [12]. The Quantity One software (Bio-Rad, Hercules, CA) was used to quantify individual protein bands from densitometrically scanned Western images.
Streptavidin Pull-down Assay
Streptavidin pull-down assay (SAPA) was performed as described previously with few modifications [19,28,29]. HaCaT cell nuclear extracts were prepared using the Nuclear Extract Kit (Active Motif, Carlsbad, CA). Each biotinylated DNA fragment was incubated with 100 µg of HaCaT nuclear extract by rotation at 4°C, overnight. Protein-DNA complexes were precipitated using streptavidin Dynabeads (Invitrogen). Beads were washed thoroughly and boiled in sodium dodecyl sulfate sample buffer to elute the bound proteins, which were then analyzed by Western blot analysis.
Tissue Microarray and Immunohistochemistry
Immunohistochemistry was performed with the approval of the University of Chicago Institutional Review Board. Normal retina and retinoblastoma tissue microarrays were obtained from US Biomax Inc (Rockville, MD). Normal lung, cervix, oral mucosa, cervical, HPV-positive oropharyngeal, and small cell lung cancer (SCLC) tissue micro-arrays were generated using a Beecher Instruments ATA-27 Automated Tissue Arrayer. Deparaffinized sections were microwaved in ET buffer, NOL7 (1:120; Sigma) and RB (1:100; Cell Signaling Technology) primary antibodies were applied for 1 hour at room temperature, and the rabbit or mouse EnVision+ Kit (DAKO, Carpinteria, CA) was used for detection. All sections were counterstained with hematoxylin and scored on a 0 to 3 scale, with 0 being the lowest staining and 3 the greatest.
Statistical Analysis
The error bar values are reported as mean ± SEM. A two-tailed Student's t test was used to compute P values, and the graphs were made using SigmaPlot software version 10 (Systat, Chicago, IL).
Results
pRB Repression Causes Down-regulation of NOL7 Expression
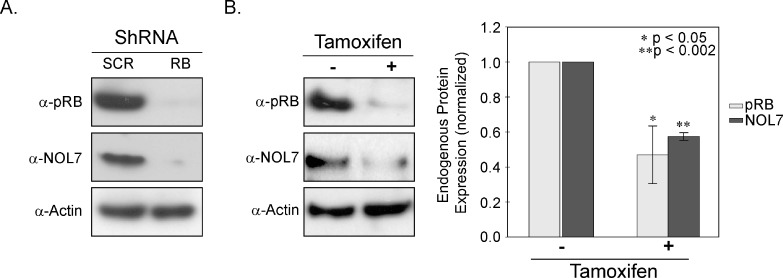
NOL7 expression is significantly downregulated in CC where RB is functionally inactivated by the HPV E7 oncoprotein [8,30]. To examine the effect of RB knockdown on NOL7 expression, we stably transfected HaCaT cells with either control or RB-specific shRNA. These cells have a nonfunctional p53 and therefore resistant to p53-dependent cell death on RB loss, thereby simulating CC that has both Rb and p53 inactivated [31,32]. Western blot analysis demonstrated that RB knockdown in HaCaT cells resulted in significantly reduced levels of NOL7 protein (Figure 1A), suggesting that pRB positively regulates NOL7 expression. To determine if pRB is a direct upstream modulator of NOL7 expression, we generated cells stably expressing a tamoxifen-inducible HPV E7 construct. This analysis helped us assess NOL7 protein expression after conditional and transient RB loss. As Figure 1B demonstrates, tamoxifen treatment decreased endogenous pRB expression by 50%. Furthermore, cells with reduced pRB levels also exhibited reduced endogenous NOL7 protein expression (Figure 1B). Taken together, these data suggest that pRB is a positive upstream modulator of NOL7 expression.
Figure 1.
pRB repression downregulates NOL7 protein expression. (A) Representative Western image illustrating endogenous pRB and NOL7 expression in pRB knocked down HaCaT cells. (B) HaCaT-HPV E7 cells were treated with media containing 1 µM tamoxifen for 20 hours. Representative Western image depicting expression of endogenous pRB and NOL7 +/- treatment with tamoxifen and quantification of endogenous pRB and NOL7 protein levels.
pRB Positively Regulates NOL7 at the Transcriptional Level
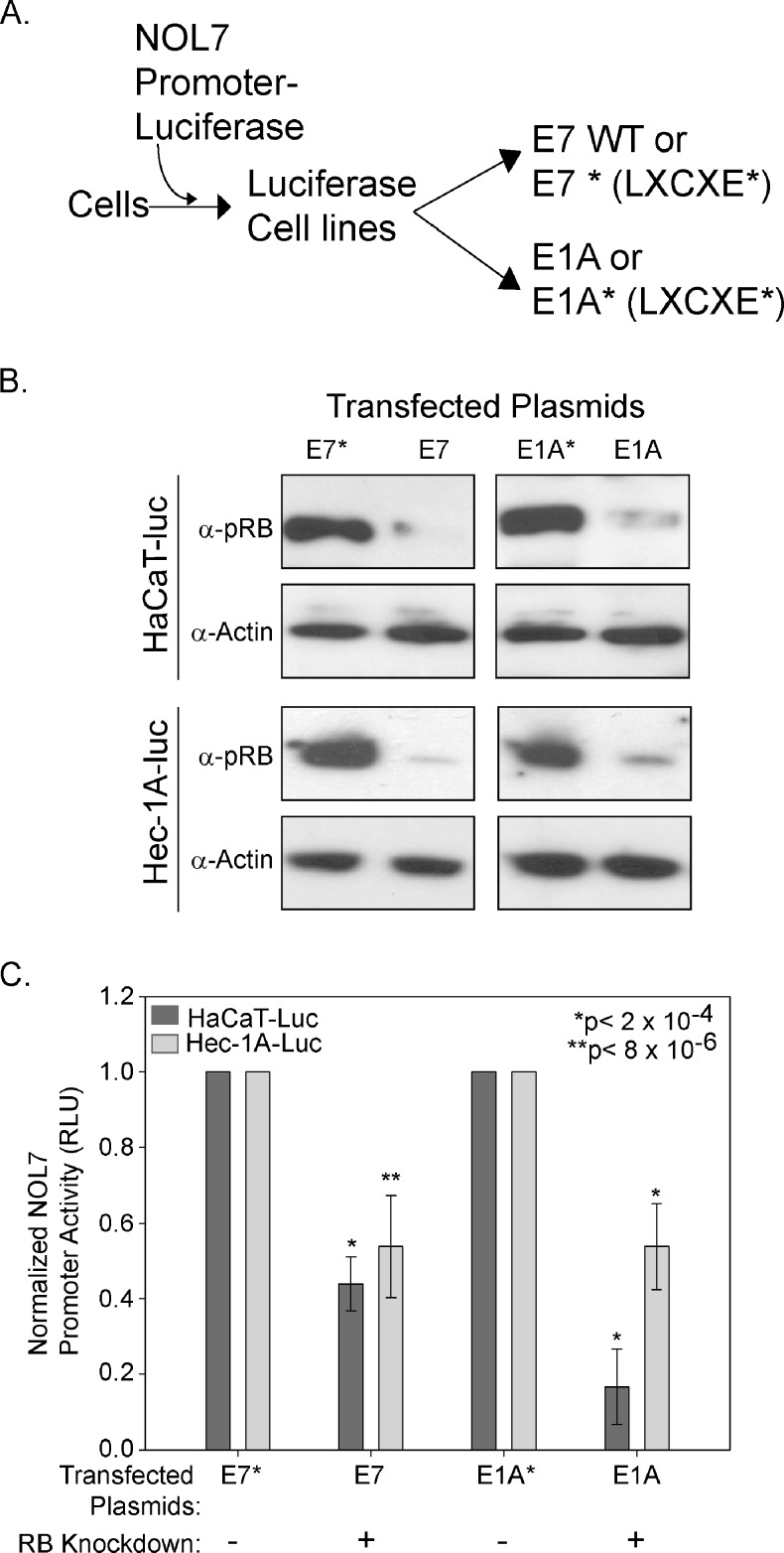
As pRB loss negatively affected NOL7 protein expression, we sought to determine the effect of RB loss on NOL7 transcription. We generated NOL7 promoter-luciferase construct expressing cell lines, which were then transiently transfected with either wild-type HPV E7 or E1A or their respective mutant forms that are unable to bind to and degrade pRB (Figure 2A) [33]. In addition to HaCaT cells, we used Hec-1A cells, which express a variant p53 protein that is less efficient in inducing cell death [34,35]. Endogenous pRB levels were reduced exclusively in wild-type E7- and E1A-transfected cells (Figure 2B). NOL7 promoter activity was significantly reduced (P < 2 x 10-4) in Hec-1A-luc and HaCaT-luc cells where pRB expression was downregulated (Figure 2C). These data demonstrate that pRB positively regulates NOL7 transcription.
Figure 2.
pRB positively regulates NOL7 transcription. (A) Schematic illustrating generation of stable cell lines expressing the NOL7 promoter-luciferase, transfected with either wild-type or mutant (*) forms of HPV E7 or E1A proteins. (B) Western blot analysis to evaluate endogenous pRB expression in these cells. (C) Relative NOL7 promoter-luciferase activity represented as a percentage of the wild-type E7 or E1A transfectants. The figure represents triplicate experiments with error bars corresponding to SEM.
pRB-interacting Proteins E2F3 and SP1 Are Enriched on the NOL7 Promoter
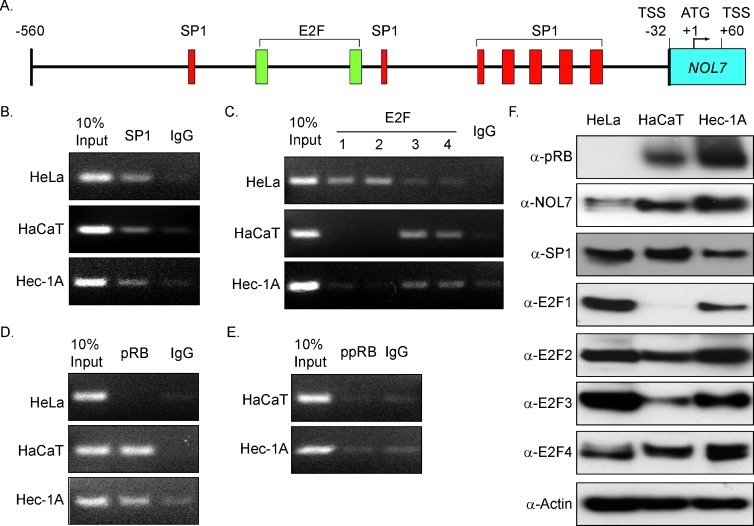
pRB lacks a DNA-binding domain and therefore is localized to gene promoters through interaction with site-specific TFs including the E2F family proteins [36,37]. Recent reports have shown that pRB positively regulates gene transcription by interacting with SP1 and E2F1 [19,20]. In silico analysis using promoter prediction programs identified seven SP1 and two E2F binding sites within the NOL7 promoter (Figure 3A) [38,39]. Using ChIP analysis, we determined that SP1 associated with the NOL7 promoter in all cell lines tested (Figure 3B), while E2F1 and E2F2 showed greater association with the NOL7 promoter in HeLa cells (Figure 3C). In contrast, E2F3 and E2F4, which also interact with pRB, associated with the NOL7 promoter to a greater extent in HaCaT and Hec-1A cells compared to HeLa cells (Figure 3C) [37]. Together, these data demonstrated that pRB-interacting TFs associated with the NOL7 promoter, thereby suggesting tha tpRB may localize to the NOL7 promoter through interaction with these TFs.
Figure 3.
pRB and pRB-interacting TFs associate with the NOL7 promoter. (A) NOL7 promoter schematic depicting predicted SP1 and E2F binding sites. (B–E) ChIP analysis to evaluate SP1, E2F, pRB, and ppRB association on the NOL7 promoter in HeLa, HaCaT, and Hec-1A cell lines as described previously. (F) Western blot analysis depicting expression of TFs in these cell lines.
Hypophosphorylated pRB Stimulates NOL7 Transcription by Associating with Its Promoter
Because pRB-interacting TFs associated with the NOL7 promoter (Figure 3, B and C), a ChIP assay was performed to determine if pRB positively regulated NOL7 transcription by associating with its promoter. This analysis demonstrated that pRB specifically associated with the NOL7 promoter in HaCaT and Hec-1A cells but not in HeLa cells, which have reduced pRB expression because of inhibition by HPV E7 (Figure 3, D–F). This suggested that pRB may induce NOL7 transcription by interacting with TFs, E2Fs, or SP1 that were also enriched on its promoter (Figure 3, B and C).
pRB is hyperphosphorylated and inactive following the G1-S transition of mammalian cell cycle [40]. Therefore, we assessed the enrichment of hyperphosphorylated RB (ppRB) on the NOL7 promoter by repeating the ChIP analysis using antibodies that recognize key phosphorylation sites (Ser 780-795-807-811) on pRB [41–45]. As shown in Figure 3E, ppRB did not associate with the NOL7 promoter, suggesting that only hypophosphorylated pRB interacts with the NOL7 promoter. Next, we evaluated the effect of cell cycle phase-specific changes in hypophosphorylated pRB for effects on NOL7 expression. We treated cells with aphidicolin and nocodazole to arrest them at S phase or M phase of cell cycle, respectively (Figure W1A). NOL7 protein expression was downregulated in G2 and early M phases, which have increased ppRB levels, compared to G1 and early S phases (Figure W1B). These data suggest that hypophosphorylated pRB drives NOL7 transcription by associating with its promoter and cell cycle phase-specific pRB phosphorylation downregulates NOL7 expression by limiting hypophosphorylated pRB availability.
To evaluate the enrichment of pRB and pRB-interacting proteins on the NOL7 promoter through an alternate approach, SAPA analysis was performed. This is a semiquantitative assay that is extremely useful in analyzing multiple TFs that bind to a promoter from a non-cross-linked nuclear extract [28,29]. SAPA analysis was performed using a biotinylated NOL7 promoter fragment that was incubated with non-cross-linked nuclear extract, followed by precipitation of the promoter-bound protein complexes with streptavidin beads and analysis of the bound proteins. A biotinylated p21 promoter was used as a positive control for this assay, as p21 is positively regulated by pRB and SP1 [19], and a random nongenic region (R02) was used as a negative control. SAPA analysis demonstrates enrichment of pRB, SP1, and E2F3 on the NOL7 promoter (Figure W2). In addition, RNA Pol II was enriched on the NOL7 promoter region (Figure W2), suggesting that pRB activates NOL7 transcription by interacting with its promoter along with the proteins E2F3, SP1, and RNA Pol II.
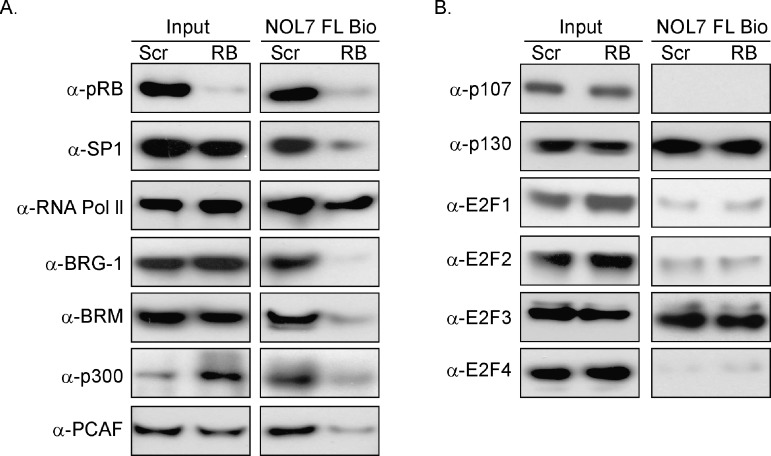
pRB Interacts with TFs, RNA Pol II, Transcriptional Coactivators, and Chromatin Remodeling Enzymes at the NOL7 Promoter
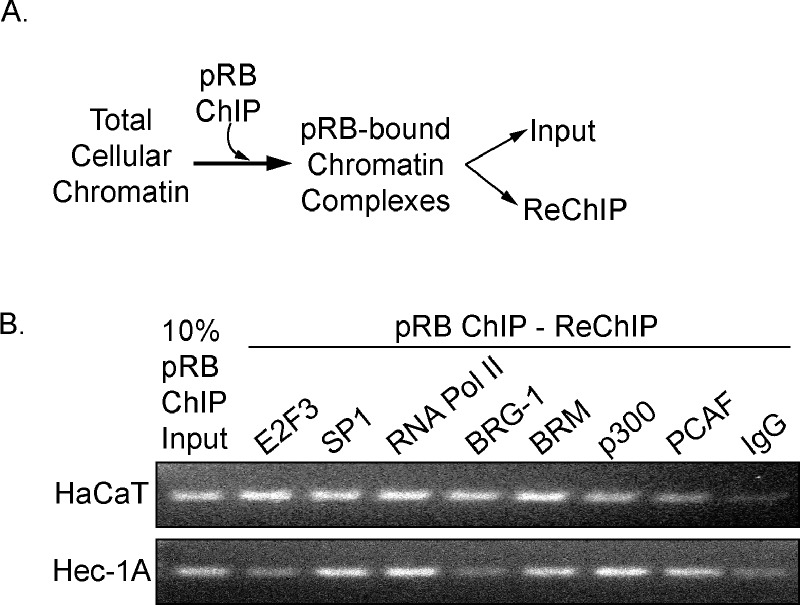
pRB associates with DNA through interactions with TFs and chromatin remodeling proteins [20,25]. Therefore, we hypothesized that pRB is part of a protein complex that assembles on NOL7 promoter. To assess this possibility, we performed a ChIP-ReChIP assay (Figure 4A). Strong interaction between SP1, RNA Pol II, and pRB was observed in the HaCaT and Hec-1A cell lines, whereas E2F3 interaction with pRB was more pronounced in the HaCaT cells (Figure 4B). In addition, the SWI/SNF component BRM and coactivators p300 and PCAF interacted with pRB at the NOL7 promoter in both cell lines, while the pRB-BRG-1 interaction was more specific to HaCaT cells (Figure 3B). These data indicate that pRB interacts with TFs, RNA Pol II, coactivators, and chromatin remodeling proteins, thereby suggesting that it is part of a transcriptional complex that is enriched on the NOL7 promoter
Figure 4.
pRB interacts with TFs, RNA Pol II, coactivators, and chromatin remodeling proteins at the NOL7 promoter. (A) Schematic depicting the steps in the ReChIP assays. (B) Representative ReChIP PCR image for HaCaT and Hec-1A cells. Each ReChIP experiment was repeated twice.
pRB Recruits SP1, Transcriptional Coactivators, and Chromatin Remodeling Proteins to the NOL7 Promoter
Our analysis indicated that pRB was part of a complex of proteins that bound the NOL7 promoter. To functionally characterize pRB's role in the assembly of this complex, we evaluated the effect of RB loss on the enrichment of these proteins on the NOL7 promoter by performing SAPAs in cells where RB expression was knocked down. We observed that knockdown of RB significantly lowered pRB enrichment on the NOL7 promoter (Figure 5A). Interestingly, RB loss also reduced SP1, BRG-1, BRM, p300, and PCAF binding to the NOL7 promoter, suggesting that pRB was essential for recruitment of these proteins to the NOL7 promoter (Figure 5A). These data suggested that pRB recruits multiple proteins, including components of the basal transcriptional machinery to drive NOL7 transcription.
Figure 5.
pRB activates NOL7 transcription by recruiting SP1, co-activators, and chromatin remodeling proteins to its promoter. (A and B) SAPA analysis was performed on cells expressing control (SCR) or RB-specific shRNA to assess the enrichment of proteins on the NOL7 promoter on RB loss. Each SAPA experiment was repeated twice. A representative Western image depicting NOL7 promoter-enriched proteins in control or pRB knockdown cells is shown here.
NOL7 Repression Is Not Mediated by Binding of an Alternative pRB-E2F Complex to Its Promoter
In addition to binding to pRB, E2F family members can also complex with the other pocket protein family members, p107 and p130, to repress gene transcription [37]. In particular, “pocket protein shuffling” occurs when pRB is knocked down or out, such that new complexes between the “activator” class of E2Fs (E2F1, E2F2, and E2F3) and p107 or p130 are formed [46]. We assessed whether an alternative repressor complex downregulated NOL7 transcription in the absence of pRB. Overall, no significant change in binding of the pocket proteins, p107 and p130, and various E2Fs was observed on pRB loss (Figure 5B). p107, E2F1, E2F2, and E2F4 continued to demonstrate very weak or no enrichment on the NOL7 promoter, suggesting that pRB loss does not result in the recruitment of an alternative pocket protein-E2F complex to the NOL7 promoter.
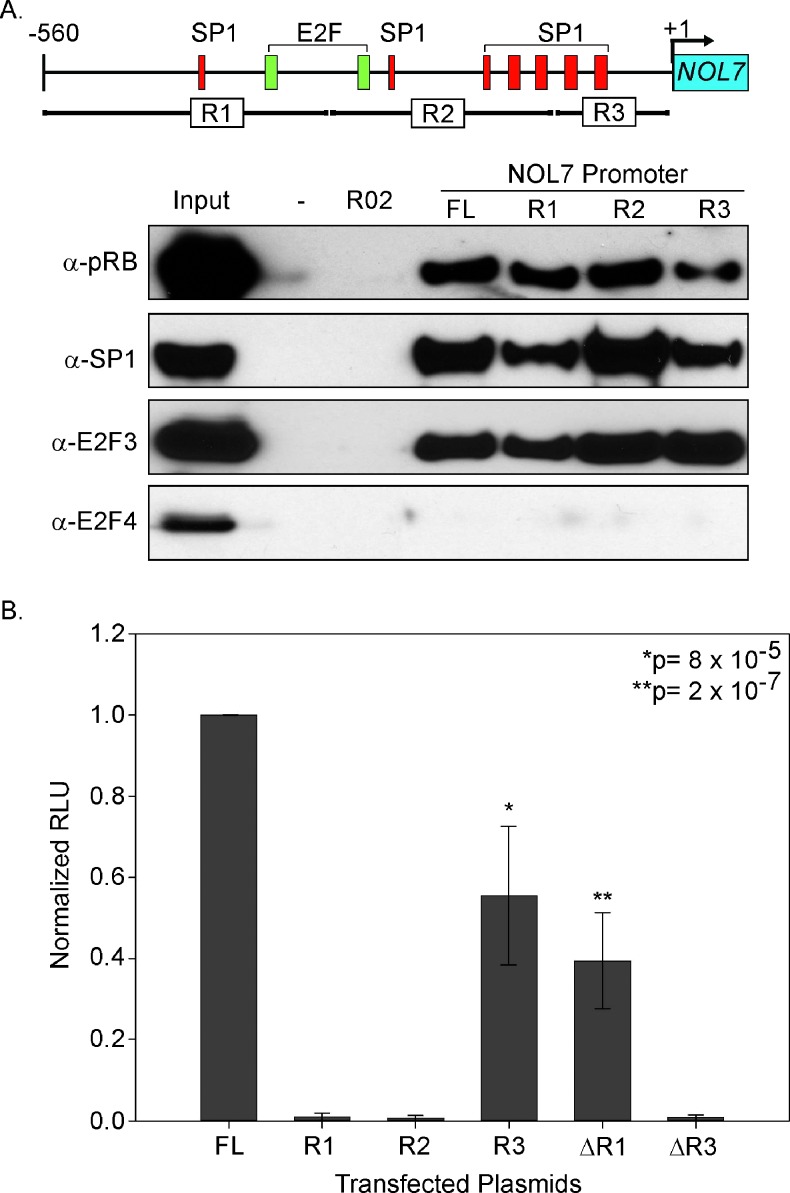
The NOL7 Promoter Contains Multiple Binding Sites for pRB-TF Complexes
Next, we sought to further define the functional pRB-TF complex binding region within the NOL7 promoter. For this purpose, the NOL7 promoter was divided into three nonoverlapping subregions (R1–R3; Figure 5A). SAPA analysis with nonoverlapping NOL7 promoter subregions demonstrated comparable pRB, E2F3, and SP1 enrichment on each subregion (Figure 6A), suggesting that the NOL7 promoter contains multiple binding sites for pRB-TF complexes. To assess the functional contribution of each of these subregions to NOL7 transcriptional activation, we cloned promoter subregions into luciferase reporter constructs and compared the activity of each subregion to the FL. R3 exhibited the greatest luciferase reporter gene expression compared to the other subregions (Figure 6B). However, it had 45% lower promoter activity compared to the FL (P = 8 x 10-5; Figure 6B), suggesting that while R3 contained necessary elements for basal promoter activity, optimum transcriptional activation of NOL7 required the interaction of the pRB-TF complexes at all three regions.
Figure 6.
pRB-TF complexes have multiple binding sites in the NOL7 promoter. (A) NOL7 promoter schematic depicting predicted E2F, SP1 binding sites, and NOL7 promoter subregions R1 to R3. SAPA analysis of the proteins coprecipitated with each promoter subregion by Western blot analysis. (B) Relative luciferase activity of the NOL7 promoter subregions R1 to R3, ΔR1 (R2 + R3), or ΔR3 (R1 + R2) represented as a fraction of the FL transfectants. The figure represents triplicate experiments with error bars corresponding to SEM.
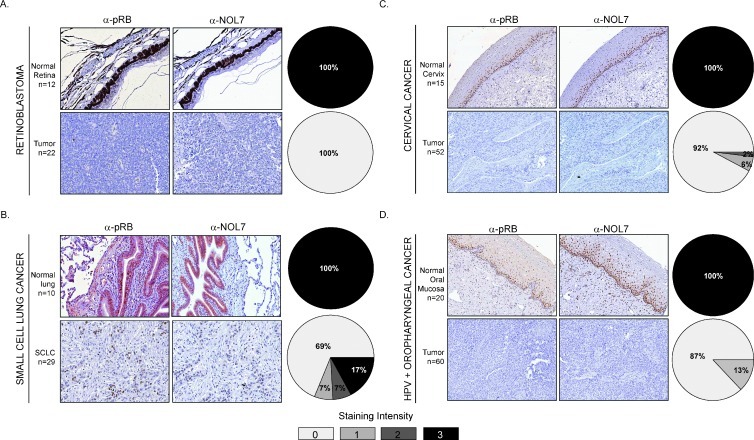
NOL7 Protein Expression Is Decreased in Human Malignancies Lacking pRB
The RB pathway is disrupted in a number of human cancers through several different mechanisms including RB gene deletions (observed in retinoblastoma and SCLC) and RB protein degradation [observed in CC and HPV-associated oropharyngeal cancer (HPV + OPC)] [15]. To determine whether RB protein loss correlated with downregulated NOL7 levels in human tumors, we evaluated NOL7 and RB protein levels in human retinoblastoma, CC, SCLC, and HPV + OPC tissues. Previous studies have observed significantly lower pRB expression in these tumors [47–50]. As expected, robust pRB and NOL7 expression was observed in 100% of normal lung, retinal, cervical, and oral mucosa tissues (Figure 7, A–D, upper panels). By contrast, retinoblastoma, CC, and HPV + OPC tissues expressed significantly lower levels of pRB (Figure 7, A, C, and D, lower panels); these results were in accordance with previous studies. Strikingly, these tumors had reduced NOL7 levels, which strongly correlated with pRB expression. While prior reports have detected low or absent pRB expression in 100% of SCLC tissue samples [49], we observed that 76% of the SCLC samples exhibited low pRB expression. However, again the NOL7 expression closely correlated with pRB levels as observed in the other RB null tumors (Figure 7). These data showed that NOL7 was significantly downregulated in tumors that lack pRB, irrespective of the mechanism of RB inactivation.
Figure 7.
NOL7 expression is downregulated in human malignancies that lack pRB. Immunohistochemistry analysis demonstrating pRB and NOL7 expression levels in (A) normal retina and retinoblastoma, (B) normal lung and SCLC, (C) normal cervix and CC, and (D) normal oral mucosa and HPV + OPC tissues. Data are represented as percentage of tumor specimens demonstrating a staining intensity ranging from 0 to 3+.
Discussion
Angiogenesis is a critical step in tumor progression and metastasis [1,2]. The diversity and redundancy of angiogenic pathways has hampered the design of effective and long-term anti-angiogenic therapies [4,51]. We identified a novel anti-angiogenic protein, NOL7, that may function as master regulator of angiogenesis by downregulating pro-angiogenic and upregulating anti-angiogenic factors [6,7]. NOL7 expression is lost in CC where HPV E7 inactivates the RB tumor suppressor protein [8,30]. Here, we demonstrate that the RB tumor suppressor positively regulates NOL7 transcription. Using multiple approaches, we show that pRB positively regulates NOL7 transcription by recruiting TFs, chromatin remodeling enzymes, and coactivator proteins to form a transcriptional activation complex at the NOL7 promoter. RB loss prevents formation of this complex, thereby resulting in down-regulation of NOL7 transcription and expression. This down-regulation of NOL7 expression is also observed in RB-deficient human tumors irrespective of the mechanism of RB inactivation.
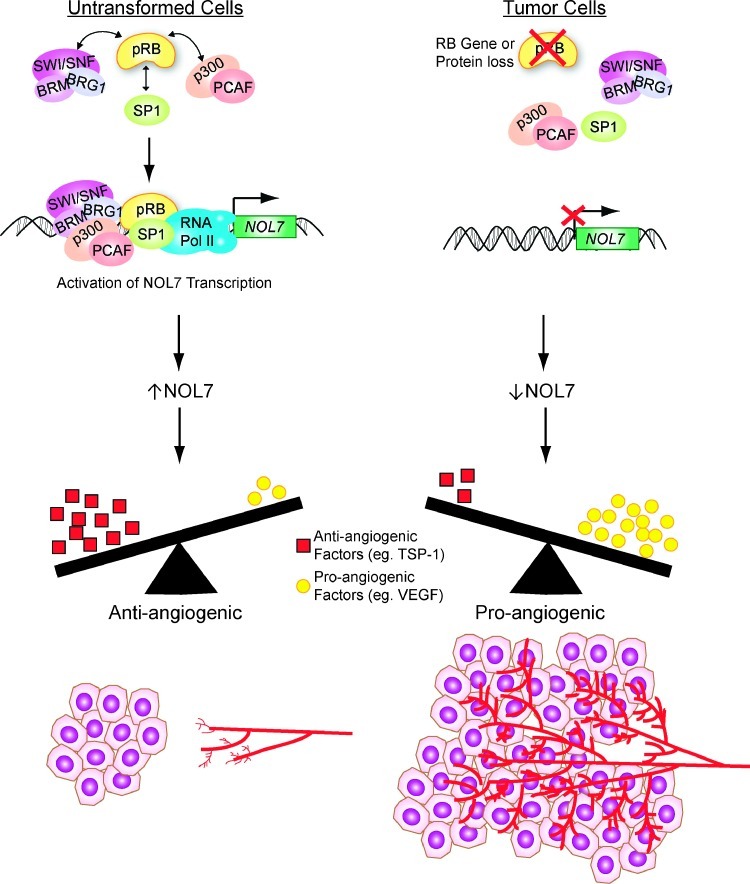
Traditionally, pRB is considered to be a repressor of E2F target gene transcription [37,52]. Recent reports have demonstrated that pRB can also synergize with site-specific TFs to activate transcription [19,20]. We have demonstrated that pRB positively regulates NOL7 transcription by recruiting transcriptional coactivators and remodeling proteins to its promoter (Figure 8). These findings are in agreement with our preliminary data showing increased acetylated histone H4 enrichment at the NOL7 promoter (data not shown). Future studies will focus on determining the changes in chromatin conformation and histone modification at the NOL7 promoter on pRB loss. We determined that the NOL7 promoter has multiple pRB-TF complex binding sites, which collectively contribute to NOL7 transcriptional activation (Figure 6). The p300 coactivator protein is known to facilitate enhancer activity for several genes [53–55]. We have shown that pRB recruits p300 to the NOL7 promoter; therefore, it may be important to characterize each of the pRB-TF binding sites to determine if they possess enhancer activity.
Figure 8.
Model illustrating the mechanism by which pRB inhibits angiogenesis through transcriptional activation of NOL7.
pRB regulates diverse cellular processes such as differentiation, apoptosis, growth arrest, cell cycle, and metabolism in a context-specific manner. However, a role for pRB in inhibiting angiogenesis has not been clearly established [15]. Studies have implicated phosphorylated forms of the pRB family proteins in promoting angiogenesis, suggesting that hypophosphorylated and active pRB may inhibit angiogenesis [56]. A recent study showed that pRB-E2F complexes repress endothelial cell vascular endothelial growth factor receptor transcription [57]. Moreover, RB-deficient tumors that have increased vascularity [58–61] exhibited significantly reduced NOL7 protein expression (Figure 7). We have previously demonstrated that reexpressing NOL7 in CC cells that have reduced pRB and NOL7 expression inhibited tumor angiogenesis [6]. This suggests that there may be a connection between RB loss-mediated reduction in NOL7 expression and increased vascularity observed in RB-deficient tumors. Our data suggest that RB-mediated activation of NOL7 transcription may be crucial for maintaining cells in a relatively anti-angiogenic state through steady NOL7 expression. Subsequently, inactivation of RB in human malignancies negatively regulates NOL7 expression, thereby shifting the tumor microenvironment to a more pro-angiogenic state that stimulates tumor growth and progression (Figure 8).
In conclusion, this study has further characterized the transcriptional activator function of pRB. Our data also provide evidence to support the hypothesis that reactivation of pRB may influence multiple antiangiogenic pathways through its regulation of NOL7. Several natural and synthetic compounds have demonstrated their anti-angiogenic activity by increasing levels of hypophosphorylated and active pRB [56,62]. This knowledge may be exploited in the future to design more comprehensive anti-angiogenic therapies that target multiple angiogenic pathways, thereby promising longer term efficacy.
Supplementary Material
Acknowledgments
We thank Kay Macleod for her valuable input and help in setting up this study.
Abbreviations
- CC
cervical cancer
- NOL7
nuclear protein 7
- VEGF
vascular endothelial growth factor
- TSP-1
thrombospondin-1
Footnotes
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J, Hanahan D. Switch to the angiogenic phenotype during tumorigenesis. Princess Takamatsu Symp. 1991;22:339–347. [PubMed] [Google Scholar]
- 3.Grépin R, Pagès G. Molecular mechanisms of resistance to tumour anti-angiogenic strategies. J Oncol. 2010;2010:835680. doi: 10.1155/2010/835680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med. 2011;17:1359–1370. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 5.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasina R, Pontier AL, Fekete MJ, Martin LE, Qi XM, Brigaudeau C, Pramanik R, Cline EI, Coignet LJ, Lingen MW. NOL7 is a nucleolar candidate tumor suppressor gene in cervical cancer that modulates the angiogenic phenotype. Oncogene. 2006;25:588–598. doi: 10.1038/sj.onc.1209070. [DOI] [PubMed] [Google Scholar]
- 7.Doçi CL, Zhou G, Lingen MW. The novel tumor suppressor NOL7 post-transcriptionally regulates thrombospondin-1 expression. Oncogene. doi: 10.1038/onc.2012.464. (in revision) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doçi CL, Mankame TP, Langerman A, Ostler KR, Kanteti R, Best T, Onel K, Godley LA, Salgia R, Lingen MW. Characterization of NOL7 gene point mutations, promoter methylation, and protein expression in cervical cancer. Int J Gynecol Pathol. 2012;31:15–24. doi: 10.1097/PGP.0b013e318220ba16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L, Zheng M, Zhou Q-M, Zhang M-Y, Yu Y-H, Yun J-P, Wang H-Y. Identification of a 7-gene signature that predicts relapse and survival for early stage patients with cervical carcinoma. Med Oncol. 2012;29:2911–2918. doi: 10.1007/s12032-012-0166-3. [DOI] [PubMed] [Google Scholar]
- 10.Rader JS, Gerhard DS, O'Sullivan MJ, Li Y, Li L, Liapis H, Huettner PC. Cervical intraepithelial neoplasia III shows frequent allelic loss in 3p and 6p. Genes Chromosomes Cancer. 1998;22:57–65. doi: 10.1002/(sici)1098-2264(199805)22:1<57::aid-gcc8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Borecki I, Nguyen L, Ma D, Smith K, Huettner PC, Mutch DG, Herzog TJ, Gibb RK, Powell MA, et al. CD83 gene polymorphisms increase susceptibility to human invasive cervical cancer. Cancer Res. 2007;67:11202–11208. doi: 10.1158/0008-5472.CAN-07-2677. [DOI] [PubMed] [Google Scholar]
- 12.Mankame TP, Zhou G, Lingen MW. Identification and characterization of the human NOL7 gene promoter. Gene. 2010;456:36–44. doi: 10.1016/j.gene.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghittoni R, Accardi R, Hasan U, Gheit T, Sylla B, Tommasino M. The biological properties of E6 and E7 oncoproteins from human papilloma-viruses. Virus Genes. 2009;40:1–13. doi: 10.1007/s11262-009-0412-8. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Li F, Mead L, White H, Walker J, Ingram DA, Roman A. Human papillomavirus causes an angiogenic switch in keratinocytes which is sufficient to alter endothelial cell behavior. Virology. 2007;367:168–174. doi: 10.1016/j.virol.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 17.Nam EJ, Kim JW, Kim SW, Kim YT, Kim JH, Yoon BS, Cho NH, Kim S. The expressions of the Rb pathway in cervical intraepithelial neoplasia; predictive and prognostic significance. Gynecol Oncol. 2007;104:207–211. doi: 10.1016/j.ygyno.2006.07.043. [DOI] [PubMed] [Google Scholar]
- 18.Johung K, Goodwin EC, DiMaio D. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J Virol. 2007;81:2102–2116. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decesse JT, Medjkane S, Datto MB, Cremisi CE. RB regulates transcription of the p21/WAF1/CIP1 gene. Oncogene. 2001;20:962–971. doi: 10.1038/sj.onc.1204169. [DOI] [PubMed] [Google Scholar]
- 20.Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, Haigis K, Gulino A, Lees JA. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell. 2009;15:184–194. doi: 10.1016/j.ccr.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howley P, Scheffner M, Huibregtse J, Münger K. Oncoproteins encoded by the cancer-associated human papillomaviruses target the products of the retinoblastoma and p53 tumor suppressor genes. Cold Spring Harb Symp Quant Biol. 1991;56:149–155. doi: 10.1101/sqb.1991.056.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Roberts J, Dakic A, Zhang Y, Schlegel R. HPV E7 contributes to the telomerase activity of immortalized and tumorigenic cells and augments E6-induced hTERT promoter function. Virology. 2008;375:611–623. doi: 10.1016/j.virol.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dirlam A, Spike BT, Macleod KF. Deregulated E2f-2 underlies cell cycle and maturation defects in retinoblastoma null erythroblasts. Mol Cell Biol. 2007;27:8713–8728. doi: 10.1128/MCB.01118-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flowers S, Beck GR, Moran E. Transcriptional activation by pRB and its coordination with SWI/SNF recruitment. Cancer Res. 2010;70:8282–8287. doi: 10.1158/0008-5472.CAN-10-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 2007;26:412–423. doi: 10.1038/sj.emboj.7601516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira KF, Levings PP, Hill MA, Crusselle VJ, Kang SH, Engel JD, Bungert J. Recruitment of transcription complexes to the β-globin gene locus in vivo and in vitro. J Biol Chem. 2004;279:50350–50357. doi: 10.1074/jbc.M408883200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng W-G, Zhu Y, Montero A, Wu KK. Quantitative analysis of binding of transcription factor complex to biotinylated DNA probe by a streptavidin-agarose pulldown assay. Anal Biochem. 2003;323:12–18. doi: 10.1016/j.ab.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Wu KK. Analysis of protein-DNA binding by streptavidin-agarose pull-down. Methods Mol Biol. 2006;338:281–290. doi: 10.1385/1-59745-097-9:281. [DOI] [PubMed] [Google Scholar]
- 30.Darnell GA, Schroder WA, Antalis TM, Lambley E, Major L, Gardner J, Birrell G, Cid-Arregui A, Suhrbier A. Human papillomavirus E7 requires the protease calpain to degrade the retinoblastoma protein. J Biol Chem. 2007;282:37492–37500. doi: 10.1074/jbc.M706860200. [DOI] [PubMed] [Google Scholar]
- 31.Lehman TA, Modali R, Boukamp P, Stanek J, Bennett WP, Welsh JA, Metcalf RA, Stampfer MR, Fusenig N, Rogan EM, et al. p53 Mutations in human immortalized epithelial cell lines. Carcinogenesis. 1993;14:833–839. doi: 10.1093/carcin/14.5.833. [DOI] [PubMed] [Google Scholar]
- 32.Macleod KF, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 33.Phelps WC, Yee CL, Munger K, Howley PM. The human papilloma-virus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell. 1988;53:539–547. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- 34.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murai Y, Hayashi S, Takahashi H, Tsuneyama K, Takano Y. Correlation between DNA alterations and p53 and p16 protein expression in cancer cell lines. Pathol Res Pract. 2005;201:109–115. doi: 10.1016/j.prp.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- 37.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 38.Werner T. Computer-assisted analysis of transcription control regions. Matinspector and other programs. Methods Mol Biol. 2000;132:337–349. doi: 10.1385/1-59259-192-2:337. [DOI] [PubMed] [Google Scholar]
- 39.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucl Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen P-L, Scully P, Shew J-Y, Wang JYJ, Lee W-H. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 41.Burke JR, Deshong AJ, Pelton JG, Rubin SM. Phosphorylationinduced conformational changes in the retinoblastoma protein inhibit E2F trans-activation domain binding. J Biol Chem. 2010;285:16286–16293. doi: 10.1074/jbc.M110.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knudsen ES, Wang JY. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Connell-Crowley L, Harper JW, Goodrich DW. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lees JA, Buchkovich KJ, Marshak DR, Anderson CW, Harlow E. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J. 1991;10:4279–4290. doi: 10.1002/j.1460-2075.1991.tb05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee EY, Cam H, Ziebold U, Rayman JB, Lees JA, Dynlacht BD. E2F4 loss suppresses tumorigenesis in Rb mutant mice. Cancer Cell. 2002;2:463–472. doi: 10.1016/s1535-6108(02)00207-6. [DOI] [PubMed] [Google Scholar]
- 47.Fiedler M, Müller-Holzner E, Viertler H-P, Widschwendter A, Laich A, Pfister G, Spoden GA, Jansen-Dürr P, Zwerschke W. High level HPV-16 E7 oncoprotein expression correlates with reduced pRb-levels in cervical biopsies. FASEB J. 2004;18:1120–1122. doi: 10.1096/fj.03-1332fje. [DOI] [PubMed] [Google Scholar]
- 48.Wiest T, Schwarz E, Enders C, Flechtenmacher C, Bosch FX. Involvement of intact HPV16 E6/E7 gene expression in head and neck cancers with unaltered p53 status and perturbed pRb cell cycle control. Oncogene. 2002;21:1510–1517. doi: 10.1038/sj.onc.1205214. [DOI] [PubMed] [Google Scholar]
- 49.Yuan J, Knorr J, Altmannsberger M, Goeckenjan G, Ahr A, Scharl A, Strebhardt K. Expression of p16 and lack of pRB in primary small cell lung cancer. J Pathol. 1999;189:358–362. doi: 10.1002/(SICI)1096-9896(199911)189:3<358::AID-PATH452>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 50.Damjanovich J, Adány R, Berta A, Beck Z, Balázs M. Mutation of the RB1 gene caused unilateral retinoblastoma in early age. Cancer Genet Cytogenet. 2000;119:1–7. doi: 10.1016/s0165-4608(99)00198-3. [DOI] [PubMed] [Google Scholar]
- 51.Cook KM, Figg WD. Angiogenesis inhibitors: current strategies and future prospects. CA Cancer J Clin. 2010;60:222–243. doi: 10.3322/caac.20075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H-Z, Tsai S-Y, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowling T, Bernadt C, Johnson L, Desler M, Rizzino A. The co-activator p300 associates physically with and can mediate the action of the distal enhancer of the FGF-4 gene. J Biol Chem. 2003;278:13696–13705. doi: 10.1074/jbc.M207567200. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gabellini C, Del Bufalo D, Zupi G. Involvement of RB gene family in tumor angiogenesis. Oncogene. 2006;25:5326–5332. doi: 10.1038/sj.onc.1209631. [DOI] [PubMed] [Google Scholar]
- 57.Pillai S, Kovacs M, Chellappan S. Regulation of vascular endothelial growth factor receptors by Rb and E2F1: role of acetylation. Cancer Res. 2010;70:4931–4940. doi: 10.1158/0008-5472.CAN-10-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossler J, Dietrich T, Pavlakovic H, Schweigerer L, Havers W, Schuler A, Bornfeld N, Schilling H. Higher vessel densities in retinoblastoma with local invasive growth and metastasis. Am J Pathol. 2004;164:391–394. doi: 10.1016/S0002-9440(10)63129-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaghloul MS, El Naggar M, El Deeb A, Khaled H, Mokhtar N. Prognostic implication of apoptosis and angiogenesis in cervical uteri cancer. Int J Radiat Oncol Biol Phys. 2000;48:1409–1415. doi: 10.1016/s0360-3016(00)00800-2. [DOI] [PubMed] [Google Scholar]
- 60.Lucchi M, Mussi A, Fontanini G, Faviana P, Ribechini A, Angeletti CA. Small cell lung carcinoma (SCLC): the angiogenic phenomenon. Eur J Cardiothorac Surg. 2002;21:1105–1110. doi: 10.1016/s1010-7940(02)00112-4. [DOI] [PubMed] [Google Scholar]
- 61.Mikulić D, Ilić I, Cepulić M, Orlić D, Giljević JS, Fattorini I, Seiwerth S. Tumor angiogenesis and outcome in osteosarcoma. Pediatr Hematol Oncol. 2004;21:611–619. doi: 10.1080/08880010490501015. [DOI] [PubMed] [Google Scholar]
- 62.Stengel KR, Thangavel C, Solomon DA, Angus SP, Zheng Y, Knudsen ES. Retinoblastoma/p107/p130 pocket proteins: protein dynamics and interactions with target gene promoters. J Biol Chem. 2009;284:19265–19271. doi: 10.1074/jbc.M808740200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.