Abstract
Acute myeloid leukemia (AML) is a genetically heterogeneous clonal disorder characterized by two molecularly distinct self-renewing leukemic stem cell (LSC) populations most closely related to normal progenitors and organized as a hierarchy. A requirement for WNT/β-catenin signaling in the pathogenesis of AML has recently been suggested by a mouse model. However, its relationship to a specific molecular function promoting retention of self-renewing leukemia-initiating cells (LICs) in human remains elusive. To identify transcriptional programs involved in the maintenance of a self-renewing state in LICs, we performed the expression profiling in normal (n = 10) and leukemic (n = 33) human long-term reconstituting AC133+ cells, which represent an expanded cell population in most AML patients. This study reveals the ligand-dependent WNT pathway activation in AC133bright AML cells and shows a diffuse expression and release of WNT10B, a hematopoietic stem cell regenerative-associated molecule. The establishment of a primary AC133+ AML cell culture (A46) demonstrated that leukemia cells synthesize and secrete WNT ligands, increasing the levels of dephosphorylated β-catenin in vivo. We tested the LSC functional activity in AC133+ cells and found significant levels of engraftment upon transplantation of A46 cells into irradiated Rag2-/-γc-/- mice. Owing to the link between hematopoietic regeneration and developmental signaling, we transplanted A46 cells into developing zebrafish. This system revealed the formation of ectopic structures by activating dorsal organizer markers that act downstream of the WNT pathway. In conclusion, our findings suggest that AC133bright LSCs are promoted by misappropriating homeostatic WNT programs that control hematopoietic regeneration.
Introduction
Different genetic causes result in variable clinical courses of acute myeloid leukemia (AML) and different responses to standard chemotherapy including stem cell transplant. Despite the genetic differences among individual patients, most AML clones display certain common features. Ample evidence exists in mouse models that AML develops through the stepwise acquisition of collaborating genetic and epigenetic changes in self-renewing LICs, which exhibit a committed myeloid immunophenotype and give rise to nonleukemogenic progeny in a myeloid-restricted hierarchy [1–3]. An important issue to understand the early events in the origin of AML is the observation that long-term hematopoietic stem cell (LT-HSC) expansion precedes the generation of committed myeloid LICs [4].
Although well-orchestrated cell intrinsic programs and environmental cues represent the main contributory factors for normal LT-HSC expansion, it is still unclear if transcriptional programs responsible for the expansion of premalignant LT-HSC populations and leukemia initiation share common embryonic or post-embryonic functions, such as stem cell renewal, tissue repair, and regeneration [5,6]. Even though recent studies have addressed the role of Hedgehog signaling for maintenance of cancer stem cells in myeloid leukemia [7], its requirement in AML remains controversial [8].
Recently, the notion that LICs are restricted only to the CD34+CD38- population has been challenged [9,10] and it has been suggested that more cell surface markers could be appropriately used to enrich the leukemia-initiating cell (LIC)-containing fraction. One of such markers is the AC133 antigen (a glycosylation-dependent epitope of CD133) that defines a desirable population of stem and progenitor cells containing in turn all the CD34brightCD38- progenitors, as well as the CD34brightCD38+ cells committed to the granulocytic/monocytic lineage [11]. In addition, AC133 represents a well-documented marker of tumor-initiating cells in a number of human cancers [12]. In this study, fluorescence-activated cell sorter (FACS) analysis demonstrates that AC133+ cell population is dramatically expanded in 25 AML cases analyzed. We carried out genome-wide transcriptional analysis of AC133+ cells isolated from newly diagnosed non-promyelocytic AML patients (n = 33) andhealthy donors (n = 10). Results obtained from a multistep analysis of the generated data defined the involvement of the ligand-dependent WNT receptor signaling pathway as the self-renewal associated signature in the AC133-enriched fraction in human AML. Furthermore, the results presented here suggested that WNT10B and other WNT genes expressed during the regenerative process of the hematopoietic system [13,14] are aberrantly upregulated in AC133bright AML cells. To obtain a localized detection of each single transcript, we first applied an in situ detection of individual mRNA molecules [15] on bone marrow (BM) sections from AML patients. By the establishment of a primary culture of AC133+ AML cells (termed A46 hereafter), we confirmed that secreted WNTs activated a β-catenin/human T-cell factor (TCF) transcription-based reporter construct. Moreover, we intend to clarify the relationship between the abnormal WNT activation in AC133+ population and the leukemic stem cell (LSC) activity. Using Rag2-/-γc-/- as immunodeficient xenotransplant model [16], AC133+ A46 cells were injected intravenously into sublethally irradiated mice.
To achieve a complete view of how AC133+ A46 cells modulated the microenvironment and given that hematopoietic regeneration converge to developmental signaling, we used zebrafish embryonic model as an in vivo biosensor.
Our results confirmed previously reported data [17] and raise new important implications for the involvement of the ligand-dependent canonical WNT pathway in AML. These suggestive findings are supported by the pivotal function of WNT in promoting self-renewal [18,19], its emerging role in myeloid leukemogenesis [20,21], and the effects of its constitutive activation through a stabilized form of β-catenin, by inducing quiescent stem cells to enter the cell cycle and arresting their differentiation [22,23].
Materials and Methods
Collection of Patient Samples and Normal Hematopoietic Cells
BM MNCs were collected from 33 newly diagnosed, unselected non-promyelocytic AML patients, according to Niguarda Hospital's Ethical Board-approved protocols (116_04/2010). According to the revised Medical Research Council risk group stratification, based on cytogenetic and molecular markers/mutations [24], samples included 14 adverse, 13 intermediate, and 6 favorable risk patients. Human adult BM cells obtained from 10 consenting healthy donors were processed as previously described [25].
Cell Sorting and Flow Cytometry
We carried out AC133+ cell separation based on MACS MicroBeads and cytofluorometric determinations, as previously described [25].
Microarray Expression Analysis
Total RNA for expression profiling was extracted using RNAqueous-4PCR kit (Ambion, Austin, TX) from AC133-selected cells. Expression profiling was performed on Affymetrix HGU133plus2.0 GeneChip arrays according to the manufacturer's procedures. The bioinformatics analysis performed in this study was realized using the R language for statistical computing (http://www.r-project.org/) and the annotation libraries provided by the Bioconductor project (http://www.bioconductor.org/). Microarray data have been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), with accession number E-MTAB-220. We performed a genome-wide analysis to select genes differentially expressed between AML AC133+ patients and AC133+ healthy donors (Welch t test, 0.05 significance level). The resulting set of differentially expressed genes has been analyzed for functional enrichment with respect to the terms of the Biological Process (BP) branch of the Gene Ontology (GO) and the pathways of the KEGG database. We relied on three different methods for functional enrichment analysis: GOStats (version 2.12.0 of the Bioconductor package, http://www.bioconductor.org/packages/release/bioc/html/GOstats.html) [26], Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/home.jsp) [27], and the iterative procedure of dysregulated pathway analysis proposed by Majeti et al. [17]. The first two tests are based on the hypergeometric distribution, whereas the last one is based on a non-parametric test and on an iterative procedure. Overrepresented GO terms and KEGG pathways have been selected at 0.05 significance level.
WNT/β-catenin Responsive Luciferase Assay
HEK293T cells grown in 24-well plates at a density of 1.7 x 105 cells per well were transfected with M50 Super 8x TOPFlash (plasmid 12456; Addgene, Cambridge, MA) and pRL-TK (Renilla luciferase; Promega, Madison, WI) using jetPEI (Polyplus, New York, NY). Cells were treated for 12 hours with A46 conditioned medium (CM) or HEK293T cells transfected with BA-WNT10B (plasmid 1831; Addgene) CM as positive control. WNT10B expression in HEK293T transfected with BA-WNT10B was evaluated by SYBR Green-based real-time reverse transcription-polymerase chain reaction (RT-PCR) using WNT10B FW-5′-GCTGTAACCATGACATGGAC-3′ and RW-5′-CTGCCTGATGTGCCATGAC-3′ specific primers. Luciferase activity measurement was performed with the Dual-Luciferase Reporter Assay System (Promega).
Immunoblot
Protein expression was assessed by immunoblot analysis using standard procedures, applying anti-active β-catenin (ABC) monoclonal mouse (anti-ABC clone 8E7; Millipore, Billerica, MA), anti-β-catenin monoclonal rabbit (E247; Abcam, Cambridge, United Kingdom), anti-WNT10B polyclonal rabbit (H-70; Santa Cruz Biotechnology, Inc, Santa Cruz, CA), anti-WNT10B monoclonal mouse (5A7; Abcam), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) polyclonal rabbit (ab97626; Abcam), and anti-Pygopus 2 polyclonal rabbit (H-216; Santa Cruz Biotechnology, Inc) antibodies. Secondary antibodies used were anti-mouse HRP, anti-goat HRP, and anti-rabbit HRP (Thermo Fisher Scientific, Waltham, MA).
In Situ mRNA Detection
In situ detection of individual mRNA molecules was performed as described [15]. One micromolar of locked nucleic acid-modified cDNA primer (WNT10B, 5′-C+A+G+G+C+CGGACAGCGTCAAGCACACG-3′; β-actin, 5′-C+TG+AC+CC+AT+GCCCACCATCACGCCC-3′; Exiqon, Vedbaek, Denmark) was added to the reverse transcriptase reaction. Ligation was then carried out with 0.1 µM of the WNT10B padlock probe or β-actin padlock probe (WNT10B, 5′-[Phos]ACCGTGCCTGTCGGACCCTCCTCTATGATTACTGACCTAAGTCGGAAGTACTACTCTCTTCTTCTTTTAGTGAAGCCCAGGCAACCCA-3′; β-actin, 5′-[Phos] GCCGGCTTCGCGGGCGACGATTCCTCTATGATTACTGACCTATGCGTCTATTTAGTGGAGCCTCTTCTTTACGGCGCCGGCATGTGCAAG-3′; Sigma-Aldrich, St Louis, MO). Rolling circle products (RCPs) were visualized using 100 nM of detection probe (WNT10B, 5′-Cy5-AGTCGGAAGTACTACTCTCT-3′ and β-actin, 5′-Cy3-TGCGTCTATTTAGTGGAGCC-3′; Sigma-Aldrich). Nuclei were counterstained with 100 ng/ml Hoechst 33258 (Sigma-Aldrich). Images of BM tissue slides were acquired using an Axioplan II epifluorescence microscope (Zeiss, Munchen, Germany) equipped with a charge-coupled device (CCD) camera (HRM, Zeiss) and a computer-controlled filter wheel with excitation and emission filters for visualization of 4′-6-diamidino-2-phenylindole (DAPI), Cy3, and Cy5. A x20 objective (Plan-Apocromat, Zeiss) was used for capturing the images. Images were collected using the Axiovision software (release 4.3, Zeiss). Images were collected as z-stacks to ensure that all RCPs were imaged, with a maximum intensity project created in Axiovision. For quantification, the numbers of RCPs and cell nuclei in images were counted digitally using CellProfiler software (www.cellprofiler.org) on three x20 microscope images. The total number of RCPs was divided by the number of nuclei for each image. The average for each sample was then calculated from the result of the three images and is reported as RCPs per cell.
Immunostaining
Direct and indirect immunostaining were performed following standard procedures. BM biopsies of AML patients, previously embedded in paraffin blocks, were cut in 5-µm-thick sections and mounted on slides. Slides were incubated with primary antibody mouse anti-CD133.1 (AC133) (1:100; Miltenyi Biotec GmBH, Bergisch Gladbach, Germany), directly labeled with 488-nm dye (Sigma), and with primary antibodies such as mouse anti-ABC (1:100; Millipore), rabbit anti-WNT10B (H70) (1:100; Santa Cruz Biotechnology, Inc), rabbit anti-Pygopus 2 (H-216) (1:100; Santa Cruz Biotechnology, Inc), goat anti-SMYD3 (F-19) (1:100; Santa Cruz Biotechnology, Inc). Samples were incubated with the secondary antibodies donkey anti-mouse Alexa Fluor 488 (1:500; Life Technologies, Carlsband, CA), donkey anti-rabbit Alexa Fluor 568 (1:500; Life Technologies), and donkey anti-goat Alexa Fluor 568 (1:500; Life technologies). Nuclei were counterstained with 100 ng/ml DAPI (Sigma-Aldrich). Cells were analyzed using the upright microscope (Leica, DM 4000B).
Immunofluorescence Image Analysis
Image analysis was performed by means of a custom automatic routine and the ImageJ program (1.43s; National Institutes of Health, Bethesda, MD). Maps of ABC+/WNT10B+ cells were obtained using an automatic threshold based on moments algorithm newly implemented in the ImageJ program and mathematical morphology plugin developed by D. Prodanov (http://rsbweb.nih.gov/ij/plugins/gray-morphology.html). DAPI staining was used to identify nuclei. Finally, images of nuclei for positive cells were obtained by Boolean AND operations between DAPI staining and cell maps. The resulting images were used to determine the percentage of ABC+/WNT10B+ cells both manually or through the Analyze articles function of ImageJ.
Cell Culture
Selected AC133+ cells from BM at AML diagnosis were cultured for 16 weeks. The culture was performed using synthetic medium StemSpam H3000 (StemCell Technologies, Vancouver, Canada) in the absence of serum and cytokines. StemSpan H3000 and HPGM media, conditioned by the cell culture, were collected after 12 weeks, refined with 0.2-µm filter and stored at -20°C. HEK293T cells were grown in Dulbecco's modified Eagle's medium (high glucose with sodium pyruvate and l-glutamine; Euroclone), supplemented with 10% FBS (Euroclone) and penicillin-streptomycin solution (100x) (Euroclone, Milano, Italy).
Mice and Xenogeneic Transplantation
Rag2-/-γc-/- BALB/c mice were bred and maintained under specific pathogen-free conditions in the mouse facility of Istituto Oncologico Veneto, and experiments were performed according to state guidelines and approved by the local ethics committee. Rag2-/-γc-/- mice at 6 weeks were sublethally irradiated with 5 Gy and transplanted via the tail vein with 1 x 106 of human AC133+ AML cells (A46).
Evaluation of Hematopoietic Chimerism by Flow Cytometry
Three weeks after transplantation, recipient mice were sacrificed and BM cells were harvested by flushing femurs and tibias. BM engraftment was evaluated using human antibodies CD34, CD38, AC133, and CD45 [BD Biosciences (Bedford, MA) and Miltenyi Biotec GmBH]. Multicolor flow cytometric analyses were performed using FACSCalibur flow cytometer (BD Biosciences) and analyzed by FlowJo software (Tree Star, Ashland, OR). Cell engraftment was determined by expression of the human panleukocyte marker CD45.
Zebrafish Models and Transplantation Procedures
Embryos were handled according to relevant guidelines. Fish of the AB strain were maintained at 28°C on a 14-hour light/10-hour dark cycle and collected by natural spawning. Transplantation of human A46 cells into zebrafish embryos was performed as previously reported [28]. Briefly, fluorescently labeled A46 cells were resuspended in 1x phosphate-buffered saline and injected into zebrafish blastulae (between 100 and 200 cells per injection) at 3 hours post-fertilization (hpf). Injected live embryos were observed under a fluorescent microscope at 30% of epiboly to ensure the presence of labeled A46 cells. Embryos were collected at the desired developmental stages, immediately fixed, and processed for whole-mount in situ hybridization according to Thisse et al. [29], using gsc, ntl, and pax2a DIG-labeled riboprobes.
Results
AC133+ Cells Are Highly Expanded in AML
AC133 antigen is restricted to a rare cell population with long-term reconstituting activity, ranging from 20% to 60% of all CD34+ cells, and resulting barely detectable in CD34-Lin- cells [11–30]. We have previously shown that AC133+ LT-HSCs are also highly enriched in colony forming units and have a stronger granulocyte/macrophage differentiation potential relative to unselected BM MNCs. However, their burst-forming units-erythroid forming potential is lower [25]. To determine the range of expansion of AC133+ cell fraction in AML, flow cytometric quantification of CD133.1 (AC133) expression either in single staining or in combination with the pan-hematopoietic marker CD45 was performed in BM on 25 primary non-promyelocytic AML samples and 10 age-matched healthy volunteer adult donors. The resulting CD133.1+ cell fraction results expanded among AML patients by a median of 31.5% [interquartile range (IQR), 16.5%-53.4%] with respect to normal donors (median, 0.54%; IQR, 0.17%-1.14%; P < .0001; Figure 1, A–C). To directly compare the gene expression profiles of purified populations highly enriched in LSCs or HSCs, positive selection of CD133.1+ cells was performed on all the 33 non-promyelocytic AML patients (25 de novo, 7 secondary to myelodysplasia, and 1 secondary therapy-related; Table W1) as well as on the 10 healthy donors recruited to this study, respectively. MNC selection in samples from AML patients led to an average of 236-fold enrichment of AC133-positive cells (IQR, 142.72–419.44, data not shown).
Figure 1.
Human AC133+ cells are strongly expanded in AML. Representative dot plots of the immunophenotype analysis from the BM of (A) a healthy donor and (B) a patient with AML (AML No. 42 in Table W1). The CD45/CD133.1 co-staining was gated on BM MNCs; percentages on total cellularity are shown for gated normal and AML populations. (C) Flow cytometry analysis of the CD133.1 antigen in BM MNCs of healthy donors (n = 10) and AML patients (n = 25). The IQR for each sample group is indicated in the dot. Mann-Whitney U test was used to calculate the P value (α = 0.001).
Identification of Dysregulated Pathways in AC133+ AML Cell Fraction
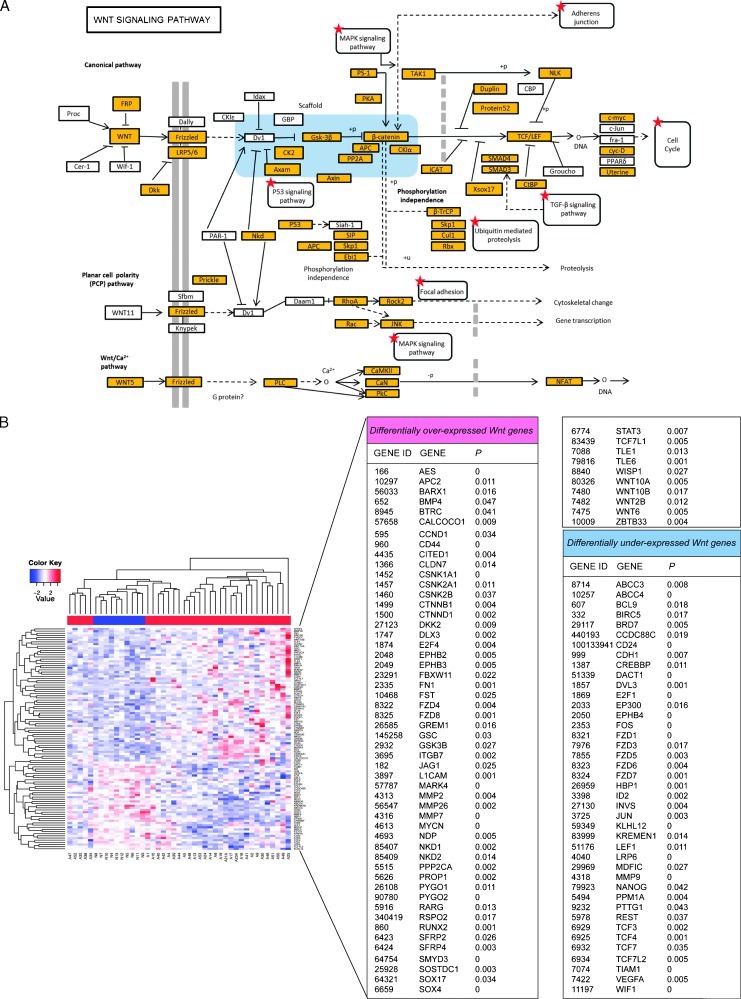
To identify the dysregulated pathways in AC133+ AML cell fraction versus normal long-term reconstituting AC133+ HSC cells, we performed a genome-wide functional enrichment analysis on gene expression microarray data of 33 AML patients and 10 healthy donors. The identification of overrepresented pathways in AC133+ AML cells was realized through three computational tools: GOStats [26], DAVID [27], and dysregulated pathway analysis according to Majeti et al. [17]. Employing the functional terms from GO BPs and the pathway information from KEGG databases, we found 212 functionally enriched GO BP terms with GOStats (P < .01), 284 GO BP terms with DAVID (P < .05), and 616 GO BP terms with the non-parametric test of Majeti et al. (P < .05). Moreover, GOStats selected 16 KEGG pathways (P < .05), DAVID selected 24 KEGG pathways (P < .05), and the dysregulated pathway analysis selected 3 KEGG pathways (P < .05). As shown in Table W2, DAVID ranks the WNT signaling pathway (P = .022) among the first 10 dysregulated pathways in AC133+ AML cells. The visualization of the WNT molecular interaction in KEGG database is presented in Figure 2A.
Figure 2.
Schematic visualization of dysregulated networks in KEGG database and representation of WNT gene expression profiles. (A) Visualization of WNT signaling pathway in the KEGG database. Highlighted genes are differentially expressed and red-starred pathways are overrepresented in AC133+ AML cells. (B) Heat map of the differentially expressed WNT-associated genes. The color bar under the patient sample dendrogram identifies AML samples (red) and healthy controls (blue). The names, accession numbers, as well as P values for the differentially overexpressed and underexpressed genes are shown in the right panels.
It is worth noting that different statistical methods agree in identifying the WNT pathway as a significant dysregulated pathway in AC133+ AML cells. Moreover, the WNT signaling pathway in KEGG is selected as overrepresented in AC133+ AML stem cells by both GOStats and DAVID. Taken together, the functional enrichment methods select the term “WNT receptor signaling pathway” (GO:0016055) as the most specific self-renewal associated dysregulated pathway.
WNT Gene Expression Profiles of Normal and Leukemia Long-term Reconstituting AC133+ Cells
To obtain insights into the WNT pathway in AC133+ leukemia cells, we focused our analysis on the probe sets annotated to “WNT receptor signaling pathway” GO class or to any of its children GO terms. The resulting probe set list was complemented with the probes mapping to genes in a manually curated list of WNT target genes and with probes mapping to other well-known WNT genes, not included in the previous two lists. The total list of analyzed WNT genes includes 480 probe sets, mapping to 193 different genes (Table_A in http://homes.dsi.unimi.it/∼re/TRdataset/). To assess differential expression of WNT genes, we employed Welch t tests (two-tailed) on the 480 probe sets, thus identifying 103 differentially expressed genes (P < .05; Figure 2B). Genes shown to be highly AML-specific include the WNT ligands WNT2B, WNT6, WNT10A, and WNT10B [14], the WNT/β-catenin signaling agonists including SMYD3 [31], DKK2 [32], SOX4 [33], PROP-1 [34], and PYGO2 [35,36], antagonists including WIF-1 [37], KLHL12 [38], LRP6 [39], KREMEN1 [40], E2F1 [41], DACT1 [42], and HBP1 [43], and the deregulated WNT targets including STAT3, MYCN, ABCC4, DLX3, MARK4, RUNX2, CD24, and CD44 [44]. Collectively, these data are consistent with ligand-dependent activation of the regeneration-associated WNT pathway [14].
We also investigated whether the expression of the WNT genes varied between the risk groups in AML (favorable, intermediate, and adverse) revised by Smith et al. [24]. Clustering results of AML cases restricted to WNT genes show that the risk groups are not clearly distinguishable in separate clusters (Figure 2B). Analysis of variance between groups confirms these results: only 3 (ABCC4, HBP1, and NDP) of 193 WNT-related genes are differentially expressed across the three categories (P < .05). Thus, our data do not support a significant distinction between risk groups in AML patients. This statement should be considered with caution, because the cardinality of the subgroups is relatively low (6 favorable, 13 intermediate, and 14 adverse samples).
Hyperactive WNT Signaling Resulted Activated in Diffuse AC133bright AML Cells Expressing the Hematopoietic Regenerative Molecule WNT10B
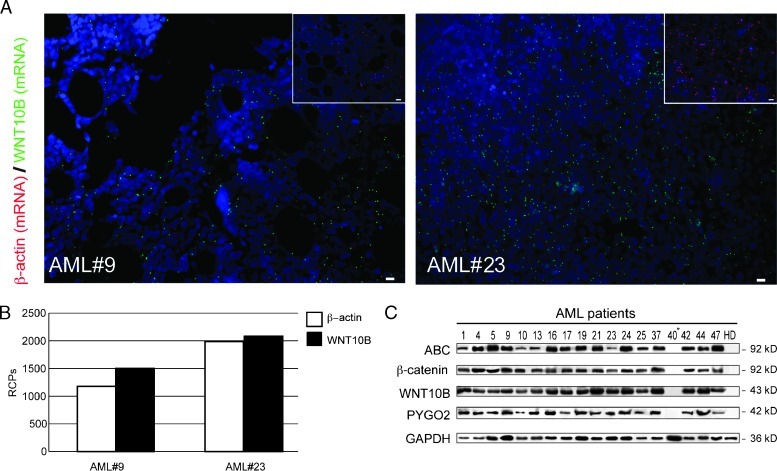
Most of what is known about hematopoietic regeneration point to WNT signaling pathway and specifically to WNT10B [13,45]; therefore, we investigated how expression of WNT10B is related to AML phenotype. To fully understand mRNA distribution of WNT10B within BM sections, we focused on the application of single molecule detection methods. In situ mRNA detection by target-primed rolling circle amplification analysis offers high sensitivity and localized detection within single cells and tissues [15]. Following this approach, we detected WNT10B-related transcript in BM sections obtained at diagnosis from two randomly selected patients. β-Actin was included as a reference transcript in consecutive sections. Visualization by using high-performance fluorescence microscopy showed a diffuse localization pattern in the tissues (Figure 3A). Signal distribution and RCP quantification showed a β-actin/WNT10B ratio close to 1, suggesting a constitutive activation of WNT10B transcription in the BM (Figure 3B). In addition, we analyzed transcriptional activation of canonical WNTs focusing on genes that have been shown to be potent regulators of stem cell functions. N-terminally dephosphorylated β-catenin (ABC) was increasingly accumulated as determined by immunoblot analysis (Figure 3C). Remarkably, we confirmed a dramatic increase in WNT10B expression in all patient samples, except for one, reanalyzed by immunoblot (Figure 3C). Interestingly, the only AML patient negative for the WNT10B expression (AML No. 40 in Table W1) was affected by therapy-related AML. According to the biologic relevance of PYGO2 in promoting the responsiveness of WNT signaling [35,36], diffuse over-expression was detected by immunoblot (Figure 3C). To better elucidate the impact of the broad WNT10B overexpression on the leukemic microenvironment, we examined its expression in histologic preparations of BM from five randomly selected AML patients at diagnosis. Double immunostaining for WNT10B and ABC, followed by ImageJ analysis, confirmed that WNT10B was expressed by a high proportion of leukemic cells. We next examined by immunostaining a number of BM biopsy sections from five randomly selected cases. In all the analyzed samples, the AC133 immunostaining revealed islands of highly positive cells (AC133bright) in an estimated proportion of 8% of cells, amid AC133dim or negative tumor blasts (Figure 4A). AC133bright cells correlated with accumulation of ABC (Figure 4, B and C). WNT10B antibody staining was also detectable in interstitial spaces, suggesting its secretion and release in the BM microenvironment (Figure 4D, top). The slides revealed that WNT10B is diffusely expressed (Figure 4, A and D) but that only the AC133bright small cells (8-10 µm diameter of the nuclei), with a clonal appearance and increased N/C ratio, shared the WNT signaling activation signature represented by accumulation of ABC [46], likely induced through an autocrine/paracrine mechanism (Figure 4, B and D, top).
Figure 3.
Altered WNT signaling in AC133+ AML cells. (A) Detection with padlock probe and target-primed rolling circle amplification of individual WNT10B transcripts on BM slides from AML patients. Green RCPs represent WNT10B transcripts, and red RCPs represent β-actin transcripts in consecutive sections. Cell nuclei are shown in blue. Images were acquired with x20 magnification. Scale bar, 10 µm. (B) The quantification of RCPs was done on three x20 images of BM biopsies of AML patients. For quantification, the numbers of RCPs of each image were counted digitally using CellProfiler software. The average of RCPs for each sample was calculated and it is reported as aratio between β-actin and WNT RCPs. (C) Immunoblot analysis of ABC, β-catenin (as detected with the N-terminal pan-β-catenin antibody), WNT10B, and Pygopus 2 protein expression in AC133+ cell fractions from 18 patient samples and 1 healthy donor introduced as control. GAPDH, loading control. HD, healthy donor; *therapy-related secondary AML.
Figure 4.
β-Catenin activation in the subpopulation of AC133bright AML cells expressing WNT10B. (A) Representative immunostaining micrographs show green fluorescence of cells expressing AC133 in a BM section of AML No. 9 (Table W1). Cell nuclei are shown in blue. Scale bar represents 10 µm. (B) Co-staining of BM from AML No. 9 adjacent serial section for expression of ABC (green) and WNT10B (red). Cell nuclei are shown in blue (DAPI). (C) False color maps of ABC/WNT10B double positive cells (blue) were obtained using an automatic threshold based on the moments algorithm implemented in the ImageJ program. DAPI staining was used to identify nuclei. Images obtained crossing ABC masks with WNT10B signals were used to count the percentage of ABC/WNT10B double positive cells over the total number of cells. The macro was validated against a trained experimenter over a sample of 830 total cells from eight different images. Differences in results were restricted to less than 0.01%. (D) Morphologic detail of cells showing intense specific staining for ABC (top panels) and WNT10B (bottom panels). All images were acquired with a x40 objective. Scale bars represent 10 µm. Representative images of at least three serial slides from five randomly selected patients.
AC133+ Leukemic Cells Express and Secrete WNT10B in a Primary Cell Culture
The notion that primary cell cultures closely mimic the in vivo state and generate more physiologically relevant data led us to establish a primary AC133+ cell culture (A46). A46 cells, with diploid karyotype, were selected from a 66-year-old male at diagnosis of AML-M2 (AML No. 46 in Table W1). Immunophenotype of MNCs before selection revealed a dominant CD133.1+CD34+CD38-CD45+CD117+ blast population (59%; Figure 5A), representing an optimal source to establish an LIC-enriched primary culture. Recently published data implicate the existence of an immunophenotypic hierarchy in AML, with a minority of CD38-CD45+ cells giving rise to CD38+CD45+ “GMP-like” cells at the apex of an LIC hierarchy [10]. The latter finding seems to suggest that CD38-CD45+ cells start to gain CD38 expression after AC133 selection procedures as observed in sorted cells (Figure 5A). Comparative analysis of spotted AC133-selected A46 leukemic and normal cells by immunostaining revealed WNT10B expression in all A46 AC133+ cells (Figure 5B, top), whereas normal BM-derived AC133+ cells resulted negative (Figure 5B, bottom). To further investigate whether the endogenous WNT production had any paracrine effect, we used A46 CM to evaluate β-catenin-mediated transcriptional activation. To this aim, HEK293T cells (H293T), transfected with Super 8x TOPFlash β-catenin/TCF transcription-based reporter construct, were exposed either to pBA-WNT10B H293T CM as control (Figure 5C) or A46 CM. This construct could be efficiently expressed in a dose-dependent manner when transiently exposed to A46 CM for 12 hours (Figure 5D).
Figure 5.
AC133+ A46 cells express and release WNT10B. Dot plots of the immunophenotype analysis from AML No. 46 BM MNCs at diagnosis and after selection. (A) Patterns of CD38/CD133.1 (top left), CD117/CD45 (top right), and CD34/CD38 (bottom left) co-staining were gated on BM AML cells before selection. Representative CD34 and CD38 expression on Ficoll-selected MNCs (bottom center) and AC133sorted cells before culture (bottom right) is shown. Percentages on total cellularity are shown for gated AML populations. (B) Immunostaining assessment for WNT10B in AC133+ populations from A46 (top panels) or healthy donor (bottom panels). Representative images of at least five serial slides. Blue, nuclei; red, WNT10B; merge, WNT10B/DAPI. All images were acquired with a x40 objective. Scale bars represent 10 µm. (C and D) TOPFlash reporter assay showing luciferase expression driven by eight TCF/lymphoid-enhancing factor (LEF) binding sites. (C) Positive control was obtained by CM of pBA-WNT10B-transfected H293T cells. Expression of WNT10B was evaluated in pBA-WNT10B-transfected H293T by real-time PCR (left). TOPFlash reporter assay shows luciferase expression induced in Super 8x TOPFlash-H293T cells by pBA-WNT10B H293T CM (right). (D) TOPFlash reporter assay showing dose-dependent luciferase expression induced in Super 8x TOPFlash-H293T cells by A46 CM. Significance was evaluated by the unpaired Student's t test: *P < .05; **P < .001. Data represent the mean ± SD of triplicate reactions and are representative of three independent experiments.
AC133 Is Expressed on Functional A46 AML LSC
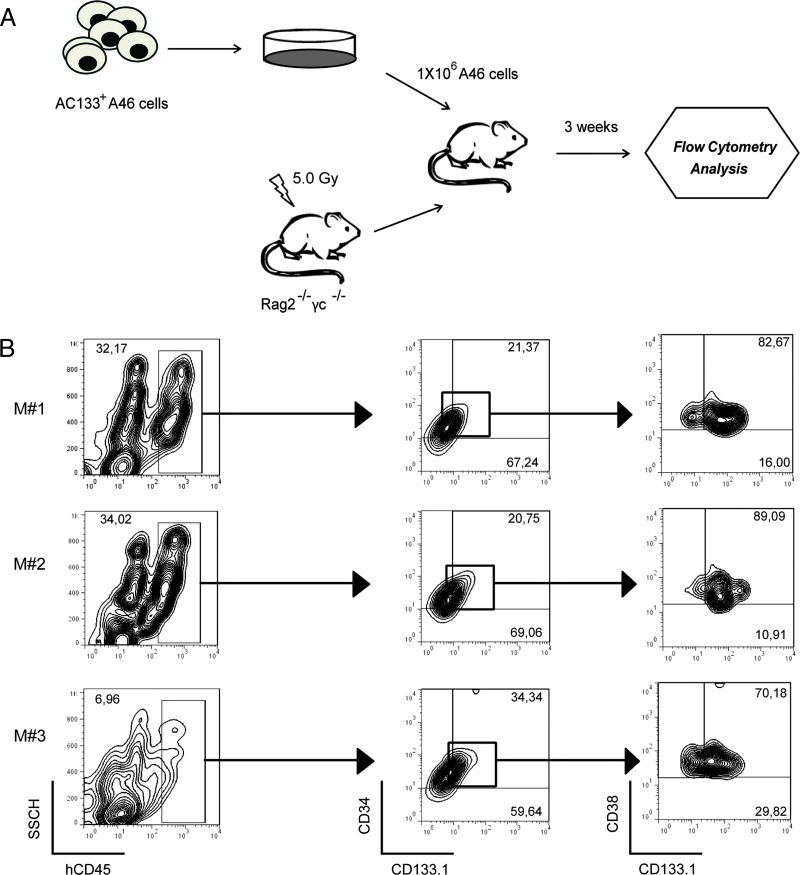
To complement the in vitro observations, we tested whether AC133 was expressed on functional A46 AML LSC. We transplanted A46 cells into sublethally irradiated (5 Gy) 6-week-old Rag2-/-γc-/- mice via the tail vein. This highly immunodeficient mouse strain lacks mature B, T, and natural killer (NK) cells, supporting efficient engraftment of human AML [16]. Transplanted mice were killed at 3 to 8 weeks after transplantation and analyzed for engraftment of human leukemia cells in BM (Figure 6A). The results of a typical experiment from three transplanted mice (M1–M3) are shown in Figure 6, with AC133+ A46 cells showing engraftment of human CD45 (hCD45) cells. We confirmed that engrafted hCD45+ cells were human myeloid leukemia blasts by measuring CD34/CD38/CD133 expression (Figure 6B).
Figure 6.
AC133+ A46 cell transplantation in Rag2-/-γc-/- mice. (A) Overview of the experimental design: 1 x 106 AC133+ A46 cells were injected into sublethally irradiated Rag2-/-γc-/- mice through the tail vein. Three weeks after transplantation, BM cells were collected and analyzed by flow cytometry. (B) Expression profiles of three recipient mice (M1–M3) are shown. hCD45+ population was identified in BMs of mice, and within hCD45+, the expression patterns of hCD34 and hCD133.1 (AC133) were analyzed. Cells were separated in subpopulations according to the expression of hCD34 and hCD133.1 and then analyzed for hCD38 and hCD133.1 expressions.
Transplantation of A46 Induces Ectopic Axial Structure Formation in Zebrafish Embryo by WNT Signaling Activation
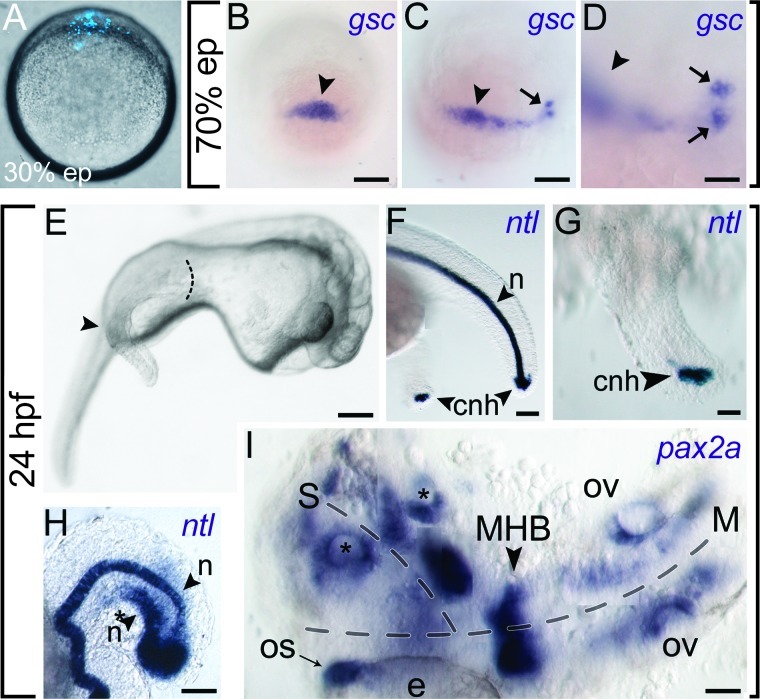
To bring our functional analyses full circle, we explored the physiological relevance of tumor-derived WNT signals by using the developing zebrafish as a biosensor. We hypothesized that WNT-secreting A46 cells transplanted into developing zebrafish embryos might act as ectopic sources of maternal WNT ligands. Cell-grafted embryos, injected at or before the mid-blastula transition stage (∼3 hpf) with Hoechst 33342 fluorescently labeled A46 cells (Figure 7A), showed the expression of the organizer-specific gene goosecoid (gsc). Expression of gsc is typically initiated by the nuclear translocation of maternal β-catenin triggered by the activation of the WNT signaling. While normal AC133+ cells did not alter the normal expression of the gene (Figure 7B), approximately 30% of the embryos grafted with A46 cells (n = 208) displayed both the expansion of the gsc endogenous domain and the activation of the gene in ectopic positions (Figure 7, C and D). Consistent with our hypothesis, the A46 cells retain a dorsal organizer- inducing activity possibly correlated with their strong WNT signaling activation. In addition, grafted embryos developed secondary axial structures, ranging from additional tail tissues (Figure 7, E–H) to an almost fully formed ectopic head (Figure 7I). Control embryos grafted with normal BM-derived AC133+ cells, devoid of any WNT signaling activity, did not display alterations of the normal phenotype (data not shown). The identity of the additional tail structures was confirmed by in situ hybridization staining for the notochord and tail bud marker ntl [47], whereas the emergence of ectopic head structures was highlighted by pax2a labeling optic stalk, midbrain-hindbrain boundary, and otic vesicles. These results imply that A46 cells might determine the establishment of an additional source of signal, with a Nieuwkoop center-like activity, able to induce an extra dorsal organizer, similarly to the endogenous situation at mid-blastula transition, when maternal WNT/β-catenin signaling initiates the formation of the dorsal organizer [48]. In zebrafish, secondary axis can be also induced by Nodal, a highly evolutionarily conserved morphogen belonging to the transforming growth factor-β superfamily and able to activate gsc expression [49]. Therefore, we investigated the possible involvement of the Nodal signaling pathway in the dorsal organizer induction mediated by A46 cells. The complete lack of Nodal expression observed after microarray analyses was confirmed by qualitative RT-PCR in A46 and samples from six different AML patients (Figure W1), allowing to exclude any Nodal contribution from A46 cells to the formation of the secondary axis and induction of gsc expression.
Figure 7.
A46 AML cells induce ectopic gene expression and secondary body axis formation upon transplantation in zebrafish embryos. (A) Fluorescence microscopy of a live zebrafish embryo at 30% of epiboly (lateral view) transplanted at 3 hpf at the animal pole region with A46 cells previously blue-stained with Hoechst 33342. (B–D) Dorsal side view of 70% epiboly-stage embryos hybridized with a gsc-specific probe. Embryos have been injected with (B) normal AC133+ cells as control or (C and D) A46 AML cells. The arrowheads indicate the gsc endogenous signal, while arrows specify the position of zebrafish cells expressing ectopic gsc. (E) Bright-field microscopy of a 24-hpf zebrafish embryo injected with A46 AML cells (lateral view). The arrowhead and the dotted line indicate the secondary trunk/tail induced by A46 cells. (F, G) The embryo in E has been hybridized with a probe specific for the notochord and tail bud marker ntl. (F) The probe labels the notochord (n) in the endogenous trunk and the chordoneural hinge (cnh) in both tails (G, higher magnification). (H) Tail of a 24-hpf embryo hybridized with ntl-specific probe. The endogenous (n) and ectopic (*n) ntl signals run parallel along the axis of the embryo, indicating the presence of additional axial structures. (I) Dorsal view of a 24-hpf embryo that developed an ectopic head on the side of the endogenous one, as indicated by the expression of the brain marker gene pax2a. The dotted lines indicate the main (M) and secondary (S) axes. The optic stalk (OS) in close vicinity to the eye (e), the midbrain-hindbrain boundary (MHB), and the otic vesicles (OV) of the embryo are stained with the pax2a riboprobe, as well as several areas of the ectopic head (the asterisks indicate the two clearly recognizable additional otic vesicles). The image is composed of different pictures corresponding to several focal planes, since the embryo is not flat, and a single focal plane cannot comprise all the labeled structures belonging to the main and secondary axes. Scale bars represent 125 µm (A–D), 150 µm (E), 40 µm (F), 15 µm (G and I), or 25 µm (H).
Discussion
The deceivingly homogeneous, undifferentiated morphology of the AML blasts is now known to mask a heterogeneous collection of cells that recapitulate the hierarchy of precursor cells that characterize the normal process of blood-cell differentiation. The concept that LIC properties occur in a self-renewing non-HSC progenitor cell population, preceded by the expansion of a preleukemic LT-HSC, has been recently reinforced [4,10]. However, the molecular functions responsible for the preleukemic LT-HSC expansion and the acquisition of self-renewal ability in AML remained poorly defined.
The WNT/β-catenin pathway has been show to play a critical role in the regulation of cell proliferation, differentiation, and apoptosis of different malignant entities. It is highly regulated in AML [50] and also involved in the self-renewal process of HSCs. WNT/β-catenin pathway requirement for LIC development in AML has emerged in mouse model [21]. Recent studies revealed aberrant WNT signaling in AML cells that is independent from the occurrence of AML-associated fusion proteins or mutations in tyrosine kinase receptors [reviewed in 20].
The results presented here using gene expression microarrays and pathway analysis provide direct evidence that the WNT/β-catenin signaling is diffusely activated in the AC133+ AML population, with a specific transcriptional signature involving overexpression of the WNT pathway agonists and down-modulation of the major antagonists.
Although the long-term reconstituting human HSC marker AC133 has been detected in a majority of CD34+ AML [51], no extensive data concerning the role of AC133 in AML were available.
Analysis of freshly fractionated cells from AML patients showed that active WNT signaling was predominant in the population highly enriched for the AC133 marker. Notably, WNT2B, WNT6, WNT10A, and WNT10B, known to promote hematopoietic tissue regeneration [13,14], are the WNT mediators specifically upregulated in the AC133+ AML cells. In addition, there is evidence that in the hematopoietic system WNT10B is specifically and significantly upregulated following an injury and that WNT10B acts to enhance the growth of HSCs [13]. It is also worth noting that greater fold expansion of murine HSC progenitors was obtained after WNT10B CM exposure [52]. Consistent with the latter observations, we showed a dramatic increase of WNT10B expression and protein release within the microenvironment in the large majority of samples from AML patients recruited to this study, with the exception of the unique therapy-related AML patient. In accordance with previous reports [52], we have not detected WNT10B gene expression in normal AC133+ hematopoietic cells. Moreover, our data also point to the strong expression of the WNT target gene CD44 and the loss of CD24, according to the expression of CD44+CD24-AC133+ phenotype that defines a putative cancer stem cell population also in breast tumors [53]. Interestingly, Dick et al. have shown that CD44 is a key regulator of AML LSC function and that targeting it eradicates LSCs [54]. In light of the higher homeostatic range of WNT/β-catenin signaling occurring upon an acute injury [55], our study demonstrated the involvement of the regeneration-associated WNT signaling in AC133bright AMLcellfraction. Theterm “regeneration” has been used to define the physiological phenomena of reconstitution from damage due to injury or disease. Hematopoietic regenerative-associated WNT ligand (WNT10B) is expressed at mRNA and protein levels on both leukemic blasts and stromal-like cells, indicating a possible autocrine/paracrine involvement of WNT in the BM microenvironment. Conversely, activation of WNT signaling marked by expression of the dephosphorylated β-catenin was restricted to the smaller population of AC133bright leukemic cells. The reasons for these differences are unclear, but it is possible that β-catenin activation by WNTs requires the expression of specific Fzd receptors, conferring a “responsive” phenotype, only restricted to a rare population of cells. In the HSC biology, a fundamental question is how self-renewal is controlled; several lines of evidence point to Notch for a WNT-mediated maintenance of undifferentiated HSCs [56]. Recent studies, examining the molecular targets of WNT10B, have highlighted the nuclear factor κB and Notch pathways as downstream targets of WNT10B [57]. It is tempting to speculate that WNT10B and other regeneration-associated WNTs (i.e., WNT2B, WNT6, and WNT10A) integrate with other signals, such as Notch, to drive oncogenic renewal. Because activation of WNT signaling can increase the ability of HSCs to reconstitute the hematopoietic system of lethally irradiated mice, we transplanted the AC133+ A46 cells into irradiated Rag2-/-γc-/- mice. Different papers have been recently published in which comparisons in the engraftment ability of different immunodeficient murine models were made. The variability of the reconstitution seemed to be mainly linked to the degree of immunodepression of the murine host. Rag2-/-γc-/- represents a powerful model to verify the biologic and malignant characteristics of human tumor cells. It is characterized by immunodeficiency of T and B cells caused by Rag2 knockout, coupled with the NK deficit mediated by the absence of the γc interleukin receptor chain [16]. The results of our experiments indicate that AC133 is expressed on AML LSC in the A46 primary cells, suggesting that regeneration-associated WNT expression signature is enriched in primary human AML LSC-containing fraction.
Regeneration requires the rapid expansions of HSCs; this process is often mediated by reactivation of developmental signal transduction pathways, such as BMP and WNT [45,58]. In the early embryo, the WNT factors induce the nuclear translocation of β-catenin, which triggers the formation of the dorsal organizer through the activation of zygotic dorsal-specific genes [59]. Such WNT-induced cascade of events results in the establishment of the embryonic dorsoventral axis [47]. To test the hypothesis that regeneration-associated WNT signaling is involved in AML LSC expansion, we used a zebrafish embryonic model as a tool by examining the ability of A46 primary leukemia cells to modulate embryonic microenvironment. Therefore, we used zebrafish embryos to show that A46 cells prompt secondary axis development, inducing the formation of a dorsal organizer-like structure, possibly through the secretion of different WNT ligands. The mechanisms promoting organizer formation are known to involve cooperation between Nodal and WNT signaling. However, implanted cell lines with different origin induce the dorsal organizer independently from the Nodal signaling [60]. Here, we demonstrated that the A46-dependent alteration of zebrafish development and activation of the organizer-specific gene gsc are not reliant on Nodal activity.
In summary, the findings we report here implicate, for the first time, that regeneration-associated ligand-dependent WNT signaling exceeds the homeostatic range in the majority of human AML cases and affects responsive AC133bright cells whose renewal is promoted by WNT pathway activity in vivo. These results not only support that AC133 is expressed on functional leukemia stem cells, but because developmental signal transduction pathways are often reactivated during regeneration, we also show that AC133+ AML cells induce the formation of a dorsal organizer-like structure in zebrafish embryos.
Finally, these studies suggest that the regenerative WNT signaling is a stem cell-associated function altered, as a common feature, in AC133bright AML leukemia stem cell fraction. Future studies will be required to demonstrate a pathogenic association of AC133bright LSC regeneration response with AML, in terms of tumorigenicity, clinicopathologic features, and patient outcomes.
Supplementary Material
Acknowledgments
We thank the patients with AML and their families. We also thank
E. Cattaneo, G. Simonutti, R. Bacchetta, and N. Santo for support with microscopy facilities, U. Landegren and O. Soderberg for supporting our activities at Rudbeck Laboratory, B. Scarpati and E. Calzavara for assistance with flow cytometry, R. Brusamolino and L. Pezzetti for assistance with patients' samples, L. Prosperi for support with zebrafish procedures, and S. Pozzi for helpful discussion.
Abbreviations
- AML
acute myeloid leukemia
- LSC
leukemic stem cell
- LT-HSCs
long-term hematopoietic stem cells
- CFU
colony forming units
- CFU-GM
granulocyte/macrophage
- BM MNCs
bone marrow mononuclear cells
- LIC
leukemia-initiating cell
Footnotes
This work was supported in part by PUR 2008 (to A.B.), Progetto Integrato Oncologia 2006 (RO 4/2007), Associazione Malattie del Sangue Onlus (AMS), and Piano Regionale Sangue-Regione Lombardia 2006 (DDG 7917). The authors declare that they have no competing financial interests.
This article refers to supplementary materials, which are designated by Table W1 and W2 and Figure W1 and are available online at www.neoplasia.com.
References
- 1.Cozzio A, Passegué E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2006;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E, Theilgaard-Mönch K, Månsson R, Pedersen TA, Pabst T, et al. Modeling of C/EBPα mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Bereshchenko O, Mancini E, Moore S, Bilbao D, Månsson R, Luc S, Grover A, Jacobsen SE, Bryder D, Nerlov C. Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPα mutant AML. Cancer Cell. 2009;16:390–400. doi: 10.1016/j.ccr.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 5.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 6.Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL, Weidinger G, Puder M, Daley GQ, Moon RT, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao C, Chen A, Jamieson CH, Fereshteh M, Abrahamsson A, Blum J, Kwon HY, Kim J, Chute JP, Rizzieri D, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann I, Stover EH, Cullen DE, Mao J, Morgan KJ, Lee BH, Kharas MG, Miller PG, Cornejo MG, Okabe R, et al. Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell Stem Cell. 2009;4:559–567. doi: 10.1016/j.stem.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taussig DC, Miraki-Moud F, Anjos-Afonso F, Pearce DJ, Allen K, Ridler C, Lillington D, Oakervee H, Cavenagh J, Agrawal SG, et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood. 2008;112:568–575. doi: 10.1182/blood-2007-10-118331. [DOI] [PubMed] [Google Scholar]
- 10.Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, Woll P, Mead A, Alford KA, Rout R, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 12.Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol. 2007;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 13.Congdon KL, Voermans C, Ferguson EC, DiMascio LN, Uqoezwa M, Zhao C, Reya T. Activation of Wnt signaling in hematopoietic regeneration. Stem Cells. 2008;26:1202–1210. doi: 10.1634/stemcells.2007-0768. [DOI] [PubMed] [Google Scholar]
- 14.Katoh M, Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3β to regulate β-catenin and SNAIL signaling cascades. Cancer Biol Ther. 2006;5:1059–1064. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 15.Larsson C, Grundberg I, Söderberg O, Nilsson M. In situ detection and genotyping of individual mRNA molecules. Nat Methods. 2010;7:395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 16.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 17.Majeti R, Becker MW, Tian Q, Lee TL, Yan X, Liu R, Chiang JH, Hood L, Clarke MF, Weissman IL. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci USA. 2009;106:3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 19.Staal FJT, Luis TC. Wnt signaling in hematopoiesis: crucial factors for self-renewal, proliferation, and cell fate decisions. J Cell Biochem. 2010;109:844–849. doi: 10.1002/jcb.22467. [DOI] [PubMed] [Google Scholar]
- 20.Mikesch JH, Steffen B, Berdel WE, Serve H, Müller-Tidow C. The emerging role of Wnt signaling in the pathogenesis of acute myeloid leukemia. Leukemia. 2007;21:1638–1647. doi: 10.1038/sj.leu.2404732. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, Leutz A. Hematopoietic stem cell and multilineage defects generated by constitutive β-catenin activation. Nat Immunol. 2006;7:1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- 23.Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell re-population and multilineage differentiation block. Nat Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- 24.Smith ML, Hills RK, Grimwade D. Independent prognostic variables in acute myeloid leukaemia. Blood Rev. 2011;25:39–51. doi: 10.1016/j.blre.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Brioschi M, Fischer J, Cairoli R, Rossetti S, Pezzetti L, Nichelatti M, Turrini M, Corlazzoli F, Scarpati B, Morra E, et al. Down-regulation of micro-RNAs 222/221 in acute myelogenous leukemia with deranged core-binding factor subunits. Neoplasia. 2010;12:866–876. doi: 10.1593/neo.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):55–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Hatta K, Takahashi Y. Secondary axis induction by heterospecific organizers in zebrafish. Dev Dyn. 1996;205:183–195. doi: 10.1002/(SICI)1097-0177(199602)205:2<183::AID-AJA9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 29.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 30.Hess DA, Wirthlin L, Craft TP, Herrbrich PE, Hohm SA, Lahey R, Eades WC, Creer MH, Nolta JA. Selection based on CD133 and high aldehyde dehydrogenase activity isolates long-term reconstituting human hematopoietic stem cells. Blood. 2006;107:2162–2169. doi: 10.1182/blood-2005-06-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao B, Niehrs C. Kremen2 modulates Dickkopf2 activity during Wnt/LRP6 signaling. Gene. 2003;302:179–183. doi: 10.1016/s0378-1119(02)01106-x. [DOI] [PubMed] [Google Scholar]
- 33.Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate β-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Moll Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson LE, Tollkuhn J, Scafoglio C, Krones A, Zhang J, Ohgi KA, Wu W, Taketo MM, Kemler R, Grosschedl R, et al. Homeodomain-mediated β-catenin-dependent switching events dictate cell-lineage determination. Cell. 2006;125:593–605. doi: 10.1016/j.cell.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 35.Mieszczanek J, de la Roche M, Bienz M. A role of Pygopus as an anti-repressor in facilitating Wnt-dependent transcription. Proc Natl Acad Sci USA. 2008;105:19324–19329. doi: 10.1073/pnas.0806098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu B, Sun P, Yuan Y, Moraes RC, Li A, Teng A, Agrawal A, Rhéaume C, Bilanchone V, Veltmaat JM, et al. Pygo2 expands mammary progenitor cells by facilitating histone H3 K4 methylation. J Cell Biol. 2009;185:811–826. doi: 10.1083/jcb.200810133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gehrke I, Gandhirajan RK, Kreuzer KA. Targeting the WNT/β-catenin/TCF/LEF1 axis in solid and haematological cancers: multiplicity of therapeutic options. Eur J Cancer. 2009;45:2759–2767. doi: 10.1016/j.ejca.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-β-catenin pathway by targeting Dishvelled for degradation. Nat Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Lu W, King TD, Liu CC, Bijur GN, Bu G. Dkk1 stabilizes Wnt co-receptor LRP6: implication for Wnt ligand-induced LRP6 down-regulation. PloS One. 2010;5:e11014. doi: 10.1371/journal.pone.0011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K, Zhang Y, Li X, Chen L, Wang H, Wu J, Zheng J, Wu D. Characterization of the Kremen-binding site on Dkk1 and elucidation of the role of Kremen in Dkk-mediated Wnt antagonism. Biol Chem. 2008;22:23371–23375. doi: 10.1074/jbc.M802376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Näär AM, Dyson NJ. E2F1 represses β-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;25:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao X, Wen J, Zhang L, Li X, Ning Y, Meng A, Chen YG. Dapper1 is a nucleocytoplasmic shuttling protein that negatively modulates Wnt signaling in the nucleus. J Biol Chem. 2008;283:35679–35688. doi: 10.1074/jbc.M804088200. [DOI] [PubMed] [Google Scholar]
- 43.Sampson EM, Haque ZK, Ku MC, Tevosian SG, Albanese C, Pestell RG, Paulson KE, Yee AS. Negative regulation of the Wnt-β-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20:4500–4511. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quéré R, Andradottir S, Brun AC, Zubarev RA, Karlsson G, Olsson K, Magnusson M, Cammenga J, Karlsson S. High levels of the adhesion molecule CD44 on leukemic cells generate acute myeloid leukemia relapse after withdrawal of the initial transforming event. Leukemia. 2011;25:515–526. doi: 10.1038/leu.2010.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowman TV, Tropouki E, Zon LI. Linking hematopoietic regeneration to developmental signaling pathways: a story of BMP and Wnt. Cell Cycle. 2012;11:424–425. doi: 10.4161/cc.11.3.19161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staal FJ, Noort Mv M, Strous GJ, Clevers HC. Wnt signals are transmitted through N-terminally dephosphorylated β-catenin. EMBO Rep. 2002;31:63–68. doi: 10.1093/embo-reports/kvf002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin X, Rinaldo L, Fazly AF, Xu X. Depletion of Med10 enhances Wnt and suppresses Nodal signaling during zebrafish embryogenesis. Dev Biol. 2007;303:536–548. doi: 10.1016/j.ydbio.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 48.Langdon YG, Mullins MC. Maternal and zygotic control of zebrafish dorsoventral axial patterning. Annu Rev Genet. 2011;45:357–377. doi: 10.1146/annurev-genet-110410-132517. [DOI] [PubMed] [Google Scholar]
- 49.Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- 50.Siapati EK, Papadaki M, Kozaou Z, Rouka E, Michali E, Savvidou I, Gogos D, Kyriakou D, Anagnostopoulos NI, Vassilopoulos G. Proliferation and bone marrow engraftment of AML blasts is dependent on β-catenin signaling. Br J Haematol. 2010;152:164–174. doi: 10.1111/j.1365-2141.2010.08471.x. [DOI] [PubMed] [Google Scholar]
- 51.Fauth F, Weidmann E, Martin H, Schneider B, Sonnhoff S, Hoelzer D. AC133 expression on acute myeloid leukemia blasts: correlation to FAB and to CD34 expression and possible implications for peripheral blood progenitor cell purging in AML. Leuk Res. 2001;25(3):191–196. doi: 10.1016/s0145-2126(00)00112-0. [DOI] [PubMed] [Google Scholar]
- 52.Austin TW, Solar GP, Ziegler FC, Liem L, Matthews W. A role for the Wnt gene family in hematopoiesis: expansion of multilineage progenitor cells. Blood. 1997;89:3624–3635. [PubMed] [Google Scholar]
- 53.Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:1–16. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 55.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 56.Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6(3):314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 57.Mödder UI, Oursler MJ, Khosla S, Monroe DG. Wnt10b activates the Wnt, Notch and NFκB pathways in U2OS osteosarcoma cells. J Cell Biochem. 2011;112(5):1392–1402. doi: 10.1002/jcb.23048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage regulators direct BMP and WNT pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu FI, Thisse C, Thisse B. Identification and mechanism of regulation of the zebrafish dorsal determinant. Proc Natl Acad Sci USA. 2011;108:15876–15880. doi: 10.1073/pnas.1106801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hashiguchi M, Shinya M, Tokumoto M, Sakai N. Nodal/Bozozok-independent induction of the dorsal organizer by zebrafish cell lines. Dev Biol. 2008;321:387–396. doi: 10.1016/j.ydbio.2008.06.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.