Abstract
Double-stranded RNA (dsRNA) viruses transcribe and replicate RNA within an assembled, inner capsid particle; only plus-sense, mRNA emerges into the intracellular milieu. During infectious entry of a rotavirus particle, the outer layer of its three-layer structure dissociates, delivering the inner, double-layered particle (DLP) into the cyotosol. DLP structures determined by x-ray crystallography and electron cryomicroscopy (cryoEM) show that the RNA coils uniformly into the particle interior, avoiding a “fivefold hub” of more structured density projecting inward from the VP2 shell of the DLP along each of the 12 fivefold axes. Analysis of the x-ray crystallographic electron density map suggested that principal contributors to the hub are the N-terminal arms of VP2, but reexamination of the cryoEM map has shown that many features come from a molecule of VP1, randomly occupying five equivalent and partly overlapping positions. We confirm here that the electron density in the x-ray map leads to the same conclusion, and we describe the functional implications of the orientation and position of the polymerase. The exit channel for the nascent transcript directs the nascent transcript toward an opening along the fivefold axis. The template strand enters from within the particle, and the dsRNA product of the initial, replication step exits in a direction tangential to the inner surface of the VP2 shell, allowing it to coil optimally within the DLP. The polymerases of reoviruses appear to have similar positions and functional orientations.
Introduction
Double-stranded RNA (dsRNA) viruses do not release their genomic RNA when they enter a cell. Instead, for each genomic segment, they package a dsRNA-dependent, RNA polymerase, which transcribes the segment into mRNA and exports the product from the particle. Like most other mammalian dsRNA viruses, rotavirus particles have a multi-layer protein shell1. The virion is a “triple layered particle” (TLP); the transcriptionally active, inner capsid particle is a “double layered particle” (DLP). The role of the outer protein layer, which includes the proteins VP4 (cleaved to VP8* and VP5*) and VP7, is to attach virions to the cell surface and to deliver the DLP into the cytosol1.
The DLP contains four protein species: VP2 (120 copies), assembled into a thin but tightly packed shell surrounding the RNA; VP6 (780 copies), arrayed as 260 trimers on the outer surface of the VP6 shell; VP1 (11 or 12 copies), the dsRNA-dependent, RNA polymerase, enclosed along with the 11 dsRNA segments within the VP2 shell; and VP3 (11 or 12 copies), the mRNA capping enzyme, likewise enclosed within the VP2 shell. Fig. 1 shows the organization of VP2 and VP6, the icosahedrally symmetric components, from the structure of the DLP as determined by X-ray crystallography2. Each icosahedral asymmetric unit contains two chemically identical VP2 subunits (designated VP2A and VP2B), having closely related conformations but making completely distinct inter-subunit contacts. The VP2A subunits encircle the fivefold positions; the VP2B subunits insert into gaps in the VP2A ring distal to the fivefold. The (VP2A-VP2B)5 decamer is probably the assembly unit of the VP2 shell2; 3; 4. The well-ordered, VP2A chain trace begins at residue 100 and continues to the end of the polypeptide chain at residue 880; the ordered VP2B chain trace begins at residue 81. The 80–100 residue N-terminal “arms” of both VP2 conformers project toward the fivefold (Fig. 2).
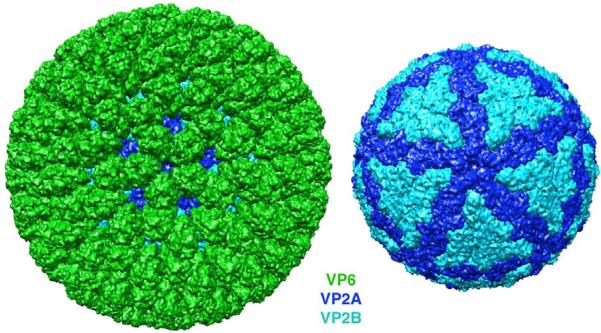
Fig. 1.
Surface views of the rotavirus DLP (left) and the VP2 inner layer (right). VP6 is in green, and the conformers of VP2, VP2A and VP2B, are in dark blue and cyan, respectively. The view is along a fivefold axis. Note that small gaps in the VP6 layer allow the VP2 layer to show from beneath.
Fig 2.
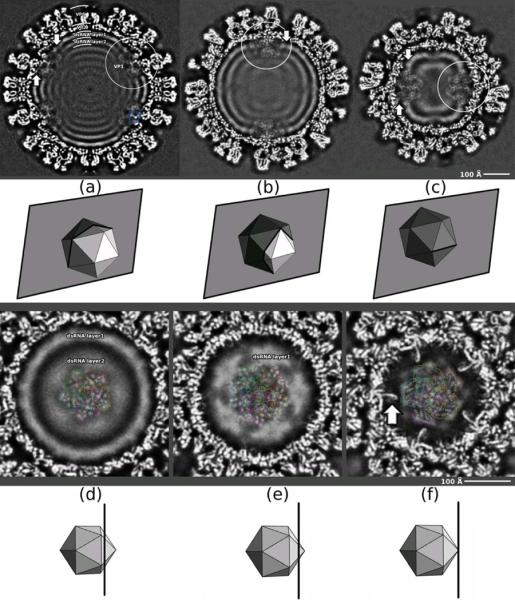
Cross-sections through reconstructed DLP density map, calculated with symmetry adapted functions to a nominal resolution of 4.2 Å (FSC=0.143 criterion). Beneath each panel is a diagram showing the orientation and position of the section with respect to icosahedral symmetry axes as a semi-transparent plane cutting an icosahedron. (a)–(c) Sections normal to a twofold axis. (a) Section passes through DLP center; (b) section cuts two vertically aligned fivefold axes; (c) section normal to twofold axis and intersecting two horizontally aligned fivefold axes. Densities associated with VP1 are indicated by white circles. (d)–(f) Sections normal to a fivefold axis. The upward arrows indicate VP2 N-terminal “arms” “holding” VP1 and downward arrows indicate the contact between VP1 and the first RNA layer. In (d)–(f), coordinates of VP1 fitted to the density are shown as light-gray ribbons (overlay) at four of the five equivalent positions occupied by VP1, showing the match between secondary-structure elements of VP1 and rod-like densities in the EM map. The outer contour of each VP1 position is in color, showing overlap between neighboring VP1 positions. One position (green) left with no ribbons to show EM density more clearly.
The eleven dsRNA genomic segments appear to coil tightly inside the VP2 shell, generating a series of concentric, low-resolution density layers after icosahedral averaging2; 5; 6. These layers avoid a region around the fivefold, which contains a set of rod-like density features. In the original interpretation of the X-ray crystallographic density map, some of these features were ascribed to the N-terminal arms of VP2, with the suggestion that they contribute to a “five-fold hub”2. One of us (LE) has examined those features in an improved cryoEM reconstruction of the DLP and shown that VP1, in five equivalent orientations, fits the density with excellent fidelity, including the density close to the fivefold axis, which is a superposition of five VP1 densities (each at 20% occupancy)6. As reported here, we have re-examined the X-ray map and arrive at precisely the same interpretation. We further show that a cryoEM reconstruction of the TLP has similar, VP1-derived features, but displaced radially inward by about 10 Å, consistent with changes previously detected in the VP2 and VP6 layers, when the outer-layer proteins are added7; 8. We describe the functional consequences of the position and orientation of VP1 confirmed by examination of these various, independently derived maps. Zhang et al have drawn comparable conclusions from a closely related assignment of density to the dsRNA-dependent RNA polymerase (λ3) in a 7 Å resolution cryoEM map of an orthoreovirus9.
Results
Locating VP1 in the DLP cryoEM reconstruction
We calculated an improved reconstruction of the rotavirus DLP, at an estimated resolution of 4.2 Å, applying the “symmetry-adapted-functions” procedure6 to the images used to generate the previously reported DLP structure5. The cryoEM reconstruction shows a readily interpretable hub of density features near the fivefold axis, just within the VP2 shell (Fig. 2). The ratio of the density in this “fivefold hub” to the density in the VP2 and VP6 shells is about 0.17, consistent with the assumptions that the distinct features represent the average of a single copy of VP1 in five equivalent positions and that other contributions (from the N-terminal arms of VP2, which also occupy some of this volume, and potentially from RNA) are effectively uniform. The lower occupancy of protein in the fivefold hub and the likely overlap with disordered structures (e.g., the VP2 N-terminal arms) diminish the effective resolution in that region, which we estimate (based on the appearance of α-helices, etc.) to be between 5 and 6 Å.
There are a number of rod-like features in the fivefold hub, particularly in regions displaced from the axis. Fitting the VP1 coordinates4 into these features, while taking into account the weighted sum of contributions to the overlap volume10, yielded a convincing and unambiguous match6 (Fig.3a). The centroid of VP1 is offset from the symmetry axis, so that the five equivalent images of VP1 overlap only near the fivefold axis. The fit is particularly well defined because much of the protein does not overlap fivefold-related images of other well-defined density. Strong features near the fivefold are superpositions of secondary-structural elements in two or more of the five equivalent orientations of VP1. For example, the radially directed rods, previously interpreted as α-helices when interpreting the X-ray crystallographic density map (see Fig. 9B in reference2), are superpositions of an α-helix (residues 32–51) and a β-strand (residues 747–754), both radially oriented.
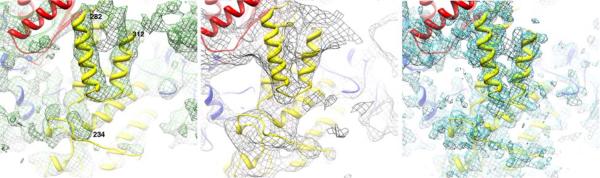
Fig. 3.
Fit of VP1 model to density. (a) DLP cryoEM map as in Fig. 2; (b) X-ray map2; (c) TLP cryoEM map8.
Locating VP1 in the X-ray crystallographic electron density map
We confirmed the fit of VP1 into the cryoEM image reconstruction by an independent analysis of the electron density map derived by X-ray crystallography2. A map calculated with terms cut-off beyond 5 Å resolution was the clearest of the those we examined. A strong helix-turn-helix feature, with strong connectivity in the turn, provided a good starting point. Analysis of the VP1 structure showed only one pair of sequence-adjacent helices that matched in length and relative orientation: one helix contains residues 282–298; the other, residues 301–312. A model placed to fit these helices into the helix-turn-helix density placed additional helices in or near suitable density features, and simple rigid-body shifts aligned all the main features of VP1 (or their fivefold averages, in the overlap region) with corresponding density (Fig. 3b).
Locating VP1 in the TLP
The TLP cryoEM image reconstruction8 has density features corresponding closely to the VP1 density in the DLP, but the entire set of features, along with the tip of VP2A, has shifted inward (Fig. 3c and 4). The displacement is consistent with previously analyzed changes in the VP2 shell that accompany binding of VP7 to the tip of VP6: addition of VP7 alters the radial tilt of peripentadal VP6 trimers, pushing inward on the loops of VP2 abutting the fivefold.
VP1–VP2 contacts
The placement of VP1 in density as just described creates a largely complementary interface between it and the VP2 shell (Fig. 4). There are three positions of minor overlap, one where the VP2A helix-loop-helix that defines the pore around the fivefold axis (residues 341–370) collides with residues 968–980 of VP1; a second, where the same segment of a fivefold related VP2A (the counter clockwise neighbor when viewed from outside the shell) overlaps residues 1022–1025 of VP1; and a third, where residues 337–8 of the latter VP2A comes close to a turn at residues 264–267 of VP1. At all three positions, modest adaptations in either partner could alleviate any clash. In addition, some part of VP2A must shift during transcription to allow RNA to exit, as the pore at the fivefold axis of the non-transcribing DLP is too narrow to accommodate an RNA strand2. The N-terminal densities of two adjacent VP2A subunits (residue 100) and a VP2B (residue 81) abut the VP1 model. The X-ray map suggests that in the other 4/5 of the VP2B subunits, the helix that begins at residue 81 in the DLP model might extend for another 2–3 turns in an N-terminal direction. Moreover, the full-strength density in the x-ray map at residue 100 in VP2A and residue 81 in VP2B shows that terminal “arms” of all ten VP2 subunits penetrate into the fivefold-hub region. The arms are evidently flexible tethers for VP1 and VP3, as they are necessary for incorporation of those components into assembling particles11.
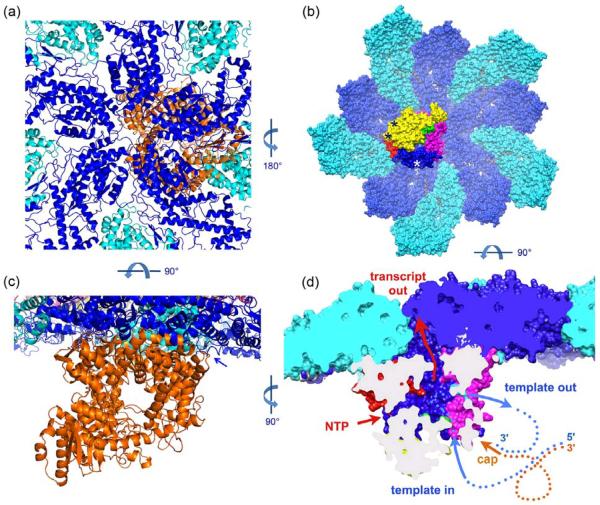
Fig. 4.
Functional orientation of VP1. (a) Ribbon representation of the placement of VP1 (orange) with respect to VP2A (blue) and VP2B (cyan), viewed along a fivefold axis from outside the VP2 shell. (b) Surface representation of ten VP2 subunits surrounding a fivefold axis, colored as in (a), and the associated VP1, viewed from inside the DLP. The domains of VP1 are colored yellow (N-terminal domain), green (fingers subdomain), red (palm subdomain), blue (thumb subdomain), and magenta (C-terminal domain. The positions at which N-terminal arms of three different VP2 subunits clearly contact the VP1 shown are marked by asterisks; the remaining N-terminal arms may also contact this VP1 molecule, but their density overlaps that of its fivefold “ghosts”. (c) As in (a), but viewed in a direction tangential to the VP2 shell, looking into the template (and dsRNA) exit channel of VP1. Arrow indicates contact between VP1 and the N-terminal arm of a VP2A subunit. (d) Representation as in (b), but viewed in a direction tangential to the VP2 shell and normal to the view in (c), cut away to show the internal cavity in VP1 that contains the catalytic site. The channels leading into this cavity are labeled, and a schematic indication (not to scale) of how the minus-sense strand of the genome (blue) threads through the polymerase, leaving a “transcription bubble” in the plus-sense strand (orange), the latter anchored to the polymerase by its 5'-cap.
Association with VP2 activates VP13; 12. A contribution to the presumed conformational change that activates the polymerase might come from the interaction of the N-terminal arm of VP2B with the surface of VP1. The contact occurs at a site that contains residues just distal to the “priming loop” (residues 492–498) and also residues just distal to the Asp-Asp sequence (residues 631–632) at the catalytic center.
Functional orientation of VP1
The catalytic center of VP1 lies within a cavity in the center of a cage-like protein4. Four channels connect the central cavity with the exterior. Structures of template-VP1 complexes and comparison with the reovirus polymerase13 show that template inserts through one of these channels and exits through a second, either as a dsRNA genomic product (when the template is the plus strand and the polymerase is in replicative mode) or as a ssRNA (when the template is the minus strand and the polymerase is in transcriptional mode). The remaining two channels appear to allow exit of the plus-strand transcript and entry and exit of small-molecule substrates and products (nucleotides; pyrophosphate; Mg2+ ions), respectively.
The position of VP1 within the DLP (Fig. 5) orients the channels so that the template enters from the interior of the particle and the dsRNA exits roughly tangential to the VP2 shell. The putative transcript exit channel opens toward VP2 beneath the position at which two fivefold-related VP2A subunits diverge from each other at the apex of a VP2B subunit, about 30 Å displaced from the fivefold axis. There is a path from the mouth of the VP1 transcript exit channel toward the fivefold, through which it is likely that transcripts emerge from the DLP14. An outward shift of the VP2 pore loop and the helices to either side of it could widen that path. Only one of the five VP2A pore loops need undergo this shift, so it might not be evident in the density. There are no strong density features that we can ascribe to VP3, which must intercept the 5' end of the nascent transcript before it exits the DLP.
Discussion
The eleven dsRNA genome segments and the 11–12 copies of VP1 and VP3 pack tightly and efficiently into the DLP interior. Their organization must meet several very restrictive functional requirements. (1) Particle assembly packages one copy each of the eleven capped, plus-sense RNA segments and discriminates against non-viral RNAs. (2) Tight packing of the dsRNA within the DLP requires that it coil efficiently. Concentric, low resolution, internal density layers suggest that each dsRNA segment coils independently around a fivefold hub. (3) Each genome segment undergoes multiple rounds of transcription; the corresponding template strand must therefore pass through a polymerase many times. (4) The capped transcripts emerge from the DLP at or near fivefold positions. The orientation and position of VP1 within the VP2 shell, as described here, adds to our understanding of each of these requirements.
DLP assembly
Association of ten VP2 subunits (5 VP2A and 5 VP2B, in a fivefold-symmetric heterodecamer) with VP1, VP3, and a segment of viral RNA initiates DLP assembly2; 3. The N-terminal arms of VP2 are necessary for inclusion of VP111. An empty DLP-like particle, lacking VP1, VP3 and RNA, assembles from VP2 (whether full length or with truncated arms) and VP6 alone15; 16. Incorporation of RNA, VP1, and VP3 into the assembling particle thus depends on the VP1–VP2 interactions described above, on RNA recruitment by VP1, and probably on association of VP1 with VP3. Recognition by VP1 of a conserved sequence element at the 3' end of plus-sense RNA can account for discrimination against non-viral RNA during packaging4, but it cannot by itself account for selection of 11 distinct RNA segments. The latter process is likely to be encoded in a set of RNA-RNA recognition events not yet evident from the segment sequences alone.
Ten VP2 N-terminal arms project toward each VP1, of which three (two from VP2A subunits and one from a VP2B subunit) appear from density features to interact with the distinct VP1 surfaces against which they lie (Fig. 4). There may be additional arm-VP1 contacts, from some of the other VP2 subunits in the heterodecamer; some sort of interaction seems likely from the disposition of the various VP2 N-terminal polypeptide segments in the vicinity of a single VP1.
RNA coiling
The most efficient configuration for tightly packed dsRNA (or any stiff but bendable rod) within a roughly spherical cavity is a more or less regular coil, with local close packing of adjacent segments2; 17; 18. Low resolution maps of the rotavirus DLP show layers of density concentric with the inner surface of the VP2 shell, becoming less pronounced toward the interior of the particle2 (see also Fig. 2). Earlier analyses of orbivirus and reovirus cores showed similar features19; 20. The layers of density avoid completely a volume around the fivefold, extending inward about 75 Å from the VP2 shell. Were RNA wrapped tightly around the single VP1 at each vertex, the density layers would continue into the hub region, overlapping the VP1 density (except in the volume close to the axis, to which VP1 in any of its five equivalent orientations will contribute). VP2 N-terminal arms and VP3 probably fill the 80% of the peri-fivefold region not accounted for by the single VP1. The volume that excludes the RNA density layers could accommodate most or all of these protein components. The clarity of the VP1 density features outside the central region of VP1-VP1 overlap (Fig. 2) suggests, however, that any contributions from VP2 arms or from VP3 create a roughly continuous background -- in other words, that neither VP3 nor any ordered segment of a VP2 arm has a fixed position relative to VP1.
The first round of RNA synthesis, which completes DLP assembly in the cytosol, generates dsRNA from the initially incorporated plus-strand RNA. The channel in VP1 for exit of dsRNA opens in a direction tangential to the inner surface of the VP2 shell (Fig. 4c,d). Thus, the polymerase in replicase mode emits its product in the orientation in which it will coil. The orientation of the polymerase suggests that it could assist this coiling, by counter-rotating as the dsRNA emerges and switching from one fivefold-related position to another. The last nucleotides of the plus strand to pass through the template channel are at the capped 5'-end. The dsRNA exit channel, through which the cap then re-emerges, is only about 10 Å from the cap-binding site on the VP1 surface4. Capture of the 5'-cap can hold the complementary, 3' end of the newly synthesized minus strand close to the template entrance channel, through which it must pass to initiate transcription (Fig. 4d).
Transcription
Release of the DLP during viral entry allows transcription to begin. The observed outward displacement of the entire transcriptional complex when the outer layer dissociates (Fig. 3a) probably contributes to activation of the polymerase. The conformational change in VP2 is not an expansion of the entire shell: only the parts of VP2A that surround the particle fivefold axis (the “apical region”) move outward. Therefore, the surface against which VP1 packs changes in contour during the TLP-to-DLP transition. Our current EM reconstructions are not sufficiently accurate, however, to show changes in VP1 that might have occurred in response to the changes in VP2. We therefore cannot rule out the possibility that activation is merely a consequence of “unplugging” the transcript exit route.
During successive rounds of transcription, the 3' end of template (minus) strand must remain close to the polymerase, so that it can re-enter the template channel when a transcript is complete. We have proposed previously, for both reovirus13 and rotavirus4 polymerases, that retention of the 5' end of the plus strand during the entire transcription process prevents the 3' end of the minus strand from “getting lost” in the tightly packed interior of the DLP.
VP1 is evidently well anchored in the non-transcribing DLP, as the density for the non-overlapped regions is relatively well defined (Fig. 3), even at 20% effective occupancy. During transcription, the template feeds into the polymerase more or less axially (from the interior of the particle), but exits laterally, parallel to the VP2 inner surface. As we suggest above for the replication step, backward thrust on VP1 from the exiting minus strand might loosen its attachment or cause it to rotate in the direction opposite to that of the emerging template, with potential reattachment at any of the fivefold equivalent positions.
Comparison with reovirus
In their analysis of both inner cores and intact virions from an orthoreovirus, Zhang et al assigned to fivefold proximal density a copy of the polymerase, λ3 (the VP1 equivalent), in an orientation very similar to the one we have determined here9. The sites of nascent mRNA capping of orthoreoviruses are in the “turrets” (protein λ2) that project from the shell layer, rather than on a protein within it. Zhang et al suggested that passage of the transcript into the turrets might be through a gap somewhat displaced from the fivefold. Careful inspection of the full atomic model of a non-transcribing core suggests that the gap at that location is largely filled by side chains, but a recent cryoEM structure of a transcribing cypovirus (like reovirus, a turreted, dsRNA virus, with capping-enzyme active sites facing the interior of the turrets) shows an outward displacement of the apical region of the shell protein, very similar to the conformational change in VP2A that accompanies release of VP7121. The displacement opens the peripentadal gap, and the reconstruction shows some evidence that the transcript does indeed pass through it into the lumen of the turret. Thus, despite differences in the location of their capping activities, the polymerases of several classes of dsRNA viruses appear to have nearly identical functional orientations, linked to their roles in both replication and transcription.
Methods
Fitting VP1 into a recalculated DLP cryoEM density map
CryoEM images of the rotavirus DLP used in this work were the same as those used previously5; 6. A new reconstruction was calculated using symmetry adapted functions (SAFs)22 and fast projection matching (FPM)23. These two methods can together determine, quickly and accurately, the orientation and center of each particle. Use of SAFs (formally equivalent to Klug's “icosahedral harmonics”24; 25) produces 3D reconstructions with excellent signal-to-noise ratio. Application of these methods to the rotavirus DLP images are described elsewhere6. A total of 7,000 of the 18,125 images available in the full DLP data-set were used to compute the 3D reconstruction, with resolution estimated by Fourier-shell correlation (FSC) to be between 5.6 Å (0.5 criterion) and 4.2 Å (0.143 criterion)6. Some high-resolution features can be recognized in non-overlapping zones (Fig. 3a). The fit of the atomic model into the 3D reconstruction was performed using the program URO and its graphical interface VEDA26. The fitting procedure took into account the difference between the densities associated to each protein given the symmetry mismatch. The Fourier coefficients of VP1 were downweighted by a factor of 5 in order to prevent VP1 from being “attracted” by the stronger density of the VP2/6 layer during the fit. The atomic model of VP1 was placed interactively into the reconstruction with the program PyMOL6 (PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC), guided by the well defined sausage-shaped densities shown in Figs. 2 and 3. The positions of the models were then optimized maximizing the correlation coefficient with the 3D reconstruction, taking into account the icosahedral symmetry and using data to 6 Å. The fit produced a final correlation coefficient of 0.7.
Fitting VP1 into the DLP X-ray density map
The initial identification of features, described in the text, was done using the program O27 to visualize map and coordinates. Rigid-body refinement was carried out with CNS28, using the refined models of VP2 and VP6. VP1 was trimmed to exclude obvious overlaps with VP2. The occupancy of VP1 was set to 0.2 to account for the presence of only one VP1 at each 5-fold axis. VP1 is too small a fraction of the total asymmetric unit to contribute noticeably to the working or free R-factor in crystallographic refinement.
Accession codes
The atomic model of VP1 attached to capsid inner core has been deposited in the PDB with accession codes 4AU6 (EM) and 4F5X (x-ray). The DLP cryoEM map calculated with symmetry adapted functions has been deposited in the 3D-EM database under accession number EMD-2100. The DLP and TLP cryoEM maps calculated with FREALIGN have been deposited previously (EMD-1460 and EMD-5199, respectively).
Highlights
Rotavirus particles contain 11 dsRNA segments, each associated with a VP1 polymerase
We have analyzed cryoEM and x-ray structures of the rotavirus inner capsid (DLP)
One VP1 polymerase is at each fivefold vertex, randomly in 5 equivalent orientations
The VP1 orientation directs the nascent transcript toward an opening in the capsid
Results show common mechanism for RNA packaging and synthesis in dsRNA viruses
Acknowledgments
We thank Michael Plevin, Stephen Cusack, Irina Gutsche, Jorge Navaza, Rob Ruigrok and Guy Schoehn for discussion and advice and the EMBL/Grenoble for financial support and computational resources. We also acknowledge NIH grants CA-13202 (to SCH) and GM-62580 (to NG and SCH). SCH and NG are Investigators in the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Estes MK, Kapikian AZ. Rotaviruses. In: Howley DMKPM, editor. Fields Virology (5th ed) Williams & Wilkins, Philadelphia; Lippincott: 2007. pp. 1918–1974. [Google Scholar]
- 2.McClain B, Settembre E, Temple BR, Bellamy AR, Harrison SC. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution. J Mol Biol. 2010;397:587–99. doi: 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallegos CO, Patton JT. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology. 1989;172:616–27. doi: 10.1016/0042-6822(89)90204-3. [DOI] [PubMed] [Google Scholar]
- 4.Lu X, McDonald SM, Tortorici MA, Tao YJ, Vasquez-Del Carpio R, Nibert ML, Patton JT, Harrison SC. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–88. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–72. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estrozi LF, Navaza J. Ab initio high-resolution single-particle 3D reconstructions: the symmetry adapted functions way. J Struct Biol. 2010;172:253–60. doi: 10.1016/j.jsb.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A. 2009;106:10644–8. doi: 10.1073/pnas.0904024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. EMBO J. 2010;30:408–16. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 A. Nat Struct Biol. 2003;10:1011–8. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navaza J, Lepault J, Rey FA, Alvarez-Rua C, Borge J. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr D Biol Crystallogr. 2002;58:1820–5. doi: 10.1107/s0907444902013707. [DOI] [PubMed] [Google Scholar]
- 11.Zeng CQ, Estes MK, Charpilienne A, Cohen J. The N terminus of rotavirus VP2 is necessary for encapsidation of VP1 and VP3. J Virol. 1998;72:201–8. doi: 10.1128/jvi.72.1.201-208.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patton JT, Gallegos CO. Rotavirus RNA replication: single-stranded RNA extends from the replicase particle. J Gen Virol. 1990;71(Pt 5):1087–94. doi: 10.1099/0022-1317-71-5-1087. [DOI] [PubMed] [Google Scholar]
- 13.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage--structural studies of reovirus polymerase lambda3. Cell. 2002;111:733–45. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 14.Lawton JA, Estes MK, Prasad BV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–21. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 15.Crawford SE, Labbe M, Cohen J, Burroughs MH, Zhou YJ, Estes MK. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5945–52. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Libersou S, Siebert X, Ouldali M, Estrozi LF, Navaza J, Charpilienne A, Garnier P, Poncet D, Lepault J. Geometric mismatches within the concentric layers of rotavirus particles: a potential regulatory switch of viral particle transcription activity. J Virol. 2008;82:2844–52. doi: 10.1128/JVI.02268-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earnshaw WC, Harrison SC. DNA arrangement in isometric phage heads. Nature. 1977;268:598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- 18.Harrison SC. Molecular packing of nucleic acids in spherical viruses. In: Eisenberg DS, Lake JA, Fox CF, editors. Biological Recognition and Assembly. Alan R. Liss; New York: 1980. [PubMed] [Google Scholar]
- 19.Gouet P, Diprose JM, Grimes JM, Malby R, Burroughs JN, Zientara S, Stuart DI, Mertens PP. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97:481–90. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 20.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404:960–7. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 21.Yang C, Ji G, Liu H, Zhang K, Liu G, Sun F, Zhu P, Cheng L. Cryo-EM structure of a transcribing cypovirus. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1200206109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navaza J. On the three-dimensional reconstruction of icosahedral particles. J Struct Biol. 2003;144:13–23. doi: 10.1016/j.jsb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Estrozi LF, Navaza J. Fast projection matching for cryo-electron microscopy image reconstruction. J Struct Biol. 2008;162:324–34. doi: 10.1016/j.jsb.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Finch JT, Holmes KC. Structural studies of viruses. In: Maramarsoch K, Koprowski H, editors. Methods in Virology. Academic Press; New York: 1967. pp. 352–474. [Google Scholar]
- 25.Jack A, Harrison SC. On the interpretation of small-angle x-ray solution scattering from spherical viruses. J Mol Biol. 1975;99:15–25. doi: 10.1016/s0022-2836(75)80155-0. [DOI] [PubMed] [Google Scholar]
- 26.Siebert X, Navaza J. UROX 2.0: an interactive tool for fitting atomic models into electron-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2009;65:651–8. doi: 10.1107/S0907444909008671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones TA, Zou JY, Cown SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]