Abstract
Neurodegenerative diseases are typically late-onset progressive disorders that affect neural function and integrity. Although most attention has been focused on the genetic underpinnings of familial disease, mechanisms are likely shared with more predominant sporadic forms, which can be influenced by age, environment and genetic inputs. Previous work has largely addressed the roles of select protein-coding genes; however, disease pathogenesis is complicated and can be modulated through not just protein-coding genes, but also regulatory mechanisms mediated by the exploding world of small non-coding RNAs. Here, we focus on emerging roles of miRNAs in age-associated events impacting long-term brain integrity and neurodegenerative disease.
Keywords: microRNA, neurodegenerative disease, aging, miRNA biogenesis, dicer, small RNAs
miRNAs and neurodegeneration
Neurodegenerative diseases are a group of typically late-onset, progressive disorders that lead to cognitive and/or movement disorders. Some of the most studied include Alzheimer's disease AD), Parkinson's disease (PD) Amyotrophic Lateral Sclerosis (ALS), and polyglutamine (polyQ) disorders such as Huntington's disease (HD) and the spinocerebellar ataxias (SCAs) [1–5]. These diseases share features such as the abnormal accumulation of protein, which includes plaques and tangles in AD, Lewy bodies in PD, bunina bodies in ALS, and nuclear and cytoplasmic accumulations in polyQ disease. In these diseases, key proteins accumulate, the genes of which are ones in which familial mutations can be found. Mechanisms that affect disease pathogenesis involve multiple fundamental cellular pathways, including protein folding and clearance processes. Thus, understanding the pathogenic mechanisms requires studying a broad spectrum of basic cellular machineries.
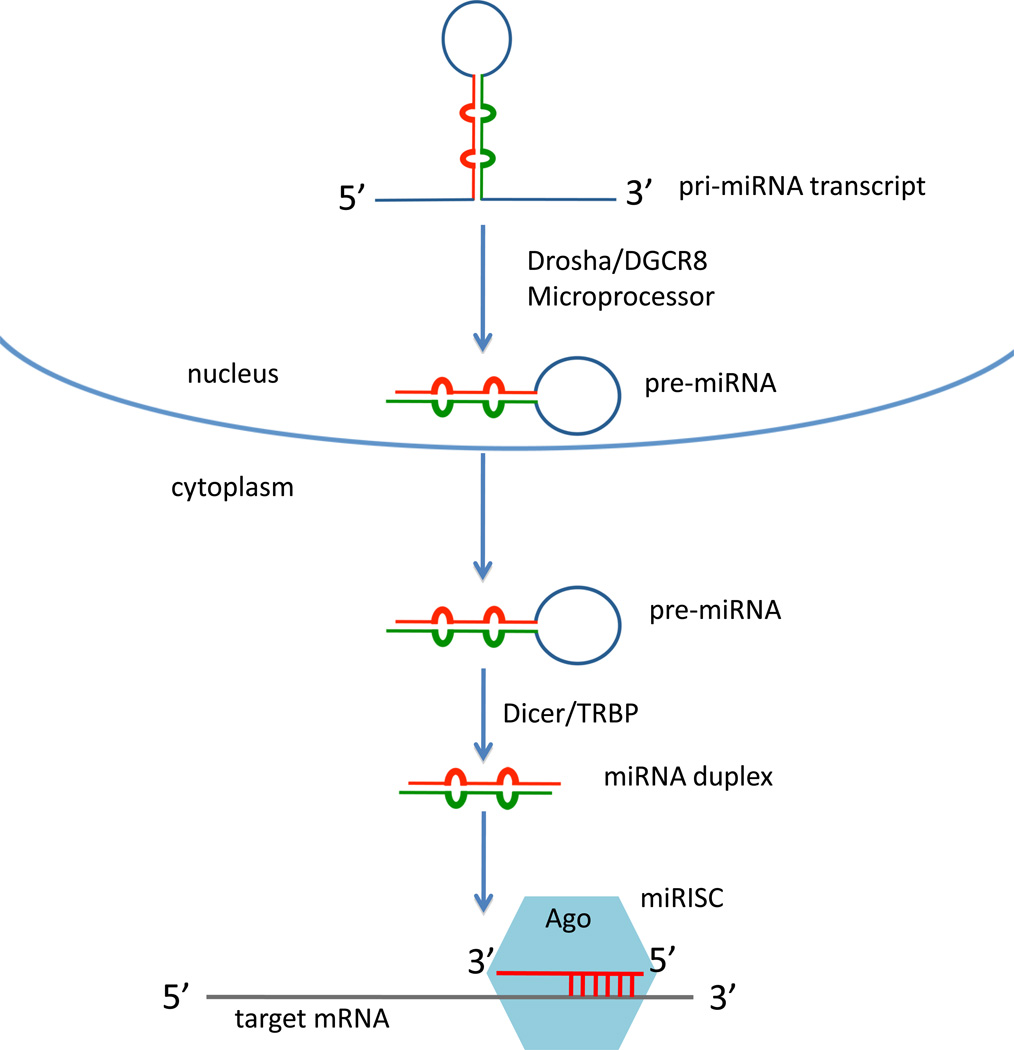
miRNAs are ~20–24 nucleotide (nt) small RNAs that regulate the translation or levels of target mRNA transcripts (Reviewed in [6–9]). Hundreds of miRNAs have been discovered from plants to animals that impact a variety of biological processes. miRNAs are generated through cleavage of a primary transcript in the nucleus (pri-miRNAs) by the Drosha/DGCR8 (Drosha/Pasha in Drosophila) microprocessor to generate a precursor miRNA (pre-miRNA) (Figure 1). The pre-miRNA is exported to the cytoplasm where Dicer cleaves it to release the double-stranded miRNA duplex. One of these strands preferentially associates with the Ago complex to form miRNA Silencing Complex (miRISC), while the other strand is usually degraded. In animals, miRNAs recognize their targets primarily through complementarity with the seed sequence at nucleotides 2–8 of the 5' end of the miRNA. Hundreds of mRNA targets could exist per miRNA family, and it is possible that most protein-coding genes are targets of miRNAs.
Figure 1. The miRNA biogenesis pathway.
The biogenesis of a miRNA starts with the transcription of the primary (pri-miRNA) by RNA Polymerase II. The pri-miRNA is cleaved by the microprocessor complex (Drosha/DGCR8 in vertebrates, Drosha/Pasha in Drosophila) to generate the precursor miRNA (pre-miRNA). The pre-miRNA is transported to the cytoplasm, then cleaved by the Dicer/TRBP (Dicer/Loqs in Drosophila) complex to generate the miRNA duplex. After incorporation into miRISC and strand selection, the mature miRNA strand induces translational repression and/or mRNA cleavage, leading to reduction of the protein.
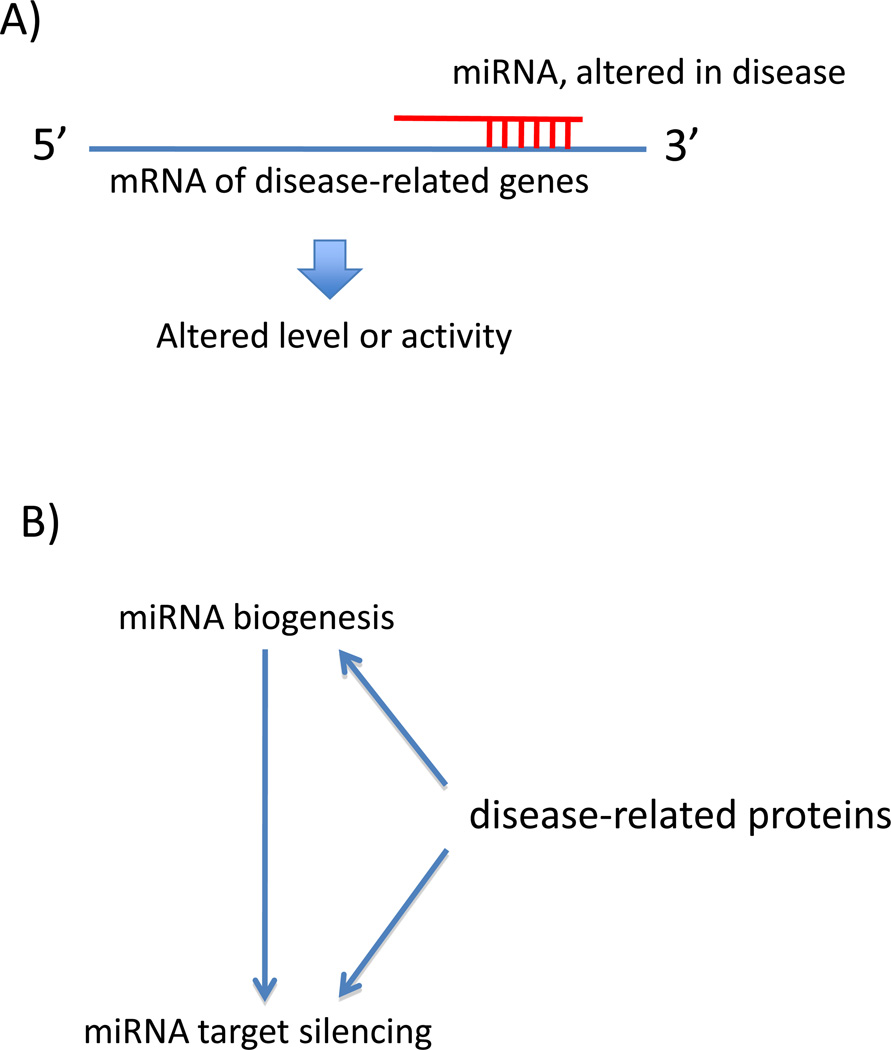
Three main approaches have been taken to study the effects of miRNAs on long-term brain integrity and neurodegenerative disease. First is the disruption of proper miRNA biogenesis followed by examination of the effect on the brain over time. Second is the identification of individual miRNAs that target specific disease genes and the impact. Third is the examination of the impact of disease-associated proteins on the miRNA pathway, such as miRNA biogenesis or mRNA-silencing function (Figure 2). Following, we will describe recent advances in each approach that reveal critical roles of miRNAs on brain integrity.
Figure 2. Ways by which miRNA pathways impact neurodegenerative disease.
(A) miRNAs, which can be altered in disease, may directly target disease-related transcripts, to alter their translation or level.
(B) Evidence also suggests that some disease-related proteins, such as HTT and TDP-43, may directly affect miRNA biogenesis or miRNA target silencing activity.
Disrupting the miRNA biogenesis pathway causes neurodegeneration
A range of approaches — including cloning of miRNAs, miRNA microarrays, and small RNA deep-sequencing analyses — have revealed expression of select miRNAs in the developing mammalian brain and primary neuronal cultures (for example, [10–13]). Analysis of the expression pattern of miRNAs using in situ hybridization with locked nucleic acid (LNA) probes in zebrafish identified their tissue-specific patterns [14]. Such patterns indicated a potential role of miRNAs in neuronal development and function. Subsequent studies revealed roles of specific miRNAs, such as miR-124, in these processes [15–17].
A functional link between miRNAs and neurodegeneration was discovered when studying the effect of global disruption of miRNA biogenesis on neuronal development. Mutants of Dicer in the mouse die early, before neurulation, precluding the ability to assess function in the brain [18]. However, disruption of zebrafish Dicer revealed an essential role in brain morphology and neural differentiation [19]. Injecting a miR-430 duplex rescued the defects in brain morphology, indicating the importance of this specific miRNA. Subsequently, conditional disruption of Dicer in different neuronal populations or cell lines revealed the effect of the miRNA pathway on proliferation, migration and differentiation, as well as long-term neural integrity [20–25]. For example, depleting Dicer in embryonic stem (ES) cells reduces the ability of the cells to differentiate into midbrain dopaminergic neurons [24]—a major neural population compromised in PD. Transfecting the small RNA fraction from embryonic mouse midbrain cells rescues the defect, assigning the role to small RNAs. Consistent with this, deleting Dicer in midbrain dopaminergic neurons in the mouse causes progressive loss of the cells, concomitant with disruption of locomotion, reminiscent of PD [24].
In another example, loss of Dicer from mouse cerebellar Purkinje cells did not impair cellular morphology or function at young ages (8–10 weeks). However, by 13 weeks of age, Purkinje cells, which are the cell type compromised in many ataxias, had progressively degenerated. Intriguingly, the older mice also developed slight tremors and mild ataxia that worsened with age [20]. Disruption of Dicer in spinal motor neurons mimics clinical and pathological features of ALS, a disease associated with loss of motor neurons, indicating a possible impact of the miRNA pathway in pathogenesis of this disease [26]. Interestingly, some key proteins associated with this disease have been shown to modulate miRNA biogenesis or function (below). Deletion of Dicer from glial cells such as astrocytes and oligodendrocytes can cause neural degeneration in the mouse [27, 28]. In addition, conditional loss of Dicer in Schwann cells in mouse revealed its importance for axonal integrity [29]. In humans, dicer protein levels have been found decreased in temporal lobe epilepsy patients with hippocampal cell loss (sclerosis), with about half of the miRNAs in the tissue reduced in levels [30].
In Drosophila, knockdown of Dicer1 is also associated with dopaminergic neural loss and climbing defects [31]. Loss of Dicer1 also enhances toxicity of human pathogenic neurodegenerative disease proteins Ataxin-3 (associated with spinocerebellar ataxia) and Tau (associated with AD and frontotemporal dementia (FTD)) [32]. Intriguingly, depleting Dicer from human HeLa cells also enhances toxicity of disease-associated pathogenic Ataxin-3 protein, and is rescued by adding back the small RNA fraction indicating a role for miRNAs. This study identified a specific miRNA, bantam, that modulates Ataxin-3 and Tau toxicity. Supporting the role of miRNAs in polyQ disease pathogenesis, study of Drosophila miR-34 reveals a potent neuroprotective function in mitigating toxicity of pathogenic forms of Ataxin-3 [33]. Beyond Dicer, haploinsufficiency of DGCR8, a component of the microprocessor complex that cleaves pri-miRNAs to generate pre-miRNAs, leads to neuronal dysfunction in the mouse [34–36].
These studies, which target disruption of components of the miRNA biogenesis pathway, strongly suggest that miRNA activity impacts long-term brain integrity. Note, however, that the identification of individual miRNAs involved is a crucial component of such work. One reason for this is the potential effect of disrupting miRNA biogenesis on the proper expression of many related or unrelated proteins. Another reason is that disrupting the major components in miRNA biogenesis may cause dysfunction or degeneration independent of an effect on miRNAs. For example, Drosha, another component of the microprocessor, is reported to regulate neurogenesis by controlling Neurogenin 2 expression independent of its role in miRNA processing [37]. This function entails Drosha binding and cleavage of a hairpin structure in the 3'UTR of Neurogenin 2 mRNA. Recent studies have also shown that DGCR8 has a much broader impact on RNA processing beyond just miRNAs [38].
Individual miRNAs target disease genes
Efforts to profile miRNAs in tissue from patients with neurodegenerative disease has identified miRNAs that are misregulated in the brain, some of which have been shown to directly target transcripts of familial disease genes. Recent reviews discuss the roles of individual miRNAs on the common neurodegenerative diseases [39–41]. In general, discovering specific miRNAs that target the 3’UTR of key disease genes, and assessing the expression pattern and level of those miRNAs can uncover the extent to which they may impact the level of the disease protein, and thus impact pathogenesis. Here, we highlight a few examples to illustrate the impact of specific miRNAs on select diseases.
AD is the most common neurodegenerative disease, and although predominantly sporadic, analysis of familial situations has identified critical genes for its etiology [2]. The pathological features of AD are the deposit of intracellular neurofibrillary tangles containing Tau protein and extracellular plaques containing Amyloid-beta (Aβ) peptides in the brain. Increased production and impaired clearance of Aβ is a likely cause of Aβ accumulation. Various Aβ peptides are produced upon the cleavage of amyloid precursor protein (APP) by β-site APP-cleaving enzyme 1 (BACE1) and γ-secretase. Some of these processing events promote amyloid formation whereas others do not [3].
Studies in AD highlight the intricate and complex loops of miRNA regulation that can occur. miR-29a/b are downregulated in a subset of AD patients that show elevated BACE1 protein expression, which is predicted to promote amyloidogenic peptide formation [42]. The 3’UTR of BACE1 contains a miR-29a/b target site, and miR-29 targets BACE1. The BACE1 3'UTR also contains sites for other miRNAs, including miR-107, miR-124, and miR-195 [43–45]. miR-107 is downregulated in AD, and targets cofilin, a component of rod-like actin structures in the AD brain. miR-15a, which belongs to miR-107/103 family, is also downregulated in AD patients [46]. Interestingly, the miR-15 family can target extracellular signal-regulated kinase 1 (ERK1), which is a Tau kinase, and this could potentially lead to abnormal Tau phosphorylation in vivo, another pathological hallmark of AD [47]. Other miRNAs implicated in AD pathology include miR-16, miR-101, miR-106a, miR-520c, and miR-153, which target APP [48–50]. Overall, these findings highlight the critical impact of select miRNAs on regulation of the expression of central proteins in AD pathogenesis and progression.
ALS is characterized by the degeneration of motor neurons in the brain and spinal cord, sharing a clinical and pathological spectrum with FTD, the second most common dementia [4]. The RNA binding proteins TDP-43 and FUS are both implicated in pathogenesis of ALS and FTD. TDP-43 is mutated in a subset of ALS patients, and TDP-43 knockdown in human cells leads to aberrant expression of some miRNAs [51]. In mouse, miR-206 deficiency accelerates disease progression in a model of ALS [52], which, together with the effect of Dicer loss in mimicking ALS pathogenesis [26], reinforces the importance of proper regulation of miRNAs and the miRNA pathway to ALS pathogenesis.
An example of discovering a miRNA-target loop that is conserved in flies and humans was revealed with studies of the miR-8 miRNA and one of its targets (Atrophin 1) in flies [53]. The Atrophin-1/Dentatorubral-pallidoluysian atrophy (DRPLA) protein is mutated and accumulates in the polyQ disease DRPLA. The DRPLA protein binds an orthologous protein RERE in vitro, and RERE overexpression causes mislocalization of the DRPLA protein [54]. miR-200b and miR-429 in human potentially target the RERE transcript [53]. Intriguingly, Drosophila miR-8, which has the same seed sequence as miR-200b and miR-429 in humans, targets the Drosophila Atrophin 1 mRNA. Further, miR-8 deletion in flies (which would lead to increased Atrophin 1 protein levels) causes a mild increase of apoptosis in larval brains and climbing defects in adults with age, reminiscent of disease features. These findings indicate a potentially conserved role of the miR-8/miR-200 family in neurodegeneration, contributing to the pathogenesis of DRPLA.
miRNAs can target pathways that impact brain integrity and disease
Many miRNAs may become misregulated in neurodegenerative disease, some of which may have a causal role in pathogenesis. However, it is often unclear what causes miRNA misregulation. One example of a miRNA with a suggested known mechanism is miR-133b. miR- 133b is downregulated in the midbrain of PD patients and in a dopaminergic neuron deficiency model mouse (Aphakia strain) [24]. The Aphakia mouse has a mutation in Pitx3, a transcription factor in which single nucleotide polymorphisms (SNPs) are associated with PD in some reports [55–57]. Pitx3 overexpression leads to upregulation of pre-miR-133b in differentiating ES cells, while miR-133b directly inhibits Pitx3 through its 3'UTR [24]. This suggests a negative feedback loop in which Pitx3 activates miR-133b expression, while miR-133b in turn represses Pitx3 expression. This study raises the possibility that an observed misregulation of miR-133b in PD may be related to the SNPs linked to PD in Pitx3, in at least some, PD situations although there are negative association reports [58–60] Although the knock-out mouse of miR-133b showed no obvious defects in midbrain dopaminergic neurons during development or aging, or in the expression of neuronal genes, including Pitx3, [61], the existence of the other miRNAs in miR- 133 family could explain the lack of effects. The role of miR-133b-Pitx3 loop in PD pathogenesis may limited to a subset of PD patients.
miR-34 defines a miRNA family that is highly conserved in human, fly and Caenorhabditis elegans. In human, the miR-34 family (miR-34a, miR-34b, miR-34c) is misregulated in many cancer types, and regulation of expression of these miRNA by p53 to impact apoptosis and cell cycle control is well defined [62, 63]. miRNA profiling in the adult mouse nervous system revealed enrichment of miR-34a in spinal cord and brainstem regions (medulla oblongata and pons) [11], and miR-34a increases with age in cortex and hippocampus by in situ hybridization [64, 65]. miR-34a is also enriched in the cerebral cortex of an Alzheimer's mouse model [66] and miR-34c increases with age in mouse and human hippocampus, AD patients, and mouse AD models [67]. Targets include SIRT1, whose regulation by miR-34c is associated with memory impairment in mice [67], and is inversely correlated with miR-34a expression in cortex and hippocampus with age [65]. miR-34c also functionally inhibits translation of Bcl-2 in cell studies, an anti-apoptotic protein whose function may also include modulating the processing of APP [66, 68]. Further, miR-34b is elevated in the plasma of Huntington's disease (HD) patients [69].
In contrast, in PD patients miR-34b/c is downregulated at early (pre-motor) stages in brain samples [70]. This study suggests that deficiency in miR-34b/c may promote mitochondrial dysfunction, concomitant with a decrease in Parkin and DJ1, two genetic loci associated with recessive parkinsonism. Overall, these studies highlight misregulation of the miR-34 family in neurodegenerative disease, although it is not clear if misregulation is a cause or consequence of disease pathogenesis.
In Drosophila, miR-34 is a brain-enriched, adult-onset miRNA. Deleting mir-34 causes early onset loss of motor behavior, susceptibility to stress, brain degeneration, and a shorter lifespan [33]. In addition, miR-34 upregulation mitigates polyQ degeneration. One miR-34 target is Eip74EF, a critical gene for the development of the animal [33]. This miRNA-target loop supports the idea of antagonistic pleiotropy; the target gene is beneficial in early life, but deleterious in later life, thus down regulation of the gene in the adult may protect the animal against its deleterious functions in the adult stage [71, 72]. Here, a single miRNA couples age- associated physiology of the animal (climbing, stress) with age-associated brain gene expression and long-term maintenance of the brain [33]. In C. elegans, miR-34 impacts lifespan through regulation of autophagy genes [73]. Given that alterations of miR-34 in mouse and human brain are associated with age and disease, although the precise role (to protect or alternatively promote loss of brain integrity and age-related functions) may be distinct or depend on precise targets, miR-34 is an intriguing a molecular link that may coordinate these age-associated biological processes in multiple organisms.
Disease proteins themselves may impact miRNA biogenesis and/or function
Beyond miRNAs affecting the level of key disease proteins or pathways, disease proteins themselves may directly affect miRNA biogenesis or miRISC target-mRNA silencing. The human Huntingtin (HTT) protein, whose CAG repeat expansion causes HD, interacts with Ago2 protein in cellular P-bodies (sites of mRNA decay) [74]. HTT depletion impairs miRNA target silencing [74], and in HD animal models, many miRNAs are misregulated [75]. This may be due, at least in part, to upregulation of Repressor Element 1 Silencing factor (REST), a transcription factor that is upregulated in HD neurons and can repress hundreds of neural genes [76, 77]. Interestingly, REST binding motifs are found in close proximity to a subset of miRNA genes in the human genome, including miR-9/miR-9*, miR-29a/b, miR-124, and miR-132 [78–81]. In addition, REST and its cofactor coREST have functional miR-9 and miR-9* target sites, respectively [82], and miR-9 is misregulated in HD patients [83]. Thus, REST may be required for the expression of many miRNA genes, although the response may depend on context, such as the differentiation state of the cells [84].
TDP-43 mislocalization and mutation is associated with ALS, and TDP-43 loss-of-function as well as gain-of-function activities may contribute to disease [4, 85]. In human cells, expression of some miRNAs is affected by TDP-43 knockdown [51]. Interestingly, TDP-43 interacts with Drosha and Dicer complexes (which function in the generation of pri-miRNAs and pre-miRNAs, respectively), and binds select pri-miRNAs and pre-miRNAs in the nucleus and the cytoplasm, respectively, through the terminal hairpin loops [86]. Furthermore, nuclear TDP-43 facilitates Drosha-dependent cleavage of select pri-miRNAs, while cytoplasmic TDP-43 promotes Dicer- dependent cleavage of select pre-miRNAs. Interesting questions for the future include the mechanisms by which TDP-43 affects the processing of only a subset of pri- and pre- miRNAs, and the precise impact of these processing defects on disease pathogenesis.
PolyQ expansions in Ataxin-2 (Atx2) are associated with spinocerebellar ataxia-2 (SCA2), parkinsonism and ALS [87, 88]. As with other disease situations, it is possible that aspects of loss-of-function as well as gain-of-function contribute to disease. In flies, Atx2 is required for the silencing activity of select miRNAs; in Atx2-deficient cells, several miRNA reporters (although not all) become upregulated [89]. The detailed mechanisms by which the Atx2 protein impacts miRNA silencing, and whether a role of Atx2 in miRNA silencing is relevant to diseases associated with altered Atx2, remain to be addressed.
These findings suggest roles of disease-relevant proteins in miRNA biogenesis or miRNA target silencing. Given the potential of these proteins to affect these general aspects of miRNAs, and the predicted large number of mRNA targets of miRNAs, it is readily conceivable that disruption of these processes might lead to misregulated expression of a large number of proteins. Such gross disruption has the potential to profoundly impact long-term neural function and integrity.
Concluding remarks
Recent technological advances in high-throughput small RNA profiling in vivo have identified changes in the small RNA population in neurodegenerative disease or with age. So far, functions of only a handful of these miRNAs have been revealed, and an important question will be identifying the roles of the many other miRNAs that change in disease or in an age-associated manner. In addressing this question, studies of C. elegans have provided crucial insights into miRNA function; not all miRNAs that change with age lead to modulated lifespan upon altered activity [90], and many miRNAs are not required for development or viability [91, 92]. Redundancy among different miRNAs is one possibility. Alternatively, the changes of such miRNAs could be a result, rather than cause, of age-associated physiological events. Another possibility is that such miRNAs might be required only in a perturbed environment or genetic background to confer ‘robustness’ on gene expression [93–95]. Considering this, sensitizing the background when studying loss-of-function of miRNAs [94, 96], or examining stressful conditions might uncover novel roles. Another important question is what initiates misregulation of miRNAs in age or disease, especially for those miRNAs that impact age- associated events or the onset or progression of neurodegeneration. We have only limited examples of such mechanisms, such as where the disease-relevant genes themselves seem to trigger the misregulation of miRNAs.
Profiling of miRNAs by deep-sequencing analyses reveals that, in addition to simple up- or downregulation, miRNAs show potential differential isoform accumulation in the HD context [83], or clearly with age such that — in the case of Drosophila miR-34 — only the shorter 21 nucleotide form accumulates [33]. Although the biological meaning of this processing is not yet defined, it is intriguing that production of different isoforms of miRNAs is regulated by a specific exonuclease in flies [97, 98]. This indicates that miRNA 3’end trimming may be a biologically critical process. Future careful analyses promise to reveal the unidentified function of many miRNAs and the impact of controlling miRNA isoform accumulation with age and/or disease.
As with Drosophila miR-34, studying the expression and function of individual miRNAs throughout adult lifespan might reveal a coordinated role of miRNAs in various aspects of age- associated processes from lifespan to long-term brain integrity. To this end, Drosophila and C. elegans remain key model organisms with their relatively shorter lifespan and ease of observing defects at the organismal level. Identifying more miRNAs that are modulated with age and detailed studies of individual miRNAs in multiple systems will bring more insights into the impact of miRNAs in age-associated processes and brain disease.
Therapeutic potential of miRNAs
Based on disease-associated changes in miRNA levels, such as the miR-34 family, one could potentially use changes in miRNA expression as biomarkers of aging and age-associated processes such as neurodegenerative disease [69, 99]. In addition, with the identification of miRNA with functions like miR-34, and the successful delivery of disease-modulating miRNAs, it is tantalizing to target the miRNA itself in vivo to mitigate disease onset or progression. Spinal and bulbar muscular atrophy (SBMA) is a polyQ disease with expansion in the Androgen Receptor (AR) gene [1]. miR-196a and miR-196b were identified as miRNAs that decrease the level of both the normal and CAG-repeat expanded AR transcripts [100]. Interestingly, these miRNAs directly target the mRNA of CUGBP Elav-like family member 2 (CELF2), whose protein positively regulates the AR mRNA level through a site on the transcript that is distinct from the CAG repeat — that is, a CTG repeat. Silencing of the CELF2 mRNA in spinal cord motor neurons by delivering miR-196a via an adeno-associated viral vector (AAV) improves the motor function of SBMA model mice. These findings, together with other examples, highlight the complex pathways and loops-within-loops of gene regulation that are impacted by miRNAs. They also offer promise that targeting and tweaking miRNAs through a variety of approaches could have therapeutic potential for neurodegenerative diseases.
ACKNOWLEDGEMENTS
We thank V. Lekova and N. Liu for helpful comments. We apologize to those colleagues whose work could not be cited due to space limitations. Research in the Bonini laboratory is supported by grants from the NIH and The Ellison Medical Foundation. NMB is an investigator of the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Ballard C, et al. Alzheimer's disease. Lancet. 2011;377:1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer's disease. Annu Rev Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferraiuolo L, et al. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7:616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- 5.Coelho M, Ferreira JJ. Late-stage Parkinson disease. Nat Rev Neurol. 2012 doi: 10.1038/nrneurol.2012.126. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krol J, et al. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 11.Bak M, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berezikov E, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapsimali M, et al. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanuki R, et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 16.Cao X, et al. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visvanathan J, et al. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 19.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 20.Schaefer A, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damiani D, et al. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi PS, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLoughlin HS, et al. Dicer is required for proliferation, viability, migration and differentiation in corticoneurogenesis. Neuroscience. 2012 doi: 10.1016/j.neuroscience.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haramati S, et al. miRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci U S A. 2010;107:13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao J, et al. Deletion of astroglial Dicer causes non-cell-autonomous neuronal dysfunction and degeneration. J Neurosci. 2011;31:8306–8319. doi: 10.1523/JNEUROSCI.0567-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin D, et al. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol. 2009;66:843–857. doi: 10.1002/ana.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira JA, et al. Dicer in Schwann cells is required for myelination and axonal integrity. J Neurosci. 2010;30:6763–6775. doi: 10.1523/JNEUROSCI.0801-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKiernan RC, et al. Reduced mature microRNA levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PLoS One. 2012;7:e35921. doi: 10.1371/journal.pone.0035921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gehrke S, et al. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilen J, et al. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 33.Liu N, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482:519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stark KL, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 35.Fenelon K, et al. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schofield CM, et al. Monoallelic deletion of the microRNA biogenesis gene Dgcr8 produces deficits in the development of excitatory synaptic transmission in the prefrontal cortex. Neural Dev. 2011;6:11. doi: 10.1186/1749-8104-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knuckles P, et al. Drosha regulates neurogenesis by controlling Neurogenin 2 expression independent of microRNAs. Nat Neurosci. 2012;15:962–969. doi: 10.1038/nn.3139. [DOI] [PubMed] [Google Scholar]
- 38.Macias S, et al. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delay C, et al. MicroRNAs in Alzheimer's disease. Neurobiol Dis. 2012;46:285–290. doi: 10.1016/j.nbd.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Mouradian MM. MicroRNAs in Parkinson's disease. Neurobiol Dis. 2012;46:279–284. doi: 10.1016/j.nbd.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 41.Gascon E, Gao FB. Cause or Effect: Misregulation of microRNA Pathways in Neurodegeneration. Front Neurosci. 2012;6:48. doi: 10.3389/fnins.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hebert SS, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang WX, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang M, et al. The miR-124 regulates the expression of BACE1/beta-secretase correlated with cell death in Alzheimer's disease. Toxicol Lett. 2012;209:94–105. doi: 10.1016/j.toxlet.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 45.Zhu HC, et al. MicroRNA-195 downregulates Alzheimer's disease amyloid-beta production by targeting BACE1. Brain Res Bull. 2012;88:596–601. doi: 10.1016/j.brainresbull.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 46.Wang WX, et al. Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hebert SS, et al. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010;19:3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 48.Long JM, Lahiri DK. Current drug targets for modulating Alzheimer's amyloid precursor protein: role of specific micro-RNA species. Curr Med Chem. 2011;18:3314–3321. doi: 10.2174/092986711796504592. [DOI] [PubMed] [Google Scholar]
- 49.Long JM, et al. MicroRNA-153 physiologically inhibits expression of amyloid-beta precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J Biol Chem. 2012 doi: 10.1074/jbc.M112.366336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang C, et al. MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 2012;1455:103–113. doi: 10.1016/j.brainres.2011.10.051. [DOI] [PubMed] [Google Scholar]
- 51.Buratti E, et al. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010;277:2268–2281. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- 52.Williams AH, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karres JS, et al. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 54.Yanagisawa H, et al. Protein binding of a DRPLA family through arginine-glutamic acid dipeptide repeats is enhanced by extended polyglutamine. Hum Mol Genet. 2000;9:1433–1442. doi: 10.1093/hmg/9.9.1433. [DOI] [PubMed] [Google Scholar]
- 55.Fuchs J, et al. The transcription factor PITX3 is associated with sporadic Parkinson's disease. Neurobiol Aging. 2009;30:731–738. doi: 10.1016/j.neurobiolaging.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 56.Le W, et al. Transcription factor PITX3 gene in Parkinson's disease. Neurobiol Aging. 2011;32:750–753. doi: 10.1016/j.neurobiolaging.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 57.Gui Y, et al. A novel synonymous SNP in PITX3 is associated with Parkinson's disease in Chinese population. Swiss Med Wkly. 2012;142:w13521. doi: 10.4414/smw.2012.13521. [DOI] [PubMed] [Google Scholar]
- 58.de Mena L, et al. Analysis of the Micro-RNA-133 and PITX3 genes in Parkinson's disease. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1234–1239. doi: 10.1002/ajmg.b.31086. [DOI] [PubMed] [Google Scholar]
- 59.Cai Y, et al. Genetic variants of the PITX3 gene are not associated with late-onset sporadic Parkinson's disease in a Chinese population. Neurosci Lett. 2011;498:124–126. doi: 10.1016/j.neulet.2011.04.073. [DOI] [PubMed] [Google Scholar]
- 60.Cai Y, et al. PITX3 polymorphism is not associated with Parkinson's disease in a Chinese population. Neurosci Lett. 2011;505:260–262. doi: 10.1016/j.neulet.2011.10.034. [DOI] [PubMed] [Google Scholar]
- 61.Heyer MP, et al. Normal Midbrain Dopaminergic Neuron Development and Function in miR-133b Mutant Mice. J Neurosci. 2012;32:10887–10894. doi: 10.1523/JNEUROSCI.1732-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong MY, et al. microRNA-34 family and treatment of cancers with mutant or wild-type p53 (Review) Int J Oncol. 2011;38:1189–1195. doi: 10.3892/ijo.2011.970. [DOI] [PubMed] [Google Scholar]
- 63.He X, et al. The guardian's little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–11101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 64.Khanna A, et al. Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging (Albany NY) 2011;3:223–236. doi: 10.18632/aging.100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, et al. Circulatory miR34a as an RNAbased, noninvasive biomarker for brain aging. Aging (Albany NY) 2011;3:985–1002. doi: 10.18632/aging.100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, et al. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer's disease, inhibits bcl2 translation. Brain Res Bull. 2009;80:268–273. doi: 10.1016/j.brainresbull.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 67.Zovoilis A, et al. microRNA-34c is a novel target to treat dementias. EMBO J. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rohn TT, et al. Lack of pathology in a triple transgenic mouse model of Alzheimer's disease after overexpression of the anti-apoptotic protein Bcl-2. J Neurosci. 2008;28:3051–3059. doi: 10.1523/JNEUROSCI.5620-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaughwin PM, et al. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington's disease. Hum Mol Genet. 2011;20:2225–2237. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- 70.Minones-Moyano E, et al. MicroRNA profiling of Parkinson's disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 71.Hughes KA, Reynolds RM. Evolutionary and mechanistic theories of aging. Annu Rev Entomol. 2005;50:421–445. doi: 10.1146/annurev.ento.50.071803.130409. [DOI] [PubMed] [Google Scholar]
- 72.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J, et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Dordr) 2011 doi: 10.1007/s11357-011-9324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Savas JN, et al. Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci U S A. 2008;105:10820–10825. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee ST, et al. Altered microRNA regulation in Huntington's disease models. Exp Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 76.Johnson R, et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zuccato C, et al. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington's disease. J Neurosci. 2007;27:6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Conaco C, et al. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson R, et al. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 81.Johnson R, Buckley NJ. Gene dysregulation in Huntington's disease: REST, microRNAs and beyond. Neuromolecular Med. 2009;11:183–199. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- 82.Packer AN, et al. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marti E, et al. A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235. doi: 10.1093/nar/gkq575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao Z, et al. Profiling of REST-Dependent microRNAs Reveals Dynamic Modes of Expression. Front Neurosci. 2012;6:67. doi: 10.3389/fnins.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cohen TJ, et al. TDP-43 functions and pathogenic mechanisms implicated in TDP-43 proteinopathies. Trends Mol Med. 2011;17:659–667. doi: 10.1016/j.molmed.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lastres-Becker I, et al. Spinocerebellar ataxia 2 (SCA2) Cerebellum. 2008;7:115–124. doi: 10.1007/s12311-008-0019-y. [DOI] [PubMed] [Google Scholar]
- 88.Bonini NM, Gitler AD. Model organisms reveal insight into human neurodegenerative disease: ataxin-2 intermediate-length polyglutamine expansions are a risk factor for ALS. J Mol Neurosci. 2011;45:676–683. doi: 10.1007/s12031-011-9548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McCann C, et al. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc Natl Acad Sci U S A. 2011;108:E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Lencastre A, et al. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol. 2010;20:2159–2168. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 94.Kato M, Slack FJ. Ageing and the small, non-coding RNA world. Ageing Res Rev. 2012 doi: 10.1016/j.arr.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brenner JL, et al. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu N, et al. The exoribonuclease Nibbler controls 3' end processing of microRNAs in Drosophila. Curr Biol. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han BW, et al. The 3'-to-5' exoribonuclease Nibbler shapes the 3' ends of microRNAs bound to Drosophila Argonaute1. Curr Biol. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pincus Z, et al. MicroRNA predictors of longevity in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002306. doi: 10.1371/journal.pgen.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miyazaki Y, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012 doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]