Abstract
Kissing interactions in RNA are formed when bases between two hairpin loops pair. Intra- and intermolecular kissing interactions are important in forming the tertiary or quaternary structure of many RNAs. Self-cleavage of the wild-type Varkud satellite (VS) ribozyme requires a kissing interaction between the hairpin loops of stem-loops I and V. In addition, self-cleavage requires a rearrangement of several base pairs at the base of stem I. We show that the kissing interaction is necessary for the secondary structure rearrangement of wild-type stem-loop I. Surprisingly, isolated stem-loop V in the absence of the rest of the ribozyme is sufficient to rearrange the secondary structure of isolated stem-loop I. In contrast to kissing interactions in other RNAs that are either confined to the loops or culminate in an extended intermolecular duplex, the VS kissing interaction causes changes in intramolecular base pairs within the target stem-loop.
RNA kissing interactions, also called loop–loop pseudoknots, occur when the unpaired nucleotides in one hairpin loop base pair with the unpaired nucleotides in another hairpin loop (1). When the hairpin loops are located on separate RNA molecules, their intermolecular interaction is called a kissing complex. These interactions generally form between stem-loops containing extensive complementarity; however, stable complexes have been observed containing only two intermolecular Watson–Crick base pairs (2).
Intramolecular kissing interactions are observed in the native structures of a variety of RNAs including Varkud satellite (VS) RNA, tRNA, and group I introns (3–6). These kissing interactions contribute to the assembly and stabilization of their respective RNA structures by joining and orienting helices. Kissing interactions may stabilize both native and nonnative interactions during tertiary folding, which can affect the rate at which the native structure is formed (7). Kissing interactions often form distorted structures that can serve as recognition sites for proteins, RNAs, metal ions, or other ligands (8–10). As a result, kissing interactions contribute to the stability of an RNA structure by affecting both global and local RNA interactions.
The transient formation of an intermolecular kissing complex is required for RNA dimerization during the life cycle of retroviruses (11–15), and for the formation of some antisense-target complexes (16, 17). Kissing complexes in which the loop nucleotides are complementary can form stable dimers that contain intermolecular base pairs between the loop nucleotides only. Other stem-loops with more extensive complementarity sometimes form unstable kissing complexes, which are quickly remodeled into stable duplex or cruciform isoforms (18–20). Secondary structure remodeling involves breaking intramolecular base pairs in each hairpin and using the bases to form intermolecular helices.
The VS ribozyme contains a kissing interaction that is required for self-cleavage of the wild-type RNA in vitro (6). This interaction involves Watson–Crick base-pairing between loops I and V, and most likely constrains the orientation of these helices in the native structure (Fig. 1A). The self-cleavage and ligation reactions of wild-type VS RNA also require a secondary structure rearrangement of stem-loop I (Fig. 1A; ref. 22). We refer to the secondary structure of stem-loop I required for activity as “shifted,” because the 3′ side of stem I is shifted “up” one base compared with the structure of stem-loop I in the absence of magnesium or ribozyme (“unshifted”; ref. 22).
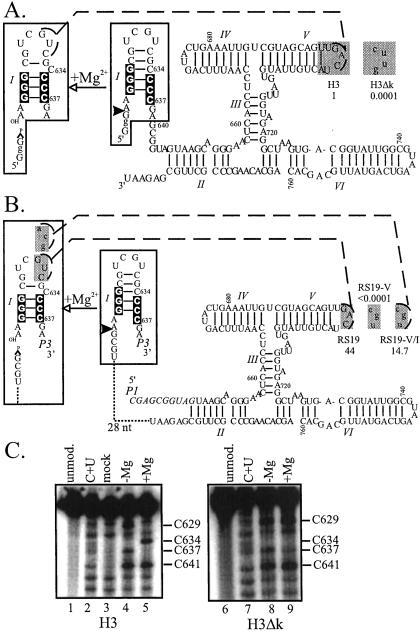
Figure 1.
Secondary structure schematics of VS RNA constructs: H3 (21) and H3Δk (A) and RS19 (ref. 22; B). VS positions are numbered as in ref. 23, and helices are numbered as in ref. 24. P1 and P3, in italics, are primer-binding sites attached where indicated (22). A functionally important kissing interaction is indicated by a dashed line (6). The unfilled boxed region indicates the secondary structure rearrangement and site-specific cleavage that occur in the presence of magnesium. Nucleotides in the lower part of stem-loop I involved in the conformational change are indicated by black boxes (22). The sequences in the gray boxes in H3 or RS19 were substituted by those in the corresponding gray boxes to construct the individual mutants. Numbers below each mutant name represent the observed cis-cleavage rate constant (per minute) of each mutant RNA. Non-VS sequences are indicated by lowercase letters or a dotted line. The cleavage site is indicated by a filled arrowhead. (C) DMS modification of 3′ end-labeled H3 D and H3Δk D RNAs (the region of the RNAs from nt 618 to nt 647 is shown). Modifications were performed on RNA in the presence (+) or absence (−) of 50 mM MgCl2, as indicated. The input material (unmod.), a chemical sequencing C + U ladder, and a control sample not exposed to DMS (mock) are included.
In this study we demonstrate that the kissing interaction between stem-loops I and V is necessary to rearrange wild-type stem-loop I. Site-directed mutagenesis showed that the requirement of the kissing interaction for self-cleavage can be partially bypassed in RNAs with a mutant shifted stem-loop I. The partial suppression of the self-cleavage defect of mutants with a disrupted kissing interaction further supports a functional role of this interaction in the rearrangement of stem-loop I. Because the suppression is only partial, the kissing interaction likely performs additional functions. Lastly, isolated stem-loop V presented in trans forms a kissing complex with, and rearranges, an isolated stem-loop I. Therefore, even though complementarity between stem-loops I and V does not extend beyond the loop nucleotides, the local interactions within the VS kissing complex are sufficient for the intramolecular rearrangement of base pairs within stem I.
Materials and Methods
Clones.
H3 has been described (Fig. 1A; ref. 21). H3Δk (disrupted kissing interaction) was derived from clone H3 by replacing nucleotides 695–701 in stem-loop V with a CUUG tetraloop (Fig. 1A). SH (for Shifted Helix) has a C634g substitution in stem-loop I of H3 (Fig. 2A). SHΔk contains the C634g mutation and the CUUG substitution in loop V.
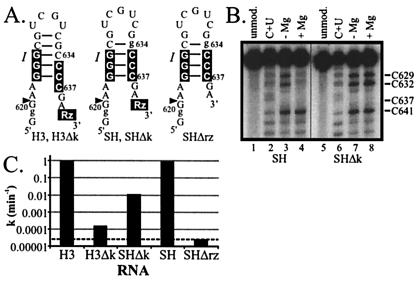
Figure 2.
One functional role of the kissing interaction is to rearrange stem-loop I. (A) Secondary structures of the stem-loop I region of H3, H3Δk, SH, SHΔk, and SHΔrz. Helices II to VI are indicated by a filled rectangle marked Rz. Non-VS sequences, the cleavage site, and nucleotides in the lower part of stem-loop I involved in the conformational change are indicated as in Fig. 1A. (B) DMS modification of 3′ end-labeled SH D and SHΔk D RNAs (the region of the RNA from nt 618 to nt 647 is shown). Modifications were performed as described in Fig. 1C. (C) A mutant constitutively shifted stem-loop I partially suppresses the self-cleavage defect of a disrupted kissing interaction mutant. The first order rate constants are shown on the y axis (averaged from at least two experiments), and the VS RNA mutant on the x axis. The dotted line indicates the rate of background cleavage. See Materials and Methods for details.
RS19 is a circular permutation of H3 in which stem-loop I has been moved to the 3′ end of the ribozyme (22). RS19-V was derived from RS19, and has G697c, A698 g, and C699u substitutions in loop V. RS19-V/I was derived from RS19-V, and additionally has G630a, U631c, and C632g substitutions in loop I.
Synthesis of RNAs.
Precursor (Pre) RNAs of H3 or RS19 (or mutant derivatives) were synthesized by T7 RNA polymerase transcription (25) from plasmid DNAs linearized at the SspI site for H3 and mutant derivatives, or at the EcoRI site for RS19 and mutant derivatives. Internally labeled, radioactive Pre RNAs used for self-cleavage assays (see below) were obtained by minimizing self-cleavage during the in vitro transcription reactions, as described (26). H3 downstream cleavage product (D) and SH D were obtained by self-cleavage during the in vitro transcription reaction. H3Δk D and SHΔk D were obtained through trans cleavage by a ribozyme derived from VS RNA (27).
Substrate for trans cleavage assays, containing stem-loop I, was obtained by T7 RNA polymerase transcription from H3 plasmid DNA digested with AvaI (nt 639). Stem-loop I for chemical structure probing was obtained from self-cleavage of RS19 Pre RNAs (or mutant derivatives) during in vitro transcription of RS19 plasmids (or mutant derivatives) digested with EcoRI (22). Wild-type RS19 D contains stem-loop I (VS nt 621–639), followed by primer binding site P3 (22). Mutant RS19-V/I D contains three substitutions within stem-loop I: G630a, U631c, and C632 g. Wild-type stem-loop V, purchased from Dharmacon Research (Boulder, CO) contains nucleotides 688–708 of VS RNA. Mutant stem-loop V was synthesized by T7 RNA polymerase transcription by using an oligonucleotide template (28). RNAs were purified by electrophoresis through a polyacrylamide/8.3 M urea gel of the appropriate percentage, and recovered from the gel as described (22).
Self-Cleavage Assay and Kinetic Analysis.
RS19 Pre or mutant derivatives were incubated at 37°C for 2 min in 40 mM Tris⋅HCl (pH 8.0)/50 mM KCl and self-cleavage was initiated by the addition of MgCl2 to a final concentration of 10 mM. The self-cleavage activity of H3 and mutant derivatives was analyzed in the buffer used for chemical structure probing. H3 or SH Pre was incubated at 37°C for 10 min in 200 mM Hepes (pH 8.0)/50 mM KCl and self-cleavage was initiated by addition of MgCl2 to a final concentration of 100 mM. H3Δk or SHΔk Pre did not self-cleave following this protocol. Instead, these RNAs were heated at 90°C for 5 min in 200 mM Hepes (pH 8.0)/50 mM KCl. MgCl2 was added to a final concentration of 100 mM, and the RNAs were cooled on ice for 5 min. No cleavage occurred during the heating and cooling steps. Self-cleavage of H3Δk and SHΔk was initiated by incubation at 37°C. Reactions were terminated at various time points by removing aliquots into three volumes of formamide loading dye (80% formamide/50 mM EDTA/22.5 mM Tris-borate/0.1% bromophenol blue/0.1% xylene cyanol). Cleavage products were separated from precursor by electrophoresis through a 4% polyacrylamide/8.3 M urea gel. RNA was visualized by exposing to a PhosphorImager screen (Molecular Dynamics).
Experimental data from self-cleavage time courses fit to the single exponential equation (r2 > 0.976)
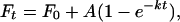 |
where Ft is the fraction of RNA cleaved at time t, F0 is the fraction of RNA cleaved at t = 0, A is the fraction of RNA capable of cleavage, and k is the first order rate constant. A varied from 0.66–0.9, depending on the RNA sequence. The parameters were determined by nonlinear regression.
Chemical Structure Probing.
H3 D or RS19 D (or mutant derivatives) RNAs were labeled at the 3′ end by using 5′-[32P]pCp and RNA ligase (29). RNAs were modified with dimethyl sulfate (DMS) as described (30, 31). Modifications of <20 nM stem-loop I RNAs were performed in the presence of 200 mM Hepes (pH 8.0) and 50 mM NaCl at 30°C, and the concentrations of MgCl2 shown in the figure legends. Modifications of <20 nM D RNAs were performed in the presence of 200 mM Hepes (pH 8.0)/50 mM KCl at 37°C and the concentrations of MgCl2 indicated in the legends. The sites of cytosine modification were determined by treatment with 50% (vol/vol) anhydrous hydrazine/water followed by aniline cleavage. Uridine + cytosine ladders were obtained as described (32). The RNAs were electrophoresed through an 8% (D) or 12% (stem-loop I) polyacrylamide/8.3 M urea gel and exposed to a PhosphorImager screen.
Trans Cleavage Assay.
Substrate for trans cleavage was labeled at the 3′ end as described above, and <20 nM was incubated at 30°C for 10 min in 200 mM Hepes (pH 8.0)/50 mM NaCl. Stem-loop V, VS ribozyme, or water was added to the concentrations indicated in the figure legend, followed by MgCl2 to a final concentration of 50 mM. Reactions were terminated by removing an aliquot into three volumes of formamide loading. Cleavage products were separated from substrate by electrophoresis through a 20% polyacrylamide/8.3 M urea gel and visualized as described above.
Results
Chemical Modification Reveals the Kissing Interaction Is Required for the Secondary-Structure Rearrangement of Stem-Loop I.
To extend our previous investigation of the role of the loop I-V kissing interaction in the self-cleavage of VS RNA (6), we performed mutational analysis of constructs H3 and RS19 (Fig. 1 A and B). Replacing loop V with a 5′-CUUG-3′ tetraloop sequence disrupts the kissing interaction by abolishing Watson–Crick base-pairing with wild-type loop I. This loop V substitution significantly reduced the self-cleavage activity of H3 (H3Δk; Fig. 1A), consistent with the previously observed effect of this substitution in the G11 construct (26). Self-cleavage of H3Δk was about 10,000-fold slower than H3, barely detectable above background uncatalyzed cleavage (see Fig. 2C). The kissing interaction was disrupted in RS19, a circular permutation of H3, by substituting three positions within loop V (Fig. 1B, RS19-V). These substitutions reduced the self-cleavage activity at least 400,000-fold. Activity was restored, almost to the rate of RS19 wild-type RNA, when the loop V substitutions were combined with complementary substitutions in loop I that would restore the kissing interaction (RS19-V/I). These data reiterate the functional importance of the kissing interaction in different circular permutations of VS RNA, and enable analysis of intra and intermolecular versions of this kissing interaction.
To determine whether the loop I-V kissing interaction is required for secondary structure rearrangement of wild-type stem-loop I, we performed DMS modification analysis of H3 and H3Δk. Chemical modification was performed on the downstream cleavage products (D) of these RNAs, rather than on the precursors (Pre). H3 Pre self-cleaves in the presence of magnesium; H3Δk Pre was cleaved in trans by a wild-type VS ribozyme (27). Analysis of D avoids the analysis of a mixed population of Pre and D in the presence of magnesium (24). In addition, D differs from the minimal self-cleaving domain by only one nucleotide at the 5′ end, the identity of which is unimportant (21). In the absence of magnesium C629 and, to a lesser extent, C637, of H3 D were accessible to DMS modification and C626, 634, 635, and 636 were protected, as expected for the unshifted conformation (Fig. 1C, lane 4). The partial protection of C637 could be due to its interaction with A622 in a non-Watson–Crick pair, as observed by NMR in an isolated, model stem-loop I in the unshifted conformation (33). In the presence of magnesium (lane 5), C629 was protected from modification by a yet unknown interaction, as observed (24). Also, C637 was protected from modification, consistent with its pairing with G623 in the shifted conformation of stem-loop I. Importantly, C634 was accessible to modification only in the presence of magnesium (compare lanes 4 and 5), indicating that this nucleotide becomes unpaired to accommodate shifting of the lower base pairs. The DMS modification of H3 D indicates that the conformation of stem-loop I is unshifted in the absence of magnesium, and shifted in the presence of magnesium.
In the absence of magnesium, the reactivities of the cytosines within stem-loop I of H3Δk D revealed the unshifted conformation: C634 was protected from modification, and only C629 and, to a lesser extent, C637 were reactive (lane 8). In the presence of magnesium, C637 of H3Δk D was even less reactive (lane 9), possibly because of stabilization of the C:A+ pair (see above). In contrast to H3 D, C634 remained completely protected from DMS modification in the presence of magnesium, indicating that stem I remains in the unshifted conformation (lane 9). Also, C629 remained reactive, indicating that its protection depends on formation of the kissing interaction. These mutational and biochemical data strongly suggest that a functional role of the loop I-V interaction in the wild-type RNA is to rearrange stem-loop I.
The Self-Cleavage Defect of the Loop V Mutant Is Partially Suppressed by a Shifted Stem I.
Previous observations indicate that both the kissing interaction and the rearrangement of stem-loop I are required for self-cleavage of wild-type VS RNA (6, 22). DMS structure probing of H3Δk indicates that the kissing interaction is required to rearrange stem-loop I. Because a functional role of the kissing interaction is to rearrange stem-loop I, the requirement for this interaction might be bypassed in RNAs that contain a mutant stem-loop I that adopts the shifted structure even in the absence of the kissing interaction. The self-cleavage defect of disrupted kissing interaction mutants (e.g., H3Δk) would be suppressed completely in such a constitutively shifted stem-loop I if the rearrangement is the only role of the kissing interaction; however, suppression would be partial, or would not occur, if the kissing interaction performs additional functions required for self-cleavage, or if any of the mutations are otherwise deleterious.
To determine whether a mutant shifted stem-loop I increases the self-cleavage rate of an RNA with a disrupted kissing interaction, and therefore possibly bypasses this interaction, we made a Shifted Helix mutant of H3Δk called SHΔk (Fig. 2A). SHΔk contains a C634g substitution (22). DMS modification analysis confirmed that stem-loop I in SHΔk D adopts the shifted structure even in the absence of magnesium (Fig. 2B). In either the absence or presence of magnesium only C632 and C629 were reactive, indicative of the expected four base pair shifted stem and the disrupted kissing interaction. SHΔk self-cleaved almost 100-fold faster than H3Δk (Fig. 2C). The activities of these RNAs are consistent with the hypothesis that mutationally shifting stem-loop I bypasses one of the roles of the kissing interaction in the wild-type RNA, the rearrangement of base pairing in stem I.
To determine whether SHΔk achieves a faster self-cleavage rate than H3Δk by bypassing a functional role of the kissing interaction, or alternatively by a mechanism independent of the kissing interaction, the same mutant shifted helix was introduced into wild-type H3 (SH, Fig. 2A). The DMS modification pattern verified that stem-loop I in SH D adopted the shifted structure in the absence of magnesium, and formed the expected tertiary interactions in the presence of magnesium (Fig. 2B). In the absence of magnesium, stem-loop I adopted the four base pair shifted conformation, revealed by the reactivities of only C629 and C632 (lane 3). In the presence of magnesium, C629 and C632 both were protected from modification (lane 4). C632 is protected because of base pairing with G697 in the loop I-V kissing interaction, whereas C629 is protected by an unidentified tertiary interaction, as previously observed (see above and refs. 22 and 24). If the faster cleavage rate of SHΔk compared with H3Δk occurs by a mechanism independent of the kissing interaction, SH might also self-cleave 100-fold faster than H3; however, we observe that SH self-cleaves at the same rate as H3 (Fig. 2C). In addition, both SH and H3 self-cleave ≈100-fold faster than SHΔk. This observation suggests that either there are multiple functions of the kissing interaction, only some of which are overcome in an RNA with a mutant shifted stem-loop I, or that the loop V substitutions in SHΔk are deleterious for reasons additional to the disruption of the kissing interaction. A functional role for the kissing interaction, other than the secondary structure rearrangement of wild-type stem-loop I, may be to stabilize the overall tertiary structure of the ribozyme (see Discussion). Taken together, these data suggest that a mutant shifted stem-loop I increases the self-cleavage rate of SHΔk by bypassing a function of the kissing interaction.
Although SHΔk partially bypasses the requirement for the kissing interaction in self-cleavage, it does not bypass the requirement for other ribozyme sequences. The self-cleavage rate of SHΔrz, which corresponds to the shifted stem-loop I lacking the rest of the ribozyme sequence (Fig. 2A), was 300-fold slower than that of full-length SHΔk (Fig. 2C). The rate of cleavage at G620 in SHΔrz was equal to the rate of background cleavage at G618 or g619 within stem-loop I (data not shown), indicating that a shifted stem-loop I alone is not capable of site-specific self-cleavage, and that the active site of the VS ribozyme must include sequences outside of stem-loop I. Therefore, an RNA with a mutant shifted stem-loop I partially overcomes the need for the kissing interaction by adopting a conformation that is necessary, but not sufficient, for catalysis.
Stem-Loop V Is Sufficient for the Rearrangement of Stem-Loop I.
We have shown that the kissing interaction is required to rearrange wild-type stem-loop I, and that an RNA with a mutant shifted stem-loop I partially bypasses the functional requirement of the kissing interaction. The kissing interaction may accomplish the secondary structure rearrangement of wild-type stem-loop I directly through local interactions between stem-loops I and V, or indirectly by facilitating tertiary interactions between stem-loop I and other regions of the ribozyme. To distinguish between these mechanisms, we investigated whether a complex forms between stem-loops I and V, and whether this complex is sufficient to rearrange stem-loop I.
Fig. 3A shows the DMS modification pattern of wild-type stem-loop I in the absence or presence of wild-type stem-loop V and/or magnesium. In the absence of stem-loop V, stem-loop I is in the unshifted conformation in the absence or presence of magnesium, deduced from C634 being protected from modification (Fig. 3A, lanes 4 and 6; ref. 22). C637 is less reactive in the presence of magnesium, possibly because of stabilization of a non-Watson–Crick base pair with A622 in the unshifted conformation of stem-loop I (33). The chemical modification pattern in the presence of magnesium and stem-loop V indicated the rearrangement of stem-loop I into the shifted conformation (lane 7): C634 was very reactive, as expected when it becomes unpaired to accommodate shifting of the lower base pairs (22). The DMS modification pattern of stem-loop I in the presence of stem-loop V, but in the absence of magnesium, was consistent with the unshifted conformation of stem-loop I: C634 was not reactive with DMS (lane 5). These biochemical data reveal that stem-loop V, in the presence of magnesium, is sufficient to rearrange the secondary structure of stem-loop I.
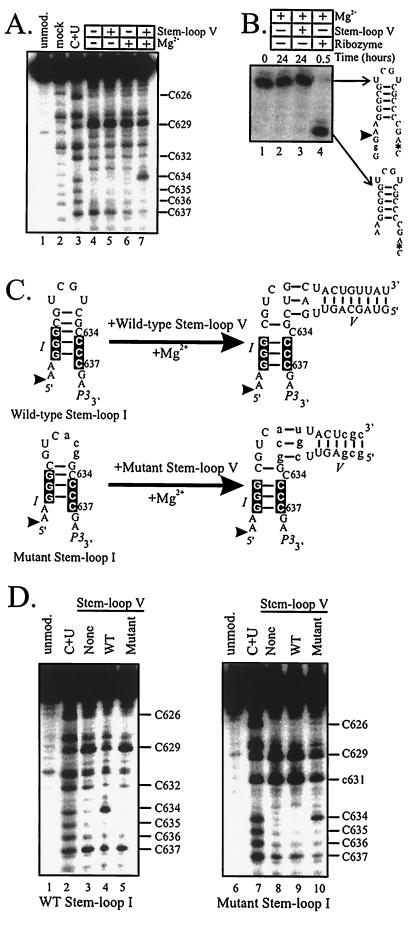
Figure 3.
Stem-loop V is sufficient to rearrange the secondary structure of stem-loop I. (A) DMS modification of 3′ end-labeled wild-type stem-loop I. Modifications were performed on stem-loop I in the presence (+) or absence (−) of 50 mM MgCl2 or 10 μM wild-type stem-loop V, as indicated. unmod., mock, and C + U are as in Fig. 1C. (B) Wild-type stem-loop V is not sufficient to cleave wild-type stem-loop I in trans. 3′ end-labeled trans cleavage substrate (nt 618–nt 639) was incubated at 30°C in 50 mM MgCl2, and in the presence (+) of 20 μM wild-type stem-loop V or 700 nM ribozyme, as indicated. Schematics of the secondary structures of the trans cleavage substrate and the 3′ cleavage product are shown. The cleavage site is marked by an arrowhead, and the 3′ end-label by an asterisk. (C) Shown are schematics of the wild-type and mutant kissing complexes. P3 is a primer-binding site (22). Nucleotides in the lower part of stem-loop I involved in the conformational change are indicated by black boxes (22). The cleavage site is indicated by an arrowhead. (D) Rearrangement of stem-loop I by stem-loop V requires native interactions, including the kissing interaction. DMS modification of 3′ end-labeled wild-type (lanes 1–5) or mutant (lanes 6–10) stem-loop I in 100 mM MgCl2, and in the presence (+) or absence (−) of 10 μM (lane 4) or 100 μM (lanes 5, 9, and 10) wild-type or mutant stem-loop V, as indicated. unmod. and C + U are as in Fig. 1C.
To confirm that C634 reactivity reflects shifting of the lower base pairs, we analyzed an “unshiftable” mutant stem-loop I in which the bottom two base pairs in stem I have been changed such that this mutant cannot adopt the shifted conformation (mutant G in figure 3B of ref. 22). DMS modification of the unshiftable stem-loop I in the presence of magnesium showed that C634 was protected from modification in the presence or absence of stem-loop V, confirming that shifting of the lower base pairs is required to unpair C634 (data not shown).
Although wild-type stem-loop V is sufficient to rearrange wild-type stem-loop I, it is not sufficient for site-specific cleavage of stem-loop I. Stem-loop I containing the cleavage site was a substrate for trans cleavage by a wild-type VS ribozyme (Fig. 3B, lane 4). This substrate alone did not cleave in the presence of magnesium (lane 2) or in the presence of stem-loop V under conditions that support the secondary structure rearrangement (lane 3). These data confirm that the shifted structure of stem-loop I is not sufficient for cleavage activity, whether achieved by mutation (SHΔrz, Fig. 2C) or by stem-loop V in trans. Therefore, a region outside of stem-loops I and V must contribute to the active site of the VS ribozyme.
To demonstrate that the secondary structure rearrangement occurs through the formation of a kissing interaction, and not through nonnative interaction(s) peculiar to the sequences of wild-type stem-loops I and V, we reconstituted the kissing interaction with different sequences. Mutant stem-loop V (Fig. 3C) has six substitutions within the helix that maintain base pairing and three substitutions within the loop, shown above to functionally disrupt the kissing interaction with wild-type loop I in RS19 (RS19-V; Fig. 1B). Mutant stem-loop I has three substitutions within loop I that have no potential for base pairing with wild-type loop V, but are complementary to, and functionally interact with, the loop V substitutions in mutant stem-loop V (Figs. 1B and 3C). Fig. 3D shows the DMS modification patterns of wild-type or mutant stem-loop I in the presence of magnesium and wild-type or mutant stem-loop V. Only the two combinations of stem-loops I and V that can form a kissing interaction (wild-type I and V, mutant I and V) resulted in secondary structure rearrangement of stem-loop I, revealed by the reactivity of C634 only in lanes 4 and 10. A higher concentration of mutant stem-loop V was required to form a complex with, and rearrange, mutant stem-loop I, suggesting the mutant kissing complex forms weaker interactions than the wild-type (Fig. 3C; data not shown). C629 is less reactive with DMS only in the two combinations of stem-loops I and V that form kissing complexes, suggesting that a stem-loop I-V complex is necessary and sufficient for the interaction with C629 that protects this base from modification (22, 24). Taken together, the mutational and chemical modification analyses indicate that stem-loops I and V interact in trans through a kissing interaction, forming a kissing complex. Moreover, this kissing complex is necessary and sufficient for secondary structure rearrangement of stem-loop I.
Discussion
Self-cleavage of VS RNA requires a kissing interaction between stem-loops I and V, and a rearrangement of the base pairs at the base of wild-type stem-loop I into a shifted conformation. Using chemical modification and mutational analyses, we observed that mutants with a disrupted kissing interaction have severe self-cleavage defects and are unable to rearrange the base pairing of stem-loop I. The self-cleavage defect of a mutant RNA with a disrupted kissing interaction can be partially suppressed by a mutant stem-loop I that constitutively adopts the shifted conformation. These data indicate that one functional role of the kissing interaction between loops I and V for cis-cleavage of the wild-type VS ribozyme is to rearrange the secondary structure of stem-loop I, which includes the cleavage site. The shifted conformation of stem-loop I creates a magnesium binding motif that may be important for function (22).
The most unexpected observation of the current work is that an isolated stem-loop V is sufficient for the magnesium-dependent secondary structure rearrangement of an isolated stem-loop I in the absence of the rest of the ribozyme (Fig. 3). Even if stem-loop I makes other tertiary interactions with the rest of the ribozyme, those interactions are not necessary for rearranging the secondary structure of stem-loop I. Secondary structure rearrangement requires a kissing interaction: disrupting the kissing interaction by changing the sequence of loops I or V prevents the secondary structure rearrangement of stem-loop I; restoring the kissing interaction with non-wild-type Watson–Crick base pairs restores rearrangement (Fig. 3D). Wild-type stem-loop V does not rearrange a mutant stem-loop I that contains a noncomplementary loop sequence, suggesting that loop V does not make other sequence-specific contacts with stem-loop I that are sufficient to stabilize the rearranged structure of stem-loop I. Also, in the absence of the kissing interaction, any other interactions within or between stem-loops I and V are not sufficient for the secondary structure rearrangement (Fig. 3D). The observation that the kissing complex derived from VS RNA is sufficient for secondary structure rearrangement of stem-loop I reveals a structural consequence of kissing interactions not observed previously: remodeling of the secondary structure within one component stem-loop.
In most ribozymes, self-cleavage requires that the cleavage site dock into another region within the RNA (34–37). An exception is the leadzyme, in which the cleavage site and active site reside within the same internal loop (38–40). In an isolated shifted VS stem-loop I (in the absence of the rest of the ribozyme) cleavage at the wild-type cleavage site occurred at the same background rate as uncatalyzed cleavage after other single-stranded nucleotides (SHΔrz, Fig. 2C). In addition, a substrate containing wild-type stem-loop I that can be cleaved in trans by the VS ribozyme is not cleaved in trans by an isolated stem-loop V (Fig. 3B). Therefore, the shifted conformation of stem-loop I, whether obtained by mutation or by secondary structure rearrangement by stem-loop V, is not sufficient for cleavage. These data strongly suggest that the active site of the VS ribozyme includes nucleotides outside of stem-loops I and V.
The observation that the self-cleavage defect of a mutant with a disrupted kissing interaction is only partially suppressed by a constitutively shifted mutant stem-loop I suggests that the kissing interaction has a functional role(s) in addition to rearranging the secondary structure of stem-loop I. Disrupting the kissing interaction in a cis-cleaved ribozyme not only prevents the secondary structure rearrangement of stem-loop I, but also appears to destabilize other tertiary interactions in the ribozyme: C665 and C687 do not become as protected from DMS modification in the presence of magnesium in H3Δk and SHΔk as in ribozymes that retain a kissing interaction (refs. 6 and 24; data not shown), which is consistent with previous diethylpyrocarbonate modification data (6) suggesting that the kissing interaction helps to stabilize the overall active structure of the ribozyme. An additional role of the kissing interaction in self-cleavage may be to facilitate docking of the internal loop containing the cleavage site into another region of the RNA by constraining helices I and V in a specific orientation.
Kissing interactions initiate the remodeling of RNA secondary structures in other well studied systems, including the dimerization initiation sites of retroviruses and antisense-target complexes (10, 41–43). In these complexes, base complementarity exists not only between the intermolecular hairpin loops, but also between the stem regions. In the case of the dimer initiation complex of the HIV-genomic RNA, a metastable intermolecular kissing interaction culminates in a linear duplex isoform stabilized by intermolecular base pairing (20, 44). A kissing interaction also initiates the binding of the antisense RNA CopA to its target, CopT, but the outcome of this interaction is an unusual four-way junction defined by the predicted base stacking arrangements of intramolecular helices and newly formed intermolecular helices (19, 45). In these types of kissing complexes, the intramolecular helices contributing to the initial kissing complex are replaced with intermolecular helices within an extended complex.
Many stable kissing complexes do not remodel the secondary structure of the contributing stems (2, 46, 47). If the contributing stems are not complementary, which is the case for the kissing complex derived from the VS ribozyme (6), extended duplexes cannot form and the intermolecular base pairs in the complex are exclusively between the hairpin loop nucleotides. In the RNA I–RNA II complex from ColE1 (8) and the tar-tar* complex from the HIV genome (47), the intermolecular interactions are exclusively between the loop nucleotides and the base pairing of the contributing stems is unchanged. The loop–loop helix in these kissing complexes is distorted from A-form, and induces a bend in the complex (8, 47). These kissing complexes are constrained by the interactions between the hairpin loops, by the necessity to maintain covalent interactions within each stem-loop, and by the A-form conformation of each contributing stem. As a result, kissing complexes can stabilize distorted conformations and provide recognition sites for cations, proteins, and/or RNAs.
The VS kissing complex is unique among known kissing complexes because secondary structure remodeling of a contributing stem occurs in the absence of complementarity between the two stems. This kissing complex contains only three intermolecular Watson–Crick base pairs, which constitute the loop–loop helix (6). Surprisingly, the kissing complex stabilizes the intramolecular rearrangement of three GC base pairs at the base of stem I and the bulging of a cytosine from the middle of stem I (Fig. 3C). The kissing complex derived from VS RNA demonstrates that secondary structures can be rearranged into alternative conformations even in kissing complexes where only the loop bases are complementary. The VS kissing interaction extends the structural and functional roles of kissing interactions to include the intramolecular secondary structure rearrangement of one component stem-loop.
Abbreviations
- D
downstream cleavage product
- Pre
precursor(s)
- DMS
dimethyl sulfate
- Δk
disrupting kissing interaction
- VS
Varkud satellite
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nowakowski J, Tinoco I., Jr Semin Virol. 1997;8:153–165. [Google Scholar]
- 2.Kim C H, Tinoco I., Jr Proc Natl Acad Sci USA. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. . (First Published August 8, 2000; 10.1073/pnas.170283697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S H, Suddath F L, Quigley G J, McPherson A, Sussman J L, Wang A H, Seeman N C, Rich A. Science. 1974;185:435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 4.Ladner J E, Jack A, Robertus J D, Brown R S, Rhodes D, Clark B F, Klug A. Proc Natl Acad Sci USA. 1975;72:4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehnert V, Jaeger L, Michel F, Westhof E. Chem Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 6.Rastogi T, Beattie T L, Olive J E, Collins R A. EMBO J. 1996;15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 7.Pan J, Deras M L, Woodson S A. J Mol Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 8.Marino J P, Gregorian R S, Jr, Csankovszki G, Crothers D M. Science. 1995;268:1448–1454. doi: 10.1126/science.7539549. [DOI] [PubMed] [Google Scholar]
- 9.Comolli L R, Pelton J G, Tinoco I., Jr Nucleic Acids Res. 1998;26:4688–4695. doi: 10.1093/nar/26.20.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jossinet F, Paillart J C, Westhof E, Hermann T, Skripkin E, Lodmell J S, Ehresmann C, Ehresmann B, Marquet R. RNA. 1999;5:1222–1234. doi: 10.1017/s1355838299990982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haddrick M, Lear A L, Cann A J, Heaphy S. J Mol Biol. 1996;259:58–68. doi: 10.1006/jmbi.1996.0301. [DOI] [PubMed] [Google Scholar]
- 12.Laughrea M, Jette L. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 13.Laughrea M, Jette L. Biochemistry. 1996;35:9366–9374. doi: 10.1021/bi960395s. [DOI] [PubMed] [Google Scholar]
- 14.Muriaux D, Fosse P, Paoletti J. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 15.Paillart J C, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 16.Malmgren C, Engdahl H M, Romby P, Wagner E G. RNA. 1996;2:1022–1032. [PMC free article] [PubMed] [Google Scholar]
- 17.Malmgren C, Wagner E G H, Ehresmann C, Ehresmann B, Romby P. J Biol Chem. 1997;272:12508–12512. doi: 10.1074/jbc.272.19.12508. [DOI] [PubMed] [Google Scholar]
- 18.Asano K, Mizobuchi K. J Biol Chem. 2000;275:1269–1274. doi: 10.1074/jbc.275.2.1269. [DOI] [PubMed] [Google Scholar]
- 19.Kolb F A, Malmgren C, Westhof E, Ehresmann C, Ehresmann B, Wagner E G, Romby P. RNA. 2000;6:311–324. doi: 10.1017/s135583820099215x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mujeeb A, Parslow T G, Zarrinpar A, Das C, James T L. FEBS Lett. 1999;458:387–392. doi: 10.1016/s0014-5793(99)01183-7. [DOI] [PubMed] [Google Scholar]
- 21.Guo H C, De Abreu D M, Tillier E R, Saville B J, Olive J E, Collins R A. J Mol Biol. 1993;232:351–361. doi: 10.1006/jmbi.1993.1395. [DOI] [PubMed] [Google Scholar]
- 22.Andersen A A, Collins R A. Mol Cell. 2000;5:469–478. doi: 10.1016/s1097-2765(00)80441-4. [DOI] [PubMed] [Google Scholar]
- 23.Saville B J, Collins R A. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 24.Beattie T L, Olive J E, Collins R A. Proc Natl Acad Sci USA. 1995;92:4686–4690. doi: 10.1073/pnas.92.10.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milligan J F, Uhlenbeck O C. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 26.Rastogi T, Collins R A. J Mol Biol. 1998;277:215–224. doi: 10.1006/jmbi.1997.1623. [DOI] [PubMed] [Google Scholar]
- 27.Guo H C, Collins R A. EMBO J. 1995;14:368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legault P. Ph.D. thesis. Boulder, CO: Univ. of Colorado; 1995. [Google Scholar]
- 29.England T E, Bruce A G, Uhlenbeck O C. Methods Enzymol. 1980;65:65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- 30.Peattie D A, Gilbert W. Proc Natl Acad Sci USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krol A, Carbon P. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- 32.Conway L, Wickens M. Methods Enzymol. 1989;180:369–379. doi: 10.1016/0076-6879(89)80112-0. [DOI] [PubMed] [Google Scholar]
- 33.Michiels P J A, Schouten C H J, Hilbers C W, Heus H A. RNA. 2000;6:1821–1832. doi: 10.1017/s1355838200001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cech T R. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 239–269. [Google Scholar]
- 35.Boudvillain M, Pyle A M. EMBO J. 1998;17:7091–7104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferre-D'Amare A R, Zhou K, Doudna J A. Nature (London) 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 37.Walter N G, Burke J M. Curr Opin Chem Biol. 1998;2:24–30. doi: 10.1016/s1367-5931(98)80032-x. [DOI] [PubMed] [Google Scholar]
- 38.Pan T, Uhlenbeck O C. Nature (London) 1992;358:560–563. doi: 10.1038/358560a0. [DOI] [PubMed] [Google Scholar]
- 39.Hoogstraten C G, Legault P, Pardi A. J Mol Biol. 1998;284:337–350. doi: 10.1006/jmbi.1998.2182. [DOI] [PubMed] [Google Scholar]
- 40.Legault P, Hoogstraten C G, Metlitzky E, Pardi A. J Mol Biol. 1998;284:325–335. doi: 10.1006/jmbi.1998.2181. [DOI] [PubMed] [Google Scholar]
- 41.Gerhart E, Wagner H, Brantl S. Trends Biochem Sci. 1998;23:451–454. doi: 10.1016/s0968-0004(98)01322-x. [DOI] [PubMed] [Google Scholar]
- 42.Altuvia S, Wagner E G. Proc Natl Acad Sci USA. 2000;97:9824–9826. doi: 10.1073/pnas.97.18.9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lease R A, Belfort M. Proc Natl Acad Sci USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. . (First Published August 22, 2000; 10.1073/pnas.170281497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mujeeb A, Clever J L, Billeci T M, James T L, Parslow T G. Nat Struct Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 45.Kolb F A, Engdahl H M, Slagter-Jager J G, Ehresmann B, Ehresmann C, Westhof E, Wagner E G H, Romby P. EMBO J. 2000;19:5905–5915. doi: 10.1093/emboj/19.21.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang K Y, Tinoco I., Jr Proc Natl Acad Sci USA. 1994;91:8705–8709. doi: 10.1073/pnas.91.18.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang K Y, Tinoco I., Jr J Mol Biol. 1997;269:52–66. doi: 10.1006/jmbi.1997.1021. [DOI] [PubMed] [Google Scholar]