SUMMARY
Cellular shape is defined by its boundary: the membrane. Extremely dynamic at small scales, biological membranes retain identifiable stationary shape as large structures, such as organelles. The lipid bilayer serves as a matrix for this shape-defining membrane, linking membrane dynamics and structural stability. Here, we analyze the central role of proteo-lipid membrane domains in the coordination of membrane dynamics and in the self-organization of membrane shape.
INTRODUCTION
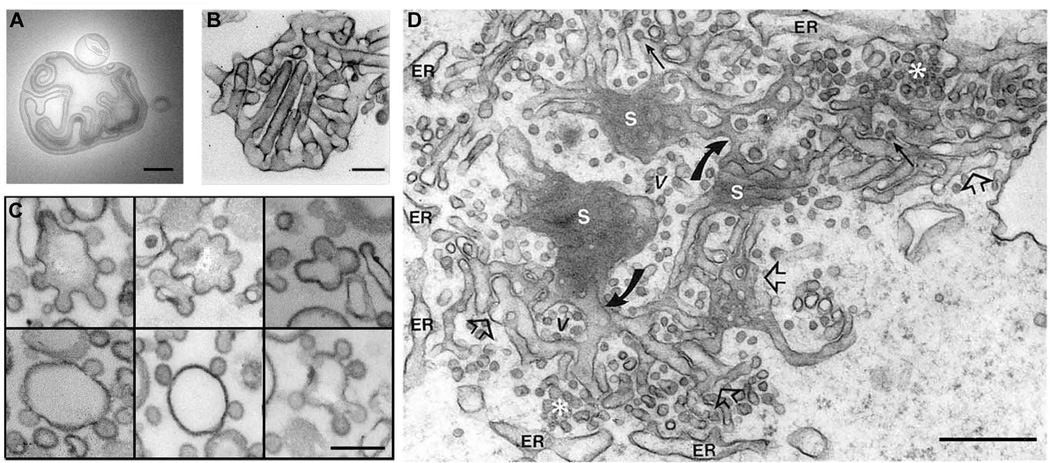
The shape of an object is determined by its boundary. In the case of a cell or its organelles the boundary is the biological membrane, structured by a bilayer of phospholipids. Aggregates of these amphiphilic molecules assemble into complex shapes even without the influence of proteins (Figure 1A; [1–3]). The ability of lipids to spontaneously self-associate into closed structures (such as vesicles) in aqueous solution, even at low concentrations, has been regarded as a basic factor in the origin of the cellular life on Earth [4–7]. A purely lipid vesicle that is formed in distilled water is a stable structure, which, however, can adapt its shape to external factors, such as the temperature, pH of light irradiation changes, direct mechanical forces, etc. [1, 4, 8, 9]. During the time-course of this adaptation, which may last from several seconds to days, the vesicle membrane appears to be “alive” until a new steady-state shape is archived. The multitude of shapes revealed in purely lipidic membrane systems covers the morphological complexity seen in reconstituted and even cellular systems (Figure 1B, C)
Figure 1.
Common tubulo-vesicular patterns of membrane shape in cellular and reconstituted systems. A. Large lipid vesicle spontaneously self-assembled upon hydration of a dried lipid film; B. Deformation of lipid vesicles produced by I-BAR proteins (from [96]). C. Deformation of lipid vesicles induced by COP I assembly (from [97]). D. Snapshot of membrane morphology of endoplasmic reticulum (from [98]). All bars 200 nm.
However, cellular shape changes perpetually, thus pointing out the difference between the living material and its construction bricks. The dynamic organization of membrane shape is vital for the functional coupling of cellular membrane transport and signaling networks [10–14], cell motility [13, 15, 16] and de novo creation of intracellular organelles during cytokinesis [17, 18]. Membrane remodeling, even in such global processes as cell motility or axon formation, is localized in small membrane domains of distinct composition. Such domains can be readily distinguished as precursors of transport vesicles [18–20], lamellapodia-associated transient protrusions [15, 21], sites of membrane fusion and fission [22] and invaginations of plasma membrane like caveolae [23, 24]. They are enriched with proteins specialized in membrane remodeling and, often, associated with particular lipid species implicated in domain formation, such as multivalent charged lipids, ceramides or cholesterol [22, 24–27]. Thus, by biological means, cellular membrane dynamics are controlled locally. This localization of dynamics allows not only preservation of the global shape of organelles and the cell as a whole, but also allows uncoupling of different membrane deformation related to different cellular functions.
Despite the apparent variety of membrane domains involved in shape regulation in cells, they share common principles of molecular organization dictated by the intrinsic properties of the lipid bilayer. The very localization of membrane transformations within domains is related to the intrinsic plasticity of lipid bilayer: if a lipid membrane is pushed or pulled in a point it will rearrange in a way that only a small part of the membrane is deformed [3, 4]. The extent of this deformation will depend on mechanical properties of the membrane, which, at the same time, will be related to the membrane composition [28, 29]
Thus, the emergence of cellular membrane shape implies intercommunication of proteins and lipids localized within membrane domains. How do cellular protein machineries cooperate with lipids to control emergence and morphology of membrane domains?
THE MORPHOLOGICAL DOMAIN
The notion of domains plays an important role in our current understanding of cellular membrane organization [30]. This notion immediately invokes membrane heterogeneity based on clustering of membrane components, proteins and lipids. Compositionally-defined spontaneous segregation of lipids was instrumental in the development of the concept of cellular lipid membrane domains [31]. The segregation of lipids is closely linked to membrane domain morphology and composition: lipid domains change their shape to minimize elastic stresses in the membrane [32–35], and vice versa, enforcing membrane curvature can cause lateral redistribution of lipids [36].
Besides forming domains on their own, by dint of the reduction in dimensionality offered by the bilayer surface, lipids provide powerful constraints and opportunities for lateral protein-protein interactions, as well as the unique opportunities for various proteo-lipid interactions. Interference of membrane deformation fields produced by proteins embedded in a lipid bilayer might lead to weak attractions between neighbor proteins, thus stimulating protein clustering (Figure 2; see also [37, 38]). In turn, charged proteins can recognize and concentrate charge (polyvalent) lipid species [39, 40]. The proteo-lipid interactions within domains may depend on the domain geometry. Protein binding to a domain can be promoted through curvature preferences of the particular protein [25, 41]. All these examples, illustrated in Figure 2, demonstrate how the lipid membrane is not a passive receptor of proteins orders, but rather an active player in its own remodeling.
Figure 2.
Schematic pathway of the MOD emergence. Creation of curved areas in the membrane, here due to thermal undulations, is coupled to the lateral redistribution of the membrane components according to their curvature preferences and to the curvature-driven binding of proteins. This coupling first leads to the enhancement of the undulations, then to the MOD emergence, if more components stabilizing membrane curvature (blue banana) or clustering on the MOD membrane (blue caps) are added. MODs can quickly disappear if the curvature-active components leaves, e.g. via desorption and disassembly coupled to GTP or ATP hydrolysis.
It is generally accepted that the interactions within the protein assemblies creating membrane shapes are hierarchically organized. This hierarchy is based on the specialization of the protein machineries and organization of protein modules in charge of different stages of shape creation [25, 42–44]. It is generally assumed that distinct protein modules are assigned to different tasks such as the initiation of membrane remodeling, coordination of the progression of membrane deformations to a certain stationary shape, and the organization of topological membrane remodeling via fusion or fission. However, recent data indicates that proteins do not obey a strict hierarchy, as a single protein species can participate in different shape transformations. For example, dynamin participates both in budding and fission of membranes in reconstituted and cellular systems [19, 45–47]. Even if the hierarchic conceptualization of the protein machinery is accepted, its modules remain united through the membrane itself. So, the modular description of membrane morphogenesis must include the lipid bilayer as an inseparable part of each of its protein modules.
To focus on the role of proteo-lipid cooperation in membrane remodeling, we generalize the modular principle of its organization by introducing the concept of the MOrphological Domain (MOD), which we define as a dynamic proteo-lipid module undergoing morphological evolution through self-organization. The MOD is not conceived of as a raft, a concept primarily based on self-assembly of lipid domains that arise through separation of lipid phases to which proteins partition. There are many examples of biological membrane domains that form with lipids that mix well with their neighbors, so we do not limit our description of cellular membrane domains to “classical“ rafts. Rather, the MOD concept features dynamic links between composition and the shape of complex proteo-lipid membranes.
DYNAMIC SELF-ORGANIZATION OF MODs: SHAPE EMERGENCE
Our understanding of membrane morphogenesis is often biased by the results obtained in reconstitution systems. There, the shape creation is generally a deterministic process based on an irreversible self-assembly of stable, equilibrium shapes, e.g. coated membrane particles whose geometry is dictated by the protein scaffold [48]. Nevertheless, recent experiments indicate that in vivo the components of the coat protein scaffolding remain dynamic, and exchange with the bulk in an energy-dependent manner. This exchange is evident in perpetual treadmilling of protein filaments involved in cell motility [16, 49, 50]. The components of clathrin and COP coats can exchange with the cytoplasmic bulk phase during the evolution of their MODs [51]. This recycling depends on membrane curvature and the presence of small GTPases (e.g. Arf1) and ATPases (e.g. HSC70) known to be involved in the regulation of membrane binding and the self-assembly of coat proteins [19, 52–56]. The involvement of GTP/ATPases in this process indicates that MOD evolution requires energy [57]. These dynamic, non-equilibrium aspects of membrane remodeling are usually missed by most of the reconstitution works.
One of the expected outcomes of the dynamic behavior of MOD components is the stochastic nature of MOD formation. For example, emergence of a clathrin-coated vesicle on the plasma membrane implicates a stochastic process. The vesicle precursors (i.e. small assemblies of the vesicle proteins) constantly appear and disappear until one of them reaches a point of dynamic instability (or “control” point) and progress further into a mature vesicle [19, 52, 56]. Thus, the appearance of MOD is unlikely to be guided by a strict template and it turns up to be a complex dynamic process that relies on energy consumption.
The coupling between membrane curvature on one side and segregation and energy consumption by proteins embedded in the lipid bilayer on the other is summarized by the conception of “active” membranes [58]. It reveals the leading role of the lipid bilayer and its intrinsic dynamics in the organization of non-equilibrium membrane deformations [21, 58–60]. According to this idea, the emergence of MOD relies on a self-enhanced process that is the accumulation of curvature-driven components in curved membrane regions (Figure 2). In an initially homogeneous membrane, this accumulation is coupled with membrane undulations which cause initial deviations of membrane curvature [16, 51, 61]. To further enforce this feedback, curvature-driven components can be “active”, i.e. providing not only additional curvature, but also energy [16, 62]. For example, accumulation of actin membrane linkers leads to a concentrated pushing force of actin filaments into peaks of membrane undulations, leading to further development of local membrane curvature and global curvature instabilities [16, 21]. Thus, dynamic curvature instabilities can lead to MOD appearance through negative feedbacks, including Turing or wave instabilities [21, 63]. MOD emergence can be described by a set of autocatalytic and negative feedbacks between the product (membrane curvature) and reactants (proteo-lipid components of MOD) that lead to self-organization of structurally stable spatial patterns [63–66].
Summarizing, direct neighbor attraction and “membrane-driven” interactions, mediated via membrane deformation fields and controlled via external stimuli, combine the main driving forces leading to emergence of early MODs. These dynamic, non-equilibrium structures can be directed toward different shapes by specialized protein machineries. Before discussing the evolution of shape by MODs, we will first classify MODs according to their geometry.
BASIC CONFIGURATIONS OF MOD
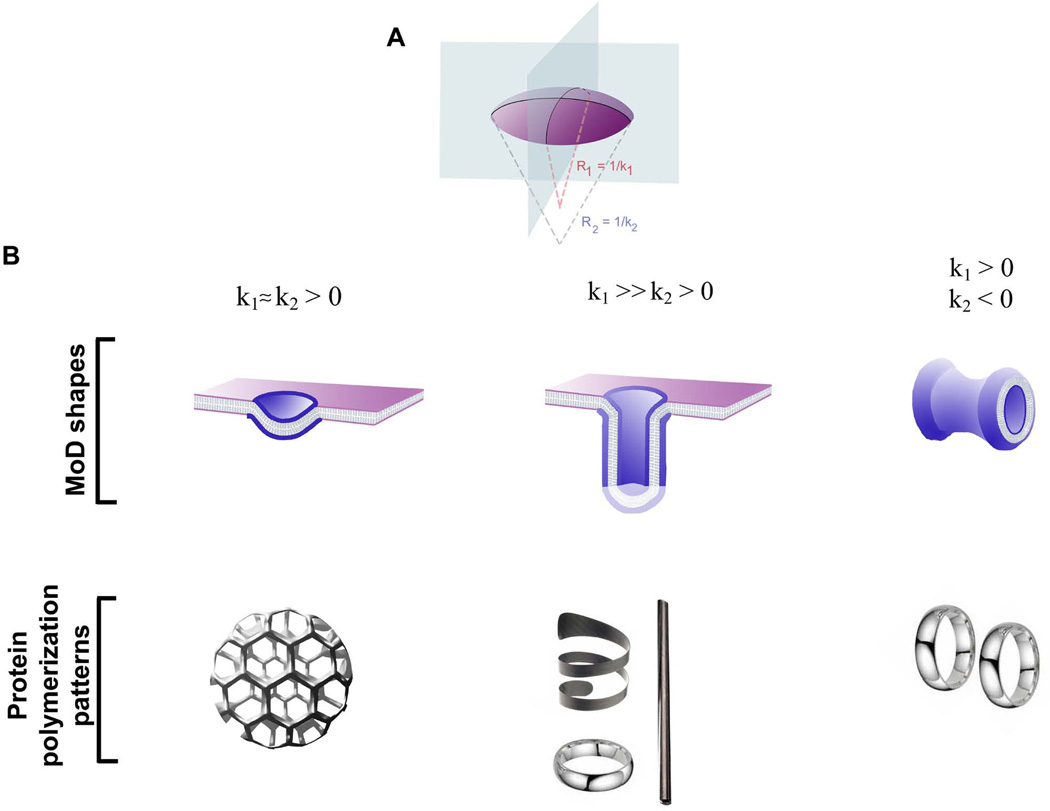
The complexity of cellular membrane shapes usually observed by electron microscopy (e.g. Figure 1D) of fixed membrane was created by the activity of dynamic MODs. What are the basic shapes that MODs may adapt? In its simplest geometrical approximation, a MOD is a piece of a surface, a two-dimensional structure. Curvature maps thus describe shapes of MODs. Curvature is defined for each point of the MOD surface, characterizing how steeply the surface deviates from a plane. In general, the surface is bent differently in different directions, meaning that the slope along every possible direction has its own curvature corresponding to the curvature of the line on the surface. The variety of slopes is limited to those between bounds: minimum and maximum curvature. If the two are similar, then the surface is uniformly bent. Otherwise, two perpendicular directions corresponding to the min and max sloped define two principal curvatures of the surface point (Figure 3A).
Figure 3.
Basic shapes of MOD. A. Principal curvature of a surface, K1,2=1/R1,2 ; B. MOD shapes, spherical, cylindrical and saddle-like, corresponding to different sets of principal curvatures indicated; C. Protein polymerization patterns corresponding to simple MOD geometries shown in B; the images illustrate interaction patterns leading to stabilization of a protein scaffolding in space.
The curvature value and sign indicate the extent and direction of the surface bending along the corresponding principal direction. The sign definition is arbitrary. For lipid monolayers it is traditionally defined from the side of the hydrophobic tails: both curvatures are negative for a membrane pit, positive for a bump and have opposite signs on a saddle point (Figure 3A, B upper panel). The value of the curvature determines the extent of membrane bending alone the principal direction. For example, for a sphere both curvatures are equal to 1/R. This way the local geometry of the surface is completely defined by the two curvatures.
The principal curvatures change from one point on a surface to another and the way they change determines the surface shape. We can illustrate how the shape builds up with simple examples. If both curvatures preserve the same sign, the surface ultimately closes into a sphere-like shape. If the curvatures keep opposite signs (or one remains zero), the surface will not close. Thus, these surfaces, such as cylinders or saddle necks, have edges. The same shape patterns, spherical, cylindrical and saddle-like, are evident in reconstituted and cellular membrane systems (Figure 1B,C), indicating their general involvement in morphogenesis of cellular membranes [67, 68].
We restrict our analysis to this prototype MODs (Figure 3B, upper panel). Importantly, the images in Figure 3 emphasize not the values of membrane curvature, but the relationship between the two principle curvatures. As we discuss below, these relationships correspond to the basic symmetries imposed by the prototype proteins on the MOD shape during its evolution.
EVOLUTION OF MODs
Self-shaping of MOD: principal curvatures dictated by membrane composition
MOD composition is the main factor determining its shape. Proteins accumulating in MOD generally possess intrinsic curvature activity, e.g. via hydrophobic insertion (e.g. N-BAR) [69]. The mechanisms by which proteins alter the intrinsic curvature of the lipid bilayer have received extensive attention recently [reviewed in [25, 43, 70]]. These mechanisms converge to the change of the intrinsic curvatures of lipid monolayers of MOD and/or creation of a stable area difference between monolayers that leads to bending due to coupling between the monolayers [43, 71].
The intrinsic curvature characterizes the shape preferences of the MOD membrane. For a monolayer consisting of a single lipid species, these preferences are defined by the molecular packing which minimizes the elastic stresses (i.e. stretching and squeezing) imposed on lipid heads and tails during self-assembly of the monolayer [43]. The monolayer bends to minimize these stresses thus adopting its intrinsic curvature. Lipids acquire different shapes dependently on such factors as steric interactions, hydrogen bonding between headgroups, charge repulsions, hydration, temperature, etc. (for review see [3, 72]). Thus, the spontaneous state of a multi-component monolayer is a complicated function of the composition. However, the direction of the deformation is evident when a dominating curvature-active agent is present. For example, so called non-bilayer lipids, characterized by extremely high values of intrinsic curvature, impose positive (e.g lyso-lipids [73]) or negative (e.g. diacylglicerol [74]) curvatures. The curvature activity of the individual protein can be represented using similar terms: e.g. hydrophobic insertion will induce positive intrinsic curvature.
Similar minimization of elastic stresses leads to the intrinsic state of the lipid bilayer. Its monolayers have opposite curvature preferences, so the resulting deformation is related to the differences (asymmetries) between monolayers. Accordingly, changes in MOD curvature are driven by the preferable accumulation of proteins, non-bilayer lipids and cholesterol in one monolayer of the MOD membrane [61, 75] (Figure 2). It is important to realize that the notion of the intrinsic curvature corresponds to the mean curvature of the bilayer; it does not discriminate between the two principal curvatures of the membrane surface (Figure 2A). However, when proteins pack tightly in the MOD area, their packing preferences, seen in the shapes these proteins assemble in solution, will impose differences between the principal curvatures (Figure 3B, lower panel). For example, the best packing of the curved and elongated BAR domains is achieved on a cylinder: BARs can cover completely a cylinder, but not a sphere [76, 77]. Hence, BARs would readily accumulate on and likely stabilize cylindrical shapes even without polymerization in a multimolecular scaffold. Similar packing preference is evident for straight filaments [78]. The packing preferences, even without direct interaction between the proteins, will determine the amount of the curvature-driven agents in the MOD and thus impact the intrinsic shape of MOD (Figure 4).
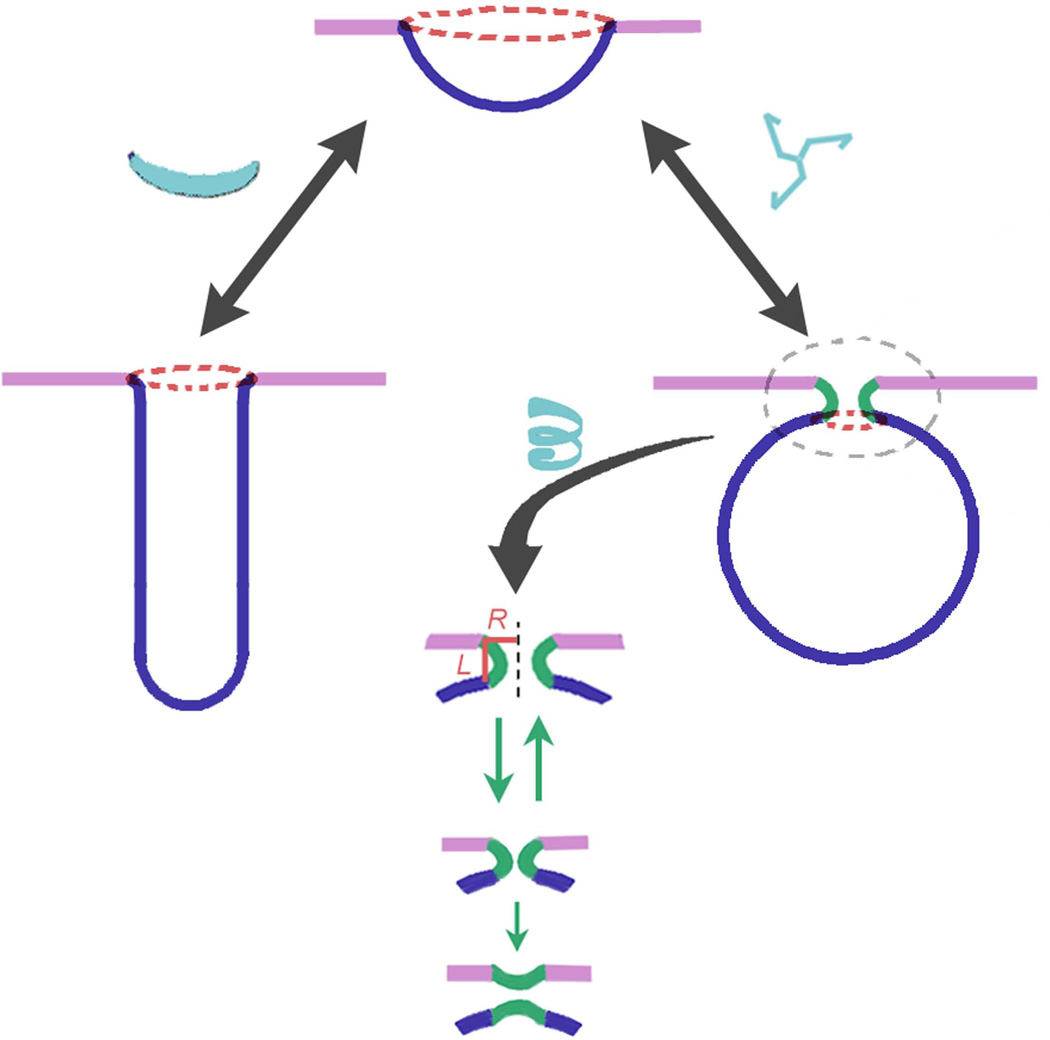
Figure 4.
Evolution of MOD. Similar cap-like precursors can develop into tubular or spherical MODs dependently on the MOD composition and forces acting on the MOD. Elongated proteins or membrane-binding protein domains, such as BAR [refs], situated along one of the principle directions on the membrane surface (see Figure 3) lead to cylindrical MOD, while more round-like proteins, e.g. clathrin triskelia, do not distinguish the principal directions leading to spherical MOD. Evolution of the spherical MOD inevitably leads to the appearance of the new MOD, the neck. For fluid-like domains, the neck formation is promoted by the line tension on the MOD edge (red ring): the line tension enforce the lessening of the edge leading to the closure of the spherical MOD. The length of the neck MOD (L) is proportional to its radius (R), this proportionality defines the evolution of this MOD towards thin and short membrane neck, an established intermediate in fusion and fission reactions.
Are the forces behind the protein packing preferences sufficiently large to ensure stability of a curved MOD? Recent works demonstrate that curvature-active membrane components can segregate spontaneously and form stable domains supported, not by direct neighbor interactions (such as protein lattice polymerization), but by the curvature these components impose on the membrane [37]. These attractive forces depend on the strength and symmetries of the membrane deformations caused by individual molecules: calculations demonstrate that only those producing sufficiently large or asymmetric deformations would cluster [37, 38].
Direct neighbor attraction can further enhance the symmetry embedded in packing: lateral interaction between F-BAR domains leads to their helical alignment on a membrane cylinder [76]. For COP the spherical symmetry is set by the interactions responsible for the assembly of the vertices of the protein lattice [48]. However, interactions with the lipid bilayer and other MOD components can dramatically alter the polymerization process (e.g. [79]). One of the clear examples is provided by the clathrin lattice, whose shape phenotype is dramatically altered upon depletion of one of the accessory proteins, CALM [80]. Formation of large holes in the lattice does not compromise membrane curvature creation [80], indicating that the integrity of the protein lattice is not a requirement for membrane remodeling.
Yet, in some cases, formation of stable protein scaffolding might be crucial for supporting membrane geometries. The tubular structures of endoplasmic reticulum (ER) are stabilized by rings of reticulons and DP1/Yop1p proteins [17, 81]. However, it remains unclear whether these rings are truly static structures as their mobility depends on the ATP [81]. Moreover, the same ring-forming proteins are required for dynamic membrane rearrangements leading to de novo formation of ER during cytokinesis [82].
If MOD components remain dynamic, altering the exchange rate of its components with the bulk phase can control the evolution of a MOD. Interestingly, this recycling might be retained through the whole evolution of MOD, providing a negative feedback between MOD composition and shape. This feedback emerges at later stages of MOD development, e.g. when the influx of the coat components through the MOD boundary becomes slower due to a diminishing boundary [54, 83]. In this way the supply of MOD components is cut off automatically upon completion of shape creation, thus demonstrating a feature of MOD evolution: self-regulation [83]. Similarly, MOD evolution can be altered by cargo proteins, which, upon accumulation on the MOD membrane, hinder the exchange of MOD components [84].
Besides recycling of the MOD components, other factors affect MOD dynamics moving it from the stationary states dictated by its composition. These factors are primarily associated with the forces acting on the MOD membrane.
Forces shaping MOD
Besides tuning the MOD composition to a particular geometry, MOD shapes can be enforced by application of an external force. The obvious examples involve pulling or pushing tubular membrane protrusions by filament polymerization or molecular motors [60, 85]. The shape of this force-induced MOD depends on the magnitude and the action time of the force, as well as on the membrane resistance, i.e. membrane elasticity and friction forces appearing during the changes of the membrane shape [29, 86, 87]. A simple example is when an external force is applied to pull a membrane tube. The force tends to thin the tube (to minimize the amount of membrane material drugging out of the parent membrane), while the bending rigidity of the membrane opposes this thinning. The balance of these two forces determines the tube diameter [87, 88].
For heterogeneous membranes, the situation becomes more complex [36]. The composition of the tubular extension can differ from that of the parent membrane, being this difference determined by the curvature preferences of the membrane components. Thus, the tubular MOD can further optimize its shape (even while being pulled) by exchanging its components with the parent membrane. However, the composition might not keep up with the curvature change imposed by the force. Then, if the force is diminished or abolished, the membrane tends to retract to the initial state. The competition between the external force and the intrinsic wishes of the MOD dictated by its composition should determine the shape dynamic of MODs such as filopodia [86].
Another force factor acting at the MOD boundary is associated with the lateral tension of the parent membrane. The effect of the lateral tension depends crucially on the MOD geometry. For cylindrical MOD, the tension acts as the pulling force applied along the cylinder axes, inducing squeezing of this MOD and promoting corresponding changes in its composition [87–89]. However, in the case of a spherical MOD, the membrane tension pulls on the MOD boundary (Figure 4), thus increasing the energy required to shrink its edge. High lateral membrane tension can completely inhibit formation and growing of spherical MODs [56, 90]. This effect of the lateral tension is important to understand how multiple spherical MODs in the same membrane can be coordinated by a single force.
Finally, there is a force inevitably associated with the MOD boundary. This force is intrinsically linked to the partial “unhappiness” of the molecules with tendency to be in the domain but localized on its edge. Therefore this force tends to diminish the domain boundary [32, 91]. For fluid-like membrane domains, this force is determined by the line tension, which is the energy per unit of domain boundary length [32]. This edge energy can be minimized by decreasing the length of the edge through transformation of a planar or slightly curved domain with a large edge into a spherical bud with a small edge (Figure 4). Thus line tension stimulates evolution of MODs and generally implies spherical morphology. Formation of membrane buds leads to the appearance of the special MOD generally involved in topological membrane remodeling, fusion and fission, the neck MOD (Figure 4).
The neck MOD
Morphology of this MOD should be considered separately. The neck MOD is a generic saddle-like intermediate (Figure 3B, upper panel; Figure 4, lower panel). Such MODs form only in the context of membrane transport, so that their appearance is closely linked to formation of other MODs. For example, the neck appearance can finalize the evolution of a spherical MOD (Figure 4).
The neck MOD is usually associated with ring-like assemblies of proteins in boundary regions. Such an assembly enforces of the membrane deformations involved in membrane remodeling in a small membrane area encircled by the rings. This way, the action of the protein machinery assembled in this MOD is focused on creation of small and highly bent intermediates commonly involved in membrane fusion and fission. Subsequently, the saddle-like geometry of the neck MOD is essential to minimize the bending energy of highly curved membranes [92]. The minimization of the bending energy relies on the fact that the mean curvature of this MOD (defined as the sum of the two principal curvatures depicted in Figure 3) remains small. The interrelation between the principal geometrical curvatures defines the evolution pathway for this MOD, as its length (L) should remain proportional to its radius (R) (Figure 4) to keep the mean curvature close to zero [92], even for very narrow necks, the dynamic intermediates for fusion and fission [93]. Thus, the sizes of protein assemblies on the neck’s boundaries are determined by the geometrical preferences of the neck MOD.
Importantly, the neck MOD remains very dynamic, as its dimensions have been estimated by measuring the ionic conductivity through the internal lumen of the neck using electrophysiology [88]. Reconstitution of dynamin-driven membrane neck remodeling on lipid nanotubes reveals that shape dynamics is coupled to the cyclic assembling and disassembling of short dynamin scaffolds fueled by GTP hydrolysis. This observation further corroborates the notion on the dynamic and stochastic nature of MODs in cellular membranes.
Summarizing, accumulation of curvature-active molecules, proteins and lipids, is not the only factor governing the MOD shape evolution. The choice between the basic geometries outlined in Figure 3 also depends on the external forces applied to the bulk part and boundary of MOD and on the spatial constrains, such as surface to volume ratio. In cellular MOD, all these factors are dynamically regulated, so that the concentration and activity of curvature- and force-creating proteins can vary in time, resulting in very dynamic evolution of MOD, often involving stochastic processes [45, 56, 88, 89].
Conclusive remarks
Biological membranes are both dynamic and organized, so they may change constantly, but they keep their identity intact. These requirements are not in contradiction. Cellular membrane morphology illustrates exceptionally well how extremely complex and dynamic systems can be organized in time and space in a way that their perpetual changes do not compromise the overall structural stability of cellular membrane systems. Cellular membranes organize through domains [30]. Each domain has a mission closely linked to its composition: cluster of channels or receptors, precursors of transport vesicles or adhesion points are only few examples [10, 94, 95]. Domains are generically involved in membrane remodeling, and domains turnover as the membrane of living cells is constantly and rapidly recycled. Thus, domains provide a crucial link between the structural and functional organization of cellular membrane. Membrane domains are the generic units of self-organization of cellular membrane morphology.
References
- 1.Lipowsky R. The morphology of lipid membranes. Curr Opin Struct Biol. 1995;5:531–540. doi: 10.1016/0959-440x(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 2.Barauskas J, Johnsson M, Tiberg F. Self-assembled lipid superstructures: beyond vesicles and liposomes. Nano Lett. 2005;5:1615–1619. doi: 10.1021/nl050678i. [DOI] [PubMed] [Google Scholar]
- 3.Mouritsen OG. Life-as a Matter of fat: The emerging science of lipidomics. New York, LLC: Springer-Verlag; 2005. [Google Scholar]
- 4.Lipowsky R. The conformation of membranes. Nature. 1991;349:475–481. doi: 10.1038/349475a0. [DOI] [PubMed] [Google Scholar]
- 5.Jin AJ, Edidin M, Nossal R, Gershfeld NL. A singular state of membrane lipids at cell growth temperatures. Biochemistry. 1999;38:13275–13278. doi: 10.1021/bi9912084. [DOI] [PubMed] [Google Scholar]
- 6.Sole RV. Evolution and self-assembly of protocells. Int J Biochem Cell Biol. 2009;41:274–284. doi: 10.1016/j.biocel.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Luisi PL, Walde P, Oberholzer T. Lipid vesicles as possible intermediates in the origin of life. Curr. Opinion Coll. Interface Sci. 1999;4:33–39. [Google Scholar]
- 8.Sackmann E. The seventh Datta Lecture. Membrane bending energy concept of vesicle- and cell-shapes and shape-transitions. FEBS Lett. 1994;346:3–16. doi: 10.1016/0014-5793(94)00484-6. [DOI] [PubMed] [Google Scholar]
- 9.Macia J, Sole RV. Synthetic Turing protocells: vesicle self-reproduction through symmetry-breaking instabilities. Philos Trans R Soc Lond B Biol Sci. 2007;362:1821–1829. doi: 10.1098/rstb.2007.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lajoie P, Goetz JG, Dennis JW, Nabi IR. Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. J Cell Biol. 2009;185:381–385. doi: 10.1083/jcb.200811059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmid EM, McMahon HT. Integrating molecular and network biology to decode endocytosis. Nature. 2007;448:883–888. doi: 10.1038/nature06031. [DOI] [PubMed] [Google Scholar]
- 12.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 13.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 14.Bannykh SI, Balch WE. Membrane dynamics at the endoplasmic reticulum-Golgi interface. J Cell Biol. 1997;138:1–4. doi: 10.1083/jcb.138.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doherty GJ, McMahon HT. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu Rev Biophys. 2008;37:65–95. doi: 10.1146/annurev.biophys.37.032807.125912. [DOI] [PubMed] [Google Scholar]
- 16.Gov NS, Gopinathan A. Dynamics of membranes driven by actin polymerization. Biophys J. 2006;90:454–469. doi: 10.1529/biophysj.105.062224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 18.Bannykh SI, Plutner H, Matteson J, Balch WE. The role of ARF1 and rab GTPases in polarization of the Golgi stack. Traffic. 2005;6:803–819. doi: 10.1111/j.1600-0854.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- 19.Loerke D, Mettlen M, Yarar D, Jaqaman K, Jaqaman H, Danuser G, Schmid SL. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 2009;7:e57. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rappoport JZ, Simon SM, Benmerah A. Understanding living clathrin-coated pits. Traffic. 2004;5:327–337. doi: 10.1111/j.1398-9219.2004.00187.x. [DOI] [PubMed] [Google Scholar]
- 21.Veksler A, Gov NS. Phase transitions of the coupled membrane-cytoskeleton modify cellular shape. Biophys J. 2007;93:3798–3810. doi: 10.1529/biophysj.107.113282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Meer G, Sprong H. Membrane lipids and vesicular traffic. Curr Opin Cell Biol. 2004;16:373–378. doi: 10.1016/j.ceb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Parton RG, Richards AA. Lipid rafts and caveolae as portals for endocytosis: new insights and common mechanisms. Traffic. 2003;4:724–738. doi: 10.1034/j.1600-0854.2003.00128.x. [DOI] [PubMed] [Google Scholar]
- 24.Martin S, Parton RG. Caveolin, cholesterol, and lipid bodies. Semin Cell Dev Biol. 2005;16:163–174. doi: 10.1016/j.semcdb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 25.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 26.Baba T, Rauch C, Xue M, Terada N, Fujii Y, Ueda H, Takayama I, Ohno S, Farge E, Sato SB. Clathrin-dependent and clathrin-independent endocytosis are differentially sensitive to insertion of poly (ethylene glycol)-derivatized cholesterol in the plasma membrane. Traffic. 2001;2:501–512. doi: 10.1034/j.1600-0854.2001.20707.x. [DOI] [PubMed] [Google Scholar]
- 27.van Blitterswijk WJ, van der Luit AH, Veldman RJ, Verheij M, Borst J. Ceramide: second messenger or modulator of membrane structure and dynamics? Biochem J. 2003;369:199–211. doi: 10.1042/BJ20021528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waugh RE. Surface viscosity measurements from large bilayer vesicle tether formation. II. Experiments. Biophys J. 1982;38:29–37. doi: 10.1016/S0006-3495(82)84527-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans E, Yeung A. Hidden dynamics in rapid changes of bilayer shape. Chem. Phys. Lipids. 1994;73:39–56. [Google Scholar]
- 30.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 31.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 32.Lipowsky R. Budding of membranes induced by intramembrane domains. J. Phys. II France. 1992;2:1825–1840. [Google Scholar]
- 33.Sackmann E, Feder T. Budding, fission and domain formation in mixed lipid vesicles induced by lateral phase separation and macromolecular condensation. Mol Membr Biol. 1995;12:21–28. doi: 10.3109/09687689509038491. [DOI] [PubMed] [Google Scholar]
- 34.Dobereiner HG, Kas J, Noppl D, Sprenger I, Sackmann E. Budding and fission of vesicles. Biophys J. 1993;65:1396–1403. doi: 10.1016/S0006-3495(93)81203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumgart T, Hess ST, Webb WW. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 36.Sorre B, Callan-Jones A, Manneville JB, Nassoy P, Joanny JF, Prost J, Goud B, Bassereau P. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc Natl Acad Sci U S A. 2009;106:5622–5626. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynwar BJ, Illya G, Harmandaris VA, Muller MM, Kremer K, Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–464. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- 38.Goulian M, Bruinsma R, Pincus P. Long-range forces in heterogeneous fluid membranes. Europhys. Lett. 1993;22:145–150. [Google Scholar]
- 39.Zimmerberg J, McLaughlin S. Membrane Curvature: How BAR Domains Bend Bilayers. Current Biology. 2004;14:R250–R252. doi: 10.1016/j.cub.2004.02.060. [DOI] [PubMed] [Google Scholar]
- 40.Mbamala EC, Ben-Shaul A, May S. Domain formation induced by the adsorption of charged proteins on mixed lipid membranes. Biophys J. 2005;88:1702–1714. doi: 10.1529/biophysj.104.048132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donaldson JG. Arfs and membrane lipids: sensing, generating and responding to membrane curvature. Biochem J. 2008;414:e1–e2. doi: 10.1042/BJ20081438. [DOI] [PubMed] [Google Scholar]
- 42.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 43.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 44.Antonny B. Membrane deformation by protein coats. Curr Opin Cell Biol. 2006;18:386–394. doi: 10.1016/j.ceb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 45.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huynh KK, Grinstein S. Phagocytosis: dynamin's dual role in phagosome biogenesis. Curr Biol. 2008;18:R563–R565. doi: 10.1016/j.cub.2008.05.032. [DOI] [PubMed] [Google Scholar]
- 47.Song BD, Schmid SL. A molecular motor or a regulator? Dynamin's in a class of its own. Biochemistry. 2003;42:1369–1376. doi: 10.1021/bi027062h. [DOI] [PubMed] [Google Scholar]
- 48.Stagg SM, LaPointe P, Balch WE. Structural design of cage and coat scaffolds that direct membrane traffic. Curr Opin Struct Biol. 2007;17:221–228. doi: 10.1016/j.sbi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 50.Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 51.Hinrichsen L, Meyerholz A, Groos S, Ungewickell EJ. Bending a membrane: how clathrin affects budding. Proc Natl Acad Sci U S A. 2006;103:8715–8720. doi: 10.1073/pnas.0600312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss M, Nilsson T. A kinetic proof-reading mechanism for protein sorting. Traffic. 2003;4:65–73. doi: 10.1034/j.1600-0854.2003.40202.x. [DOI] [PubMed] [Google Scholar]
- 53.Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 54.Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- 55.Lundmark R, Doherty GJ, Vallis Y, Peter BJ, McMahon HT. Arf family GTP loading is activated by, and generates, positive membrane curvature. Biochem J. 2008;414:189–194. doi: 10.1042/BJ20081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foret L, Sens P. Kinetic regulation of coated vesicle secretion. Proc Natl Acad Sci U S A. 2008;105:14763–14768. doi: 10.1073/pnas.0801173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid SL, Damke H. Coated vesicles: a diversity of form and function. Faseb J. 1995;9:1445–1453. doi: 10.1096/fasebj.9.14.7589986. [DOI] [PubMed] [Google Scholar]
- 58.Manneville JB, Bassereau P, Levy D, Prost J. Activity of Transmembrane Proteins Induces Magnification of Shape Fluctuations of Lipid Membranes. Phys Rev Lett. 1999;82:4356–4359. [Google Scholar]
- 59.Girard P, Prost J, Bassereau P. Passive or active fluctuations in membranes containing proteins. Phys Rev Lett. 2005;94:088102. doi: 10.1103/PhysRevLett.94.088102. [DOI] [PubMed] [Google Scholar]
- 60.Fletcher DA, Geissler PL. Active biological materials. Annu Rev Phys Chem. 2009;60:469–486. doi: 10.1146/annurev.physchem.040808.090304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shemesh T, Luini A, Malhotra V, Burger KN, Kozlov MM. Prefission constriction of Golgi tubular carriers driven by local lipid metabolism: a theoretical model. Biophys J. 2003;85:3813–3827. doi: 10.1016/S0006-3495(03)74796-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen HY. Internal states of active inclusions and the dynamics of an active membrane. Phys Rev Lett. 2004;92:168101. doi: 10.1103/PhysRevLett.92.168101. [DOI] [PubMed] [Google Scholar]
- 63.Karsenti E. Self-organization in cell biology: a brief history. Nat Rev Mol Cell Biol. 2008;9:255–262. doi: 10.1038/nrm2357. [DOI] [PubMed] [Google Scholar]
- 64.Mikhailov AS, Ertl G. Nonequilibrium microstructures in reactive monolayers as soft matter systems. Chemphyschem. 2009;10:86–100. doi: 10.1002/cphc.200800277. [DOI] [PubMed] [Google Scholar]
- 65.John K, Bar M. Alternative mechanisms of structuring biomembranes: self-assembly versus self-organization. Phys Rev Lett. 2005;95:198101. doi: 10.1103/PhysRevLett.95.198101. [DOI] [PubMed] [Google Scholar]
- 66.Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weidman P, Roth R, Heuser J. Golgi membrane dynamics imaged by freeze-etch electron microscopy: views of different membrane coatings involved in tubulation versus vesiculation. Cell. 1993;75:123–133. [PubMed] [Google Scholar]
- 68.Bard F, Malhotra V. The formation of TGN-to-plasma-membrane transport carriers. Annu Rev Cell Dev Biol. 2006;22:439–455. doi: 10.1146/annurev.cellbio.21.012704.133126. [DOI] [PubMed] [Google Scholar]
- 69.Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. Embo J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voeltz GK, Prinz WA. Sheets, ribbons and tubules - how organelles get their shape. Nat Rev Mol Cell Biol. 2007;8:258–264. doi: 10.1038/nrm2119. [DOI] [PubMed] [Google Scholar]
- 71.Sheetz MP, Singer SJ. Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc Natl Acad Sci U S A. 1974;71:4457–4461. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Epand RM. Membrane lipid polymorphism: relationship to bilayer properties and protein function. Methods Mol Biol. 2007;400:15–26. doi: 10.1007/978-1-59745-519-0_2. [DOI] [PubMed] [Google Scholar]
- 73.Fuller N, Rand RP. The influence of lysolipids on the spontaneous curvature and bending elasticity of phospholipid membranes. Biophys J. 2001;81:243–254. doi: 10.1016/S0006-3495(01)75695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leikin S, Kozlov MM, Fuller NL, Rand RP. Measured effects of diacylglycerol on structural and elastic properties of phospholipid membranes. Biophys J. 1996;71:2623–2632. doi: 10.1016/S0006-3495(96)79454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kooijman EE, Chupin V, Fuller NL, Kozlov MM, de Kruijff B, Burger KN, Rand PR. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 2005;44:2097–2102. doi: 10.1021/bi0478502. [DOI] [PubMed] [Google Scholar]
- 76.Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frolov VA, Zimmerberg J. Flexible scaffolding made of rigid BARs. Cell. 2008;132:727–729. doi: 10.1016/j.cell.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 78.Shnyrova A, Frolov VA, Zimmerberg J. ER biogenesis: self-assembly of tubular topology by protein hairpins. Curr Biol. 2008;18:R474–R476. doi: 10.1016/j.cub.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 79.Zoncu R, Perera RM, Sebastian R, Nakatsu F, Chen H, Balla T, Ayala G, Toomre D, De Camilli PV. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc Natl Acad Sci U S A. 2007;104:3793–3798. doi: 10.1073/pnas.0611733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meyerholz A, Hinrichsen L, Groos S, Esk PC, Brandes G, Ungewickell EJ. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic. 2005;6:1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- 81.Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 83.Lippincott-Schwartz J, Liu W. Membrane trafficking: coat control by curvature. Nature. 2003;426:507–508. doi: 10.1038/426507a. [DOI] [PubMed] [Google Scholar]
- 84.Aridor M, Bannykh SI, Rowe T, Balch WE. Cargo can modulate COPII vesicle formation from the endoplasmic reticulum. J Biol Chem. 1999;274:4389–4399. doi: 10.1074/jbc.274.7.4389. [DOI] [PubMed] [Google Scholar]
- 85.Leduc C, Campas O, Zeldovich KB, Roux A, Jolimaitre P, Bourel-Bonnet L, Goud B, Joanny JF, Bassereau P, Prost J. Cooperative extraction of membrane nanotubes by molecular motors. Proc Natl Acad Sci U S A. 2004;101:17096–17101. doi: 10.1073/pnas.0406598101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kress H, Stelzer EH, Holzer D, Buss F, Griffiths G, Rohrbach A. Filopodia act as phagocytic tentacles and pull with discrete steps and a load-dependent velocity. Proc Natl Acad Sci U S A. 2007;104:11633–11638. doi: 10.1073/pnas.0702449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heinrich V, Bozic B, Svetina S, Zeks B. Vesicle deformation by an axial load: from elongated shapes to tethered vesicles. Biophys J. 1999;76:2056–2071. doi: 10.1016/S0006-3495(99)77362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frolov VA, Lizunov VA, Dunina-Barkovskaya AY, Samsonov AV, Zimmerberg J. Shape bistability of a membrane neck: a toggle switch to control vesicle content release. Proc Natl Acad Sci U S A. 2003;100:8698–8703. doi: 10.1073/pnas.1432962100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rauch C, Farge E. Endocytosis switch controlled by transmembrane osmotic pressure and phospholipid number asymmetry. Biophys J. 2000;78:3036–3047. doi: 10.1016/S0006-3495(00)76842-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kohyama T, Kroll DM, Gompper G. Budding of crystalline domains in fluid membranes. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68:061905. doi: 10.1103/PhysRevE.68.061905. [DOI] [PubMed] [Google Scholar]
- 92.Fourcade B, Miao L, Rao M, Wortis M, Zia RK. Scaling analysis of narrow necks in curvature models of fluid lipid-bilayer vesicles. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1994;49:5276–5286. doi: 10.1103/physreve.49.5276. [DOI] [PubMed] [Google Scholar]
- 93.Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 94.Sieber JJ, Willig KI, Kutzner C, Gerding-Reimers C, Harke B, Donnert G, Rammner B, Eggeling C, Hell SW, Grubmuller H, Lang T. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- 95.Heinzer S, Worz S, Kalla C, Rohr K, Weiss M. A model for the self-organization of exit sites in the endoplasmic reticulum. J Cell Sci. 2008;121:55–64. doi: 10.1242/jcs.013383. [DOI] [PubMed] [Google Scholar]
- 96.Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manneville JB, Casella JF, Ambroggio E, Gounon P, Bertherat J, Bassereau P, Cartaud J, Antonny B, Goud B. COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc Natl Acad Sci U S A. 2008;105:16946–16951. doi: 10.1073/pnas.0807102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Clermont Y, Rambourg A, Hermo L. Connections Between the Various Elements of the Cis- and Mid-compartments of the Golgi Apparatus of Early Rat Spermatids. Anat Rec. 1994;240:469–480. doi: 10.1002/ar.1092400405. [DOI] [PubMed] [Google Scholar]