Abstract
Forkhead box O (FOXO) transcriptional protein family members, including FOXO1 and FOXO3, are involved in the modulation of autophagy. However, whether there is redundancy between FOXO1 and FOXO3 in the ability to induce autophagy remains unclear. In this study, we showed that FOXO3 induced a transcription-dependent autophagy, and FOXO1 was required for this process. Overexpression of wild-type FOXO3 (WT) or FOXO3 (3A), which harbors alanine mutations at residues Thr32, Ser253 and Ser315, but not transcription-inactive FOXO3 (∆DB3A), significantly induced autophagy in the human embryonic kidney cell line HEK293T and mouse embryonic fibroblast (MEF) cell lines. Interestingly, depletion of FOXO1 by siRNA attenuated FOXO3-induced autophagy. Our data also showed that FOXO3 overexpression did not increase the expression of FOXO1 at the protein level, although FOXO3 was capable of binding the promoter region of FOXO1 and inducing an increase in the transcription of FOXO1 mRNA. Furthermore, our results showed that FOXO3 promoted the translocation of FOXO1 from the nucleus to the cytoplasm, resulting in an increase in FOXO1-induced autophagy. Moreover, our results supported a mechanism whereby FOXO3 dramatically increased the expression of the class I PtdIns3K catalytic subunit PIK3CA, leading to an increase in AKT1 activity, which resulted in the phosphorylation and nuclear export of FOXO1. To the best of our knowledge, our data are the first to suggest that FOXO1 plays a central role in FOXO3-induced autophagy.
Keywords: FOXO1, FOXO3, autophagy, PIK3CA, AKT1
Introduction
Autophagy is an evolutionarily conserved cell survival pathway through which excess cytoplasmic components are degraded under normal and pathological conditions.1 Autophagy is responsible for recycling macromolecules to generate energy for intracellular renovation within the cell.1-3 The autophagic process is thus beneficial to the development and growth of normal individual cells, although the role of autophagy in cancer cells remains unclear.4-8
Stimuli, such as insulin secretion, amino acid deficiency, energy depletion or viral invasion, can stimulate autophagy.9-11 Among the factors influencing autophagy, the mechanistic target of rapamycin complex 1 (MTORC1), which is a critical nutritional sensor, plays a central negative regulatory role in autophagy by phosphorylating and inactivating ULK1/ATG1, which is required to initiate autophagy.12-15 Additionally, BECN1, the human homolog of the yeast VPS30/ATG6 gene, was among the first identified mammalian factors shown to be involved in promoting autophagy, and BECN1 functions through the BECN1- PtdIns3K complex to promote autophagosome formation.16,17
Recent evidence has revealed that the FOXO protein family members FOXO1 and FOXO3 promote autophagy.18-23 In skeletal muscle cells, FOXO3 directly upregulates autophagy-related genes, such as microtubule-associated protein 1 light chain 3 (MAP1LC3) and BCL2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3).19 FOXO1 has both transcription-dependent and transcription-independent roles in autophagy. In mouse cardiomyocytes, FOXO1 is deacetylated by the NAD-dependent deacetylase sirtuin-1 (SIRT1) that, in turn, induces the expression of the RAS-related GTP-binding protein RAB7A, which mediates the fusion of mature autophagic vesicles with lysosomes.21 In addition, FOXO1 mediates starvation-induced autophagy through a transcription-independent mechanism in human cancer cells. We have previously observed that cytoplasmic FOXO1 is required for serum starvation- or H2O2-induced autophagy in cancer cells. Under these conditions, FOXO1 became acetylated and dissociated from SIRT2 in the cytoplasm, and the acetylated FOXO1, in turn, binds to ATG7 to promote autophagy.23 Therefore, it is likely that FOXO family member proteins induce both transcription-dependent and transcription-independent autophagy.
Although FOXO3 and FOXO1 belong to the FOXO transcriptional protein family and share overlapping structure and function, they harbor several important differences.24-26 For example, FOXO3 and FOXO1 have different tissue-dependent expression patterns. Whereas FOXO1 is highly expressed in adipose tissues, FOXO3 is most highly expressed in brain tissue in mammals.26 In addition, the promoter of FOXO1, but not FOXO3, contains a consensus Forkhead response element (FHRE).27 Furthermore, in the murine liver, FOXO1 mainly regulates the expression of gluconeogenic enzymes, whereas FOXO3 plays an important role in the expression of lipogenic enzymes.28,29 More importantly, only Foxo1 knockout mice show an embryonically lethal phenotype,30,31 which implies that FOXO1 has more vital cellular and biological functions or less molecular redundancies.
Post-translational modifications of FOXO proteins, such as phosphorylation and acetylation, affect their functions.32 For example, the subcellular localization of FOXO1 in the nucleus or cytoplasm is controlled by AKT1, a kinase that catalyzes the phosphorylation of FOXO1 at Thr24, Ser256 and Ser319.33,34 Upon phosphorylation, FOXO1 translocates from the nucleus to the cytoplasm in a tyrosine 3-mono-oxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide [YWHAQ (14-3-3)] protein-dependent manner, resulting in a significant decrease in FOXO1 transcriptional activity.32,35,36 The class I PI3K enzyme is an essential upstream regulator of AKT1 and is composed of the catalytic subunit PIK3CA (p110) and the regulatory subunit PIK3R1 (p85). Class I PI3K is the main enzyme responsible for modulating FOXO1 nuclear exclusion.34,37 Class I PI3K is recruited to activated tyrosine kinase receptors where it phosphorylates phosphatidylinositol (4,5)-bisphosphate (PtdIns(4,5)P2) to form PtdIns(3,4,5)P3. PtdIns(3,4,5)P3 subsequently recruits AKT1 and 3-phosphoinositide-dependent protein kinase-1 (PDPK1) to the cell membrane, resulting in the activation of AKT1 and the subsequent phosphorylation of AKT1 targets.37,38 Thus, AKT1 activity is tightly associated with FOXO1 localization and transcriptional activity.
Recent evidence has shown that FOXO3 may transcriptionally activate FOXO1.39,40 Therefore, we investigated whether FOXO3 coordinately upregulates FOXO1 as a mechanism to induce autophagy.
In this study, we found that FOXO3 induced transcription-dependent autophagy in HEK293T cells and MEF cells, and FOXO1 was required for this process. Our data showed that FOXO3 upregulated class I PI3K-AKT1 activity by transactivating the catalytic subunit PIK3CA, which in turn phosphorylated FOXO1, resulting in the exportation of FOXO1 from the nucleus to the cytoplasm and promoting autophagy. Our data support a mechanism whereby FOXO3 coordinately regulates FOXO1 to induce autophagy.
Results
FOXO3 transcriptionally promoted autophagy
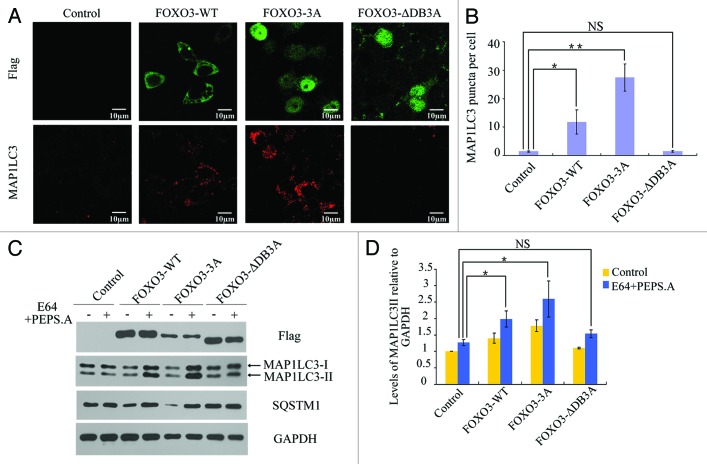
To investigate the effect of FOXO3 on the induction of autophagy in mammalian cells, plasmids expressing a Flag-tagged FOXO3 construct (WT), a nuclear-localized Flag-tagged FOXO3 construct (3A), and a nuclear-localized but DNA-binding deficient Flag-tagged FOXO3 construct (∆DB3A) were generated. Each plasmid construct was transfected into HEK293T cells, and after 48 h, the endogenous MAP1LC3 puncta were detected by confocal microscopy. MAP1LC3 is the mammalian homolog of ATG8 and is a commonly used marker for autophagosomes.41 During autophagosome formation, MAP1LC3-I is cleaved to the shorter isoform MAP1LC3-II and is conjugated to phosphatidylethanolamine (PE), which functions to anchor MAP1LC3-II into the autophagosome membrane.42 Our data showed a large increase in the number of endogenous MAP1LC3 puncta in FOXO3 (WT)-transfected cells, and an even larger increase in the number of MAP1LC3 puncta in the nucleus-localized FOXO3 (3A)-transfected cells. No difference in MAP1LC3 puncta was observed in the DNA binding-deficient, nucleus-localized FOXO3 (∆DB3A)-transfected cells (Fig. 1A and B). Proteins were extracted from cells transfected with each of the constructs, and western blotting was performed to assess changes in MAP1LC3-II accumulation and SQSTM1 (a selective autophagy substrate) turnover. Overexpression of FOXO3 (WT) or FOXO3 (3A) but not FOXO3 (∆DB3A) resulted in an increase in MAP1LC3-II levels, and this increase was more obvious in the presence of lysosomal protease inhibitors E64 and pepstatin-A (Fig. 1C and D). Similarly, SQSTM1 was degraded in HEK293T cells expressing FOXO3 (WT) or FOXO3 (3A) but not in cells expressing FOXO3 (∆DB3A) (Fig. 1C). We tested our observations in TM-ER MEF cells, which stably express a 4-hydroxytamoxifen (4-OHT)-inducible FOXO3 (3A)43 construct, and the mouse myoblast cell line C2C12 transfected with a Flag-tagged FOXO3 (3A) plasmid by assessing MAP1LC3-II accumulation and SQSTM1 turnover. Expression of FOXO3 in these cell lines induced a significant increase in MAP1LC3-II accumulation and SQSTM1 turnover (Fig. S1A and S1B) suggesting that overexpressed FOXO3 induces autophagy in multiple cell lines by acting as a transcription factor.
Figure 1. FOXO3 induced autophagy in HEK293T cells. (A) HEK293T cells were transfected with empty vector control, Flag-FOXO3 (WT), Flag-FOXO3 (3A) or Flag-FOXO3 (∆DB3A) plasmid. Forty eight hours post-transfection, the cellular localization of exogenous FOXO3 (upper panels) or endogenous MAP1LC3 puncta (lower panels) were observed by confocal microscopy. Scale bars: 10 μm. (B) Quantification of the endogenous MAP1LC3 puncta per cell. Error bars represent the standard deviation (n = 50 for three independent experiments). **p < 0.01, *p < 0.05, NS, not significant. (C) HEK293T cells were transfected with empty vector control, Flag-FOXO3 (WT), Flag-FOXO3 (3A)-expressing plasmid or Flag-FOXO3 (∆DB3A) plasmid in the presence or absence of E64 and pepstatin A. Forty-eight hours post-transfection, cell lysates were extracted and immunoblotted with anti-Flag, anti-MAP1LC3, anti-SQSTM1 or anti-GAPDH antibody. (D) Statistic analysis of the total MAP1LC3-II density relative to GAPDH levels. Error bars represent the standard deviation (n = 3). *p < 0.05, NS, not significant.
Furthermore, both confocal and western blot data confirmed that C2C12 cells underwent autophagy when cells were incubated in Hanks’ Balanced Salt Solutions (HBSS), and this autophagy was effectively suppressed by pretreatment with Foxo3 RNAi (Fig. S1C–E). These data indicated that FOXO3 is required for the induction of autophagy in response to nutrient starvation.
FOXO1 was required for autophagy mediated by FOXO3
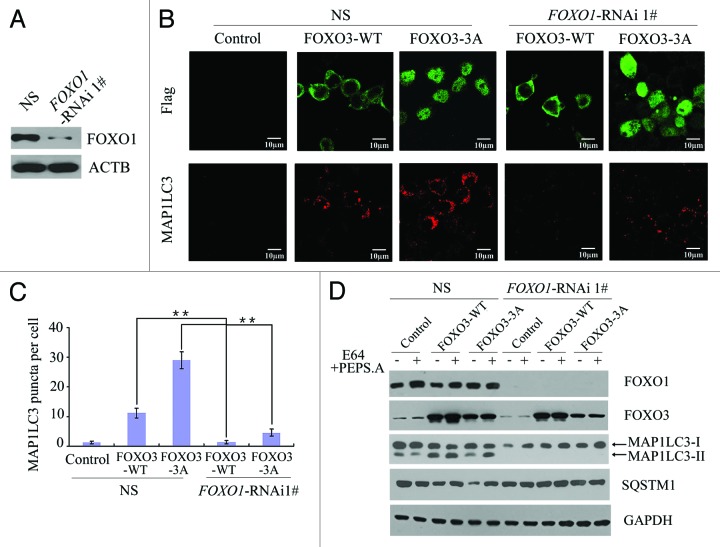
FOXO1 induces autophagy in cardiomyocytes and cancer cells,21,23,44 and FOXO3 binds to the promoter of FOXO1 to induce the expression of FOXO1 mRNA.39,40 Therefore, FOXO3 may induce autophagy by upregulating FOXO1 expression. To determine the relationship between FOXO3 and FOXO1 in the context of autophagy, a stable FOXO1 knockdown HEK293T cell line and a nonspecific HEK293T knockdown control were established by transfecting either the pGCSIL-FOXO1 RNA interference (RNAi) plasmid or a nonspecific control RNAi plasmid, respectively (Fig. 2A; Fig. S2A). FOXO3 overexpression-induced autophagy was almost completely inhibited in the FOXO1 knockdown cell line. Overexpression of FOXO3 (WT) or FOXO3 (3A) induced a significant increase in the amount of endogenous MAP1LC3 puncta in the presence of the protease inhibitor E64. However, the increase in MAP1LC3 puncta due to FOXO3 (WT) or FOXO3 (3A) overexpression was reversed when FOXO1 was knocked down (Fig. 2B and C; Fig. S2B and S2C). In addition, MAP1LC3-II accumulation and SQSTM1 turnover were also significantly decreased in the FOXO1 knockdown stable cell line when compared with stable cell lines expressing FOXO1 when assayed by western blot (Fig. 2D; Fig. S2D). These data suggested that FOXO3 requires FOXO1 to induce autophagy.
Figure 2. FOXO1 was required for FOXO3-induced autophagy. (A) Western blot analysis of FOXO1 protein levels in the stable nonspecific siRNA-expressing HEK293T cells (NS) or stable FOXO1-siRNA-expressing HEK293T cells (FOXO1-RNAi). (B) Empty control vector, Flag-FOXO3 (WT) or Flag-FOXO3 (3A) plasmid was transfected into HEK293T (NS) cells or FOXO1-RNAi cells. Expression of Flag-FOXO3 (upper panels) or endogenous MAP1LC3 puncta (lower panels) was detected by confocal microscopy. Scale bars: 10 μm. (C) Quantification of endogenous MAP1LC3 puncta per cell. Error bars represent the standard deviation (n = 50 for three independent experiments). **p < 0.01. (D) HEK293T (NS) cells or FOXO1-RNAi cells were transfected with control, Flag-FOXO3 (WT) or Flag-FOXO3 (3A) plasmid and then treated with or without E64 and pepstatin A. Forty-eight hours post-transfection, cell lysates were extracted and immunoblotted with anti-FOXO1, anti-FOXO3, anti-MAP1LC3, anti-SQSTM1 or anti-GAPDH antibody.
FOXO3 transcriptionally activated FOXO1
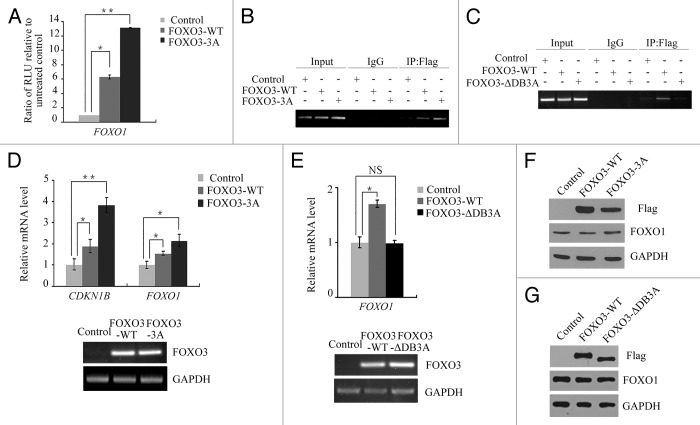
To further investigate the mechanism by which FOXO1 and FOXO3 cooperate to regulate autophagy, an empty vector control, a FOXO3 (WT) plasmid or a FOXO3 (3A) plasmid was cotransfected with a FOXO1 promoter-driven luciferase reporter construct into HEK293T cells. Then, the cells were assayed using a relative luciferase activity based on the FOXO1 promoter. As shown in Figure 3A, the relative luciferase activity of the FOXO1 promoter significantly increased when the cells were cotransfected with the FOXO3 (WT)-expressing plasmid, and luciferase activity increased further when cells were cotransfected with the FOXO3 (3A) plasmid. A chromatin immunoprecipitation (ChIP) assay was also performed to determine whether FOXO3 binds directly to the FOXO1 promoter. Our results showed that FOXO3 binding to the FOXO1 promoter increased significantly when FOXO3 was overexpressed in HEK293T cells (Fig. 3B). Due to lacking the sequence for DNA binding, FOXO3 (ΔDB3A) fails to bind to the promoter of FOXO1 (Fig. 3C). Furthermore, real-time PCR was performed to determine FOXO1 expression levels using mRNA extracted from cells transfected with an empty vector control, the FOXO3 (WT)-expressing plasmid, the FOXO3 (3A)-expressing plasmid or the FOXO3 (∆DB3A)-expressing plasmid. Cyclin-dependent kinase inhibitor 1B (CDKN1B) transcript levels, a known transcriptional target for FOXO3, was used as a positive control. Overexpression of FOXO3 transcriptionally enhanced the expression of FOXO1 mRNA (Fig. 3D and E). Together, these data demonstrated that FOXO3 transcriptionally activated FOXO1 expression by directly binding to the FOXO1 promoter.
Figure 3. FOXO3 upregulated the transcript level, but not the protein level, of FOXO1. (A) Empty control, Flag-FOXO3 (WT) or Flag-FOXO3 (3A) plasmid was cotransfected with luciferase reporter plasmid under the control of the FOXO1 promoter into HEK293T cells. After 24 h, the relative luciferase activity was measured. RLU, relative luciferase units. Error bars represent the standard deviation (n = 3). **p < 0.01, *p < 0.05. (B and C) HEK293T cells were transfected with empty control, Flag-FOXO3 (WT), Flag-FOXO3 (3A) or Flag-FOXO3 (∆DB3A) plasmid. Forty-eight hours post-transfection, DNA fragments bound to exogenous Flag-FOXO3 protein were isolated from cells by the chromatin immunoprecipitation (ChIP) assay with anti-Flag antibody. DNA fragments that copurified with the anti-FLAG antibody were amplified by PCR using primers specific for several sequences within the FOXO1 promoter. (D–G) HEK293T cells were transfected with empty control, Flag-FOXO3 (WT), Flag-FOXO3 (3A) or Flag-FOXO3 (∆DB3A) plasmid. Forty eight hours post-transfection, total RNA and total protein were extracted. Quantitative PCR (qPCR) was performed to measure the expression of CDKN1B and FOXO1 (D and E, upper panel). The transfection efficiency of Flag-FOXO3 (WT), Flag-FOXO3 (3A) or Flag-FOXO3 (∆DB3A) was measured by PCR (D and E, lower panel). A western blot was performed to detect changes in the protein levels of Flag-FOXO3, FOXO1 and GAPDH (F and G). **p < 0.01, *p < 0.05.
However, we unexpectedly did not observe an increase in FOXO1 protein level after overexpression of either FOXO3 (WT), FOXO3 (3A) or FOXO3 (∆DB3A) (Fig. 3F and G), suggesting that FOXO3 overexpression may not cause an increase in the total FOXO1 protein levels.
FOXO3 promoted FOXO1 nuclear exclusion
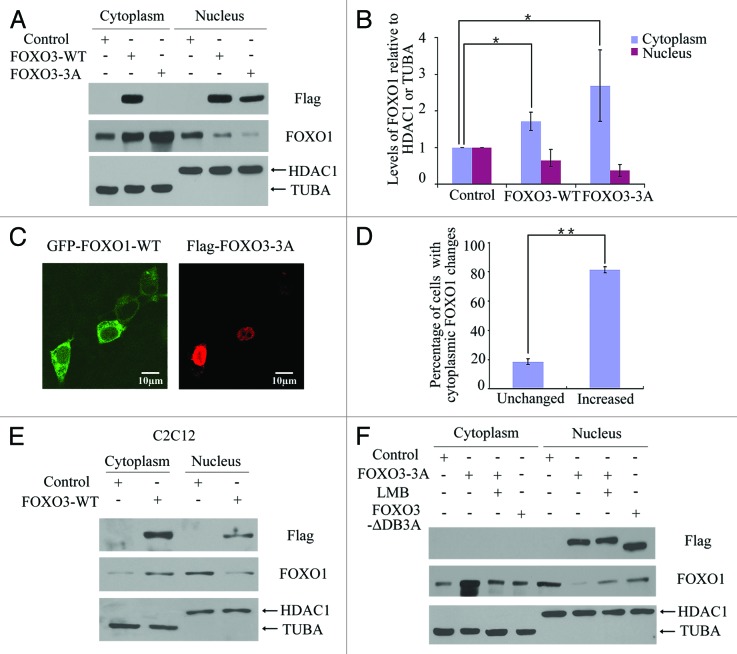
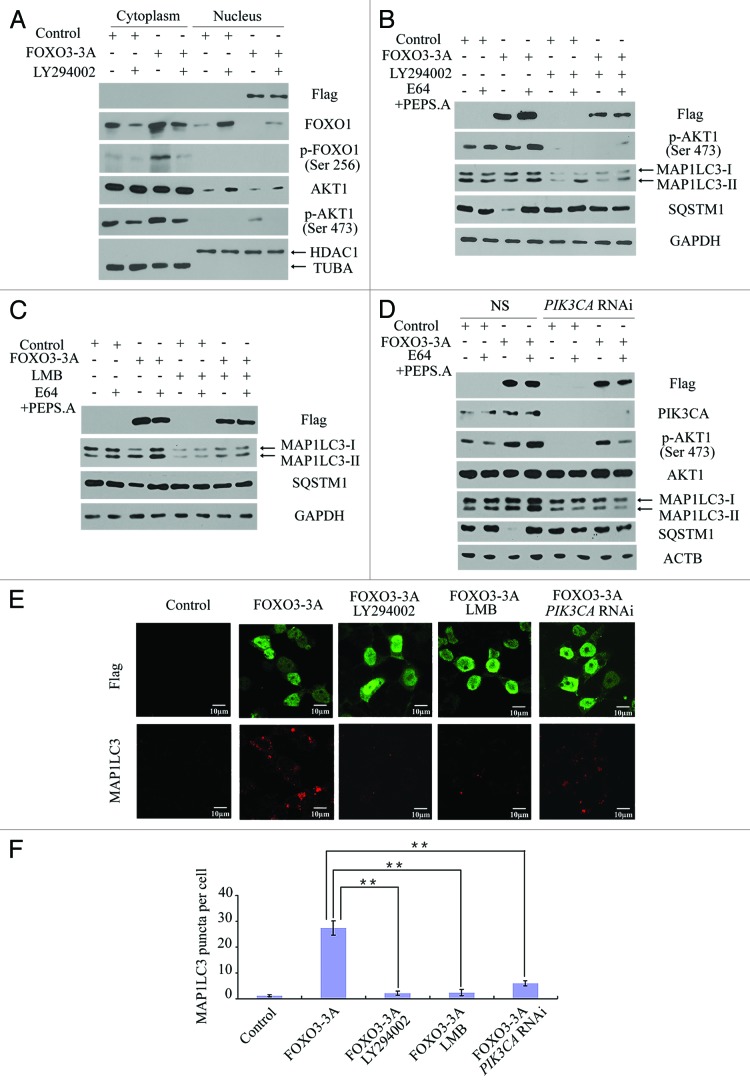
To further evaluate the role of FOXO1 in FOXO3-induced autophagy, we examined the FOXO1 protein level in the cytoplasm and in the nucleus when FOXO3 is overexpressed. An empty vector control, a FOXO3 (WT)-expressing plasmid or a FOXO3 (3A)-expressing plasmid was transfected into HEK293T cells, and the cytosolic proteins and nuclear proteins were extracted and analyzed by western blot to detect FOXO1 expression. As shown in Figures 4A and B, the level of cytosolic FOXO1 gradually increased, concordant with a decrease in nuclear FOXO1 when FOXO3 (WT) or FOXO3 (3A) was overexpressed in HEK293T cells. When HEK293T cells were co-transfected with GFP-FOXO1 (WT) and Flag-tagged FOXO3 (3A) plasmids and observed by confocal microscopy, cytoplasmic FOXO1 was generally increased in cells that overexpressed FOXO3 providing visual evidence that overexpression of FOXO3 correlates with FOXO1 cytoplasmic translocation (Fig. 4C and D). Similarly, we observed the translocation of FOXO1 from the nucleus to the cytoplasm in C2C12 cells (Fig. 4E), suggesting that the FOXO3-associated translocation of FOXO1 is a general biological phenomenon. Moreover, the FOXO3-associated change in translocation of FOXO1 appeared to be transcription-dependent. As shown in Figure 4F, FOXO3 (∆DB3A) overexpression was unable to cause FOXO1 nuclear exclusion. To investigate whether FOXO3-mediated cytosolic localization of FOXO1 is not due to FOXO1 degradation, the nuclear export inhibitor leptomycin B (LMB) was used to pretreat HEK293T cells prior to FOXO3 transfection. As shown in Figure 4F, the FOXO3-dependent FOXO1 nuclear exclusion was reversed by LMB pretreatment. Altogether, these data supported a model whereby FOXO3 causes FOXO1 nuclear exclusion through a transcriptional mechanism.
Figure 4. FOXO3 promoted FOXO1 cytoplasmic translocation. (A) Empty control, Flag-FOXO3 (WT) or Flag-FOXO3 (3A) plasmid was transfected into HEK293T cells, and cell fractionation was performed to analyze the cellular localization of endogenous FOXO1. TUBA was included as a loading control for cytoplasmic protein, and HDAC1 was included as a loading control for nuclear protein. (B) Statistical analysis of FOXO1 expression levels relative to TUBA or HDAC1. Error bars represent the standard deviation (n = 3). *p < 0.05. (C) GFP-FOXO1 (WT) plasmid and Flag-FOXO3 (3A) plasmid were cotransfected into HEK293T cells. Twenty-four hours post-transfection, localization of FOXO1 (left panel) or exogenous FOXO3 (right panel) was observed by confocal microscopy. Scale bar: 10 μm. (D) Statistical analysis of changes in cytoplasmic FOXO1 levels in response to FOXO3 overexpression. Error bars represent the standard deviation (n = 3). **p < 0.01. (E) C2C12 cells were transfected with empty control or Flag-FOXO3 (WT) plasmid. Forty eight hours post-transfection, cell fractionation was performed to analyze the cellular localization of endogenous FOXO1. TUBA was included as a loading control for cytoplasmic protein, and HDAC1 was included as a loading control for nuclear protein. (F) HEK293T cells were transfected with empty control, Flag-FOXO3 (3A) or Flag-FOXO3 (∆DB3A) plasmid in the presence or absence of LMB (0.5 ng/ml), and cytoplasmic and nuclear extracts were isolated. A western blot was performed to detect changes in FOXO1 protein levels.
FOXO3 enhanced PI3K-AKT1 activity by activating PIK3CA
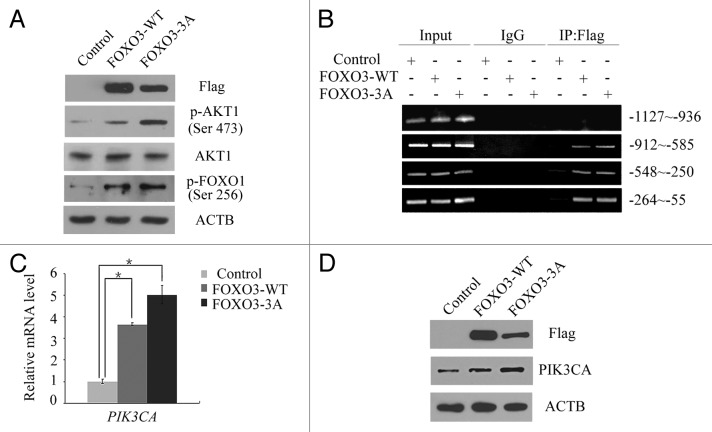
Because the phosphorylation of residue Ser256 of FOXO1 is catalyzed by AKT1 and is required for FOXO1 translocation from the nucleus to the cytoplasm,45 we tested whether FOXO3 overexpression enhanced AKT1 activity. FOXO3 (WT) or FOXO3 (3A) plasmid was transfected into HEK293T cells, and then, proteins were extracted and analyzed by western blot to assess the level of AKT1 activity. Although overexpression of either FOXO3 (WT) or FOXO3 (3A) did not induce an increase in the total amount of AKT1, both constructs induced a significant increase in phosphorylated AKT1 at residue Ser473 (Fig. 5A). FOXO1 phosphorylation at residue Ser256 was also significantly increased in response to either FOXO3 (WT) or FOXO3 (3A) overexpression, implying that FOXO3 transcriptionally activates certain genes that in turn phosphorylate AKT1, resulting in the phosphorylation of FOXO1 (Fig. 5A).
Figure 5. FOXO3 stimulated the PI3K-AKT1 pathway. (A) Empty control, Flag-FOXO3 (WT) or Flag-FOXO3 (3A) plasmid was transfected into HEK293T cells. Forty-eight hours post-transfection, total protein was extracted and was analyzed by western blot using anti-Flag, anti-p-AKT1 (Ser473), anti-AKT1, anti-p-FOXO1 (Ser256) or anti-ACTB antibodies. (B) HEK293T cells were transfected with empty control, Flag-FOXO3 (WT) or Flag-FOXO3 (3A) plasmid. Forty-eight hours post-transfection, DNA fragments binding exogenous Flag-FOXO3 protein were isolated by chromatin immunoprecipitation (ChIP) assay using anti-Flag antibody and amplified by PCR using four fragments of the PIK3CA promoter between -1127 bp and -936 bp, -912 bp and -585 bp, -548 bp and -250 bp and -264 bp and -55 bp. (C and D) HEK293T cells were transfected with empty control, Flag-FOXO3 (WT) or Flag-FOXO3 (3A) plasmid. Forty-eight hours post-transfection, total RNA and total protein were extracted. Quantitative PCR (qPCR) was performed to measure the transcript expression level of PIK3CA. *p < 0.05 (C). A western blot was also performed to detect changes in the protein levels of Flag-FOXO3, PIK3CA and ACTB (D).
Class I PI3K is primarily responsible for catalyzing the phosphorylation of AKT1;37 therefore, we investigated the relationship between FOXO3 overexpression and the expression level of PIK3CA. As shown in Figure 5B, a ChIP assay was used to show that FOXO3 was able to bind to the promoter of PIK3CA on three fragments between -912 bp and -585 bp, between -548 bp and -250 bp and between -264 bp and -55 bp. Furthermore, class I PI3K promoter binding activity was significantly enhanced upon overexpression of FOXO3. Consistently, overexpression of FOXO3 led to a significant increase in the expression of PIK3CA mRNA and protein (Fig. 5C and D), suggesting that FOXO3 increases AKT1 activity by transcriptionally activating PIK3CA and thus promoting the nuclear exclusion of FOXO1.
Translocation of FOXO1 was necessary for FOXO3-induced autophagy
To confirm whether the translocation of FOXO1 was required for FOXO3-induced autophagy, HEK293T cells were treated with the PI3K inhibitor LY294002 to abolish FOXO1 phosphorylation and nuclear exclusion, and then MAP1LC3-II accumulation and SQSTM1 turnover were assessed in FOXO3-overexpressing cells. Overexpression of FOXO3 (3A) significantly induced the translocation of FOXO1 from the nucleus to the cytoplasm, and FOXO1 translocation was inhibited by LY294002, which specifically inhibited the phosphorylation of AKT1 (Fig. 6A). Furthermore, when cells overexpressing FOXO3 (3A) were treated with LY294002, MAP1LC3-II levels decreased, and SQSTM1 turnover was significantly reduced (Fig. 6B). When cells overexpressing FOXO3 (3A) were treated with LMB, the rate of autophagy decreased as revealed by a decrease in MAP1LC3-II levels and SQSTM1 turnover (Fig. 6C). Because LY294002 was able to inhibit both class I PI3K-AKT1 activity and PtdIns3K activity,46,47 we further used a specific siRNA to target PIK3CA to confirm the role of AKT1 in FOXO3-induced autophagy. In response to PIK3CA siRNA treatment in HEK293T cells, phospho-AKT1 (Ser473) was obviously downregulated, and consequently MAP1LC3-II autophagic flux and SQSTM1 turnover caused by FOXO3 (3A) overexpression were both significantly reduced as well (Fig. 6D). Furthermore, confocal microscopy revealed that overexpression of FOXO3 induced a significant increase in endogenous MAP1LC3 puncta, and the increase in endogenous MAP1LC3 puncta was reversed upon treatment with LY294002, LMB, or PIK3CA inhibition (Fig. 6E and F). These data suggest that FOXO3-induced autophagy was mainly regulated by FOXO1 translocation from the nucleus to the cytoplasm.
Figure 6. A PI3K-AKT1 inhibitor or a nuclear export inhibitor blocked FOXO3-induced autophagy. (A) HEK293T cells were transfected with empty control or Flag-FOXO3 (3A) plasmid and were then treated with or without LY294002 (20 μg/ml). Forty-eight hours post-transfection, cell fractionation was performed to analyze cellular localization of endogenous FOXO1. TUBA was included as a loading control for cytoplasmic protein, and HDAC1 was included as a loading control for nuclear protein. (B) HEK293T cells were transfected with empty control or Flag-FOXO3 (3A) plasmid and then were treated with or without LY294002 (20 μg/ml). Forty-eight hours post-transfection, total proteins were extracted and were then analyzed by western blot with anti-Flag, anti-p-AKT1 (Ser473), anti-MAP1LC3, anti-SQSTM1 or anti-GAPDH antibody. (C) Empty control plasmid or Flag-FOXO3 (3A) plasmid was transfected into HEK293T cells and then were treated with or without LMB (0.5 ng/ml). Forty-eight hours post-transfection, cell lysates were extracted and were then analyzed by western blot with anti-Flag, anti-SQSTM1, anti-MAP1LC3 or anti-GAPDH antibody. (D) Nonspecific siRNA or PIK3CA siRNA was transfected into HEK293T cells for 48 h first. Empty control plasmid or Flag-FOXO3 (3A) plasmid was then transfected into the treated cells. Forty-eight hours post-transfection, cell lysates were extracted and were then analyzed by western blot with anti-Flag, anti-PIK3CA, anti-p-AKT1 (ser 473), anti-AKT1, anti-MAP1LC3, anti-SQSTM1 anti-ACTB antibody. (E) HEK293T cells were transfected with empty control plasmid or Flag-FOXO3 (3A) plasmid and then were treated with or without LY294002 (20 µg/ml) or LMB (0.5 ng/ml), or transfected with PIK3CA siRNA first. The cellular localization of exogenous FOXO3 (upper panels) or endogenous MAP1LC3 puncta (lower panels) was observed by confocal microscopy. Scale bars: 10 μm. (F) Quantification of endogenous MAP1LC3 puncta per cell. Error bars represent the standard deviation (n = 50 for three independent experiments). **p < 0.01.
Discussion
In this study, we identified a specific connection between FOXO1 and FOXO3 in the induction of autophagy. FOXO3 transcriptionally activated class I PI3K catalytic subunit PIK3CA, which in turn phosphorylated and upregulated AKT1 activity. Activated AKT1 catalyzed the phosphorylation of FOXO1 resulting in the translocation of FOXO1 to the cytoplasm and leading to the induction of autophagy (Fig. 7).
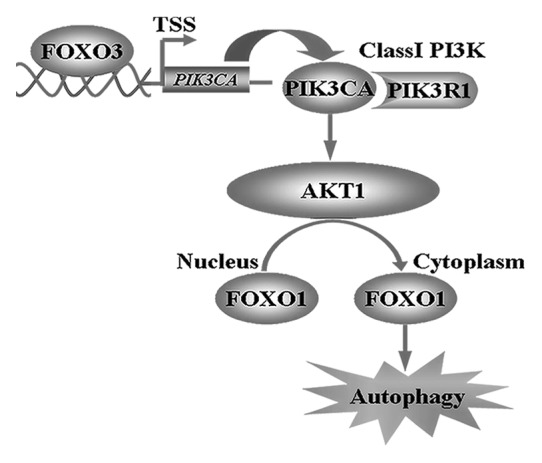
Figure 7. Schematic model illustrating the mechanism of FOXO3-induced autophagy.
FOXO3 can transcriptionally activate autophagy by upregulating autophagy-related (ATG) genes or autophagy regulatory genes, such as those encoding MAP1LC3, ATG12, GABARAPL1 (homolog of yeast ATG8) and BNIP3.19,20 However, previous reports have shown that low levels of ATGs are sufficient to induce autophagy, and thus the upregulation of ATGs may not be the only factor responsible for promoting autophagy.42,48 For example, a small amount of ATG5 is adequate to induce autophagy in mouse embryonic fibroblast (MEF) cells.48 Additionally, autophagy markers do not change when GFP-MAP1LC3 is overexpressed in F9 teratocarcinoma cells.46 However, the BCL2-related BH3-only protein BNIP3 has been shown to induce autophagy by promoting the release of BECN1 from the BECN1-BCL2 complex,49 and BNIP3 overexpression induces autophagy in HEK293T cells and malignant glioma cells.50,51 Therefore, it is possible that FOXO3 upregulates BNIP3 expression as part of the mechanism of FOXO3-induced autophagy.19,52 As a transcription factor, FOXO3 may promote autophagy by upregulating several genes in addition to BNIP3. In fact, several factors have been reported to induce autophagy through multiple pathways. For example, PRKA induces autophagy by phosphorylating several substrates, such as MTORC1, ULK1 and FOXO3.10,53,54 Our data showed that FOXO3 induced autophagy by changing the subcellular localization of FOXO1. In the presence of overexpressed FOXO3, we observed only a slight increase in autophagosome formation in the FOXO1 knockdown cells but a significant change in autophagic markers in cells with wild-type FOXO1 (Fig. 2B and C; Fig. S2B and S2C). Furthermore, in cells expressing wild-type FOXO1, FOXO3 overexpression did not induce autophagy if FOXO1 translocation was inhibited by LMB (Fig. 6C, E and F), supporting a model in which FOXO1 translocation plays a critical role in FOXO3-induced autophagy. However, whether FOXO3-mediated expression of BNIP3 or FOXO3-mediated FOXO1 nuclear exclusion is the predominant mechanism responsible for FOXO3-induced autophagy remains unclear and may vary depending on the cellular context.
Consistent with previous reports,39,40 we confirmed that FOXO3 upregulates the expression of FOXO1 mRNA (Fig. 3D and E) but does not alter FOXO1 protein levels (Fig. 3F and G). FOXO1 protein levels may be governed by a more complex mechanism, such as HUR-mediated regulation of FOXO1 mRNA stability or microRNA regulation. Precedence for an HUR type of mechanism has been observed upon H2O2 treatment of HeLa cells, which alters the protein level of SIRT155 by promoting HUR binding to the 3′UTR of SIRT1 mRNA effectively stabilizing SIRT1 transcript.55,56 Similarly, microRNAs, such as MIR182, may be responsible for regulating the FOXO1 protein level when an increase in FOXO1 mRNA is observed.57,58 MicroRNAs bind to the 3′UTR of the mRNA of specific genes and suppress their translation,59 thus preventing changes in protein expression despite changes in the mRNA level. Future studies should investigate whether microRNAs are involved in the regulation of FOXO1 translation. However, because FOXO1-dependent autophagy largely depends on its protein expression level and acetylation modification,23 upregulation of the FOXO1 level in mRNA induced by FOXO3 is not directly relevant to induction of autophagy by FOXO3.
Accumulating evidence suggests that FOXO1 translocation is dependent on its phosphorylation status.32-34,60,61 Phosphorylation of FOXO1 at different combinations of amino acids can result in different FOXO1 activity. For instance, activated AKT1 promotes the nuclear exclusion of FOXO1 by phosphorylating FOXO1 at residues Thr24, Ser256 and Ser319.33,34 In addition, cyclin-dependent kinase 2 (CDK2) mediates the nuclear exclusion of FOXO1 and inhibits FOXO1 transcriptional activity by directly phosphorylating FOXO1 at residue Ser249.60 By contrast, Ste20-like kinase (SLK) induces FOXO protein nuclear localization by phosphorylating FOXO at a conserved site, disrupting FOXO interaction with YWHAQ proteins.61 Similarly, we are the first to present data indicating that FOXO3 promotes the phosphorylation of FOXO1 by inducing AKT1 activity, which ultimately results in FOXO1 nuclear exclusion. Here, we noticed that increased AKT1 activity induced by FOXO3 overexpression also resulted in FOXO3 autophosphorylation.62 Therefore, FOXO3 (3A), which could not be phosphorylated by AKT1, was able to induce a stronger autophagy when compared with that of wild-type FOXO3 (Fig. 1A–D). We have previously published that cytosolic FOXO1 induces autophagy;23 therefore, the results of our current study are consistent by showing that autophagy increased in parallel with the nuclear exclusion of FOXO1 in response to FOXO3 overexpression.
Overexpression or somatic mutation of PIK3CA occurs in many cancers, including ovarian, colorectal and brain cancers, which correlates with increased activation of the AKT1 pathway.63-65 Previous studies have used co-immunoprecipitation assays to show that a high level of the PIK3CA subunit correlates with higher levels of the PI3K regulatory subunit PIK3R1 in ovarian cancer,63 and higher levels of PIK3CA and PIK3R1 may, in turn, upregulate the PI3K-AKT1 pathway to initiate diverse biological fuctions.37 In agreement with previous studies in drug-resistant leukemic cells,66 our data in HEK293T cells indicated that FOXO3 transcriptionally upregulated the PI3K catalytic subunit PIK3CA, which in turn activated AKT1. In addition, we identified three fragments in the promoter of PIK3CA that are bound by FOXO3. The promoter fragment from -548 bp to -250 bp is a newly identified binding site for FOXO3, which was not identified in drug-resistant leukemic cells (Fig. 5B).66 In addition to PIK3CA, FOXO3 may activate AKT1 through other pathways. A previous study in cardiomyocytes illustrated that FOXO disrupts interactions between AKT1 and PP2A as well as between AKT1 and calcineurin, with a resultant increase in AKT1 activity and decrease in insulin sensitivity.67 Furthermore, FOXO transcriptionally activates the expression of the insulin receptor, which in turn enhances AKT1 activity in the Drosophila Schneider S2 cell line.68 Future studies should investigate whether other factors contribute to AKT1 activation when FOXO3 is overexpressed.
In conclusion, our results showed that the FOXO3 transcription factor promoted autophagy through a FOXO1-dependent manner by increasing class I PI3K-AKT1 activity. Further parsing out of the various functions of the diverse members of the class I PI3K and FOXO families should shed new light on the accurate mechanism of FOXO3-dependent autophagy.
Materials and Methods
Cell culture
HEK293T and C2C12 cell line were purchased from the American Type Culture Collection, and grown in DMEM medium (GIBCO, 12800-017) supplemented with 10% fetal bovine serum (Hyclone, SV30087.02) in a 37°C incubator with a humidified, 5% CO2 atmosphere. TM-ER MEF (stably expressing FOXO3-3A MEF) cells were kindly provided by Dr. Zhengqiang Yuan. Starvation medium Hanks’ Balanced Salt Solutions (HBSS) were purchased from Ding Guo Biological Company (Ding Guo, CC0052).
Establishment of stable FOXO1 knockdown cell line
A FOXO1 stable knockdown cell line was generated by using a pGCSIL-FOXO1 RNAi plasmid (GAG CGT GCC CTA CTT CAA G). Plasmids were transfected into HEK293T cells, and the stable cell lines were established by selection with 10µg/ml puromycin (Sigma, P8833).
Plasmids and inhibitors
FOXO3 (WT) and FOXO3 (3A) expression plasmids were purchased from Addgene. FOXO3 (∆DB3A) plasmid was kindly provided by Dr. Plas. FOXO1-promoter luciferase reporter plasmid was provided by Dr. Hardingham. FOXO3 siRNA and PIK3CA siRNA were purchased from Sigma (Sigma, SASI_Mm02_00324885, SASI_Hs01_00219338).
Protease inhibitors E-64 (Sigma, E3132), pepstatin A (Sigma, P5318), nuclear export inhibitor leptomycin B (Sigma, L2913) were purchased from sigma. PI3K inhibitor LY294002 (Promega, V1201) was purchased from Shanghai Promega.
Cell transfection and luciferase assay
Lipofectamine 2000 (Invitrogen, 11668-019) was used in transfections for HEK293T cells, while GenJet (SignaGen, SL100489) was used in C2C12 transfection. Luciferase assay was performed by using luciferase assay kit (Promega, E1500) with firefly luciferase-based reporter gene activity.
ChIP assay
Briefly, 2 × 107 cells were fixed with 1% formaldehyde at 37°C for 10 min, and then they were lysed on ice for 15 min. These lysed extracts were subjected to shearing by sonication. After centrifugation at 12,000 rpm for 15 min, the soluble chromatin was subjected to immunoprecipitation with anti-Flag (Sigma, F3165). The primer sequences used in all ChIPs were as follows: promoter of FOXO1, TCG TTC AGC AAA GAC ATC GTG, AGC AAA CCA GCG TGG AGG C; promoter of PIK3CA, fragment between -264 bp and -55 bp, fragment between -548 bp and -250 bp and fragment between -1127 bp and -936 bp were as indicated,66 fragment between -912 bp and -585 bp, AAT CAC TGC TCC TAC GCT, ACT TCG TGG AGA CCT TTT.
Immunofluorescence analysis
Cells were cultured on confocal dishes to about 80% confluence. After treatment, cells were fixed with 4% paraformaldehyde and permeabilized with methanol. The dishes were incubated with blocking solution (0.8% BSA in PBS) and exposed overnight to primary antibody (1:100 dilution for all antibodies) at 4°C. After being washed three times with blocking solution, the dishes were exposed to a secondary antibody (1:200 dilution), conjugated to FITC/TRITC (fluorescein isothiocyanate/tetramethyl rhodamine isothiocyanate). Morphological alterations in the cells were observed and documented under a confocal microscope (Olympus BX-51, America Inc.).
Real-time PCR analysis of mRNA
Total RNA was isolated with TRIzol reagent (Invitrogen, 15596–026). cDNA was synthesized from 2 μg of RNA with oligo (dT)15 primers with the use of the Quantscript RT Kit (TianGen, KR103). Primer sequences of FOXO3, FOXO1, GAPDH, CDKN1B, PIK3CA for RT-PCR were as follows: FOXO3, TGA GAA GTT CCC CAG CGA CTT, TCC AAC CCA TCA GCA TCC A; FOXO1, CGC AGA TCT ACG AGT GGA TGG T, GCT CGG CTT CGG CTC TTA; GAPDH, GAA GAT GGT GAT GGG ATT TC, GAA GGT GAA GGT CGG AGT C; CDKN1B, TGG AGA AGC ACT GCA GAG AC, GCG TGT CCT CAG AGT TAG CC; PIK3CA, AAA TGA AAG CTC ACT CTG GAT TCC, TGT GCA ATT CCT ATG CAA TC.
Immunoblot analysis
Equal amounts of proteins (100 to150 µg) were size-fractionated by 7.5–15% SDS-PAGE. The antibodies used were anti-FOXO1 (Cell Signaling, 2880), anti-FOXO3 (Cell Signaling, 9467), anti-FOXO3 (Cell Signaling, 2497), anti-PIK3CA (Cell Signaling, 4249) and anti-MAP1LC3 (Cell Signaling, 2775), anti-SQSTM1 (MBL, PM045), anti-ACTB (Santa Cruz, sc-7210), anti-AKT1 (Epitomics, 1081), anti-p-AKT1 (Ser 473) (Epitomics, 2118), anti-GFP (Santa Cruz, sc-9996), and anti-Flag (Sigma, F1804). For immunoprecipitation, cells were harvested and then lysed in a Nonidet P40 buffer supplemented with a complete protease inhibitor cocktail (Roche, 04693132001). Whole-cell lysates or cytosolic proteins were used for immunoprecipitation with the indicated antibodies. Generally, 1 to 4 µg of antibody was added to 1 ml of cell lysate, which was incubated at 4°C for 8 to12 h. After the addition of Protein A/G-agarose beads, the incubation was continued for 1 h. Immunoprecipitates were extensively washed with lysis buffer and eluted with SDS loading buffer by boiling for 5 min. Main western blotting data showing MAP1LC3-II accumulation or SQSTM1 turnover. All bands of blots were scanned with a phosphorimager, and the relative intensity of each band was normalized to each band of GAPDH. Data collected came from at least three independent experiments.
Supplementary Material
Acknowledgments
We thank David R. Plas and Giles E. Hardingham for providing plasmids used in this study. We also thank Zengqiang Yuan for providing TM-ER MEF cells. This work was supported by National Key Basic Research Program of China (Grant 2011CB910100 to Y.Z., 2011CB504200 to W.Z.) and National Natural Science Foundation of China (Grant 90919030, 31070691 and 30921062 to W.Z.) and “111 project” from Minister of Education of China.
Glossary
Abbreviations:
- ATG
autophagy related
- FOXO1
forkhead box O1
- FOXO3
forkhead box O3
- GFP
green fluorescent protein
- MAP1LC3
microtubule-associated protein 1 light chain 3
- PIK3CA
class I PI3K catalytic subunit
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/21830
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/21830
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Baehrecke EH. Autophagy: dual roles in life and death? Nat Rev Mol Cell Biol. 2005;6:505–10. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 3.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 4.Eskelinen EL. Doctor Jekyll and Mister Hyde: autophagy can promote both cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1468–72. doi: 10.1038/sj.cdd.4401721. [DOI] [PubMed] [Google Scholar]
- 5.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–7. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 7.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–13. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 14.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Yang J, Zhao Y, Ma K, Jiang FJ, Liao W, Zhang P, et al. Deficiency of hepatocystin induces autophagy through an mTOR-dependent pathway. Autophagy. 2011;7:748–59. doi: 10.4161/auto.7.7.15822. [DOI] [PubMed] [Google Scholar]
- 16.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 17.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 18.Juhász G, Puskás LG, Komonyi O, Erdi B, Maróy P, Neufeld TP, et al. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007;14:1181–90. doi: 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–82. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sengupta A, Molkentin JD, Paik JH, DePinho RA, Yutzey KE. FoxO transcription factors promote cardiomyocyte survival upon induction of oxidative stress. J Biol Chem. 2011;286:7468–78. doi: 10.1074/jbc.M110.179242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–75. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 24.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/S0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 25.Obsil T, Obsilova V. Structural basis for DNA recognition by FOXO proteins. Biochim Biophys Acta 2010; 1813:1946-53. [DOI] [PubMed]
- 26.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–25. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 27.Berry FB, Skarie JM, Mirzayans F, Fortin Y, Hudson TJ, Raymond V, et al. FOXC1 is required for cell viability and resistance to oxidative stress in the eye through the transcriptional regulation of FOXO1A. Hum Mol Genet. 2008;17:490–505. doi: 10.1093/hmg/ddm326. [DOI] [PubMed] [Google Scholar]
- 28.Zhang K, Li L, Qi Y, Zhu X, Gan B, DePinho RA, et al. Hepatic suppression of Foxo1 and Foxo3 causes hypoglycemia and hyperlipidemia in mice. Endocrinology. 2012;153:631–46. doi: 10.1210/en.2011-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–5. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 30.Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–8. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- 31.Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, Cavenee WK, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–80. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Wang Y, Zhu WG. Applications of post-translational modifications of FoxO family proteins in biological functions. J Mol Cell Biol. 2011;3:276–82. doi: 10.1093/jmcb/mjr013. [DOI] [PubMed] [Google Scholar]
- 33.Nakae J, Park BC, Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem. 1999;274:15982–5. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 34.Rena G, Guo S, Cichy SC, Unterman TG, Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem. 1999;274:17179–83. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- 35.Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156:817–28. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obsil T, Ghirlando R, Anderson DE, Hickman AB, Dyda F. Two 14-3-3 binding motifs are required for stable association of Forkhead transcription factor FOXO4 with 14-3-3 proteins and inhibition of DNA binding. Biochemistry. 2003;42:15264–72. doi: 10.1021/bi0352724. [DOI] [PubMed] [Google Scholar]
- 37.Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. Biochem J. 2000;346:561–76. doi: 10.1042/0264-6021:3460561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–80. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 39.Al-Mubarak B, Soriano FX, Hardingham GE. Synaptic NMDAR activity suppresses FOXO1 expression via a cis-acting FOXO binding site: FOXO1 is a FOXO target gene. Channels (Austin) 2009;3:233–8. doi: 10.4161/chan.3.4.9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Essaghir A, Dif N, Marbehant CY, Coffer PJ, Demoulin JB. The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. J Biol Chem. 2009;284:10334–42. doi: 10.1074/jbc.M808848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M, et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med. 2006;203:1657–63. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–31. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.del Peso L, González VM, Hernández R, Barr FG, Núñez G. Regulation of the forkhead transcription factor FKHR, but not the PAX3-FKHR fusion protein, by the serine/threonine kinase Akt. Oncogene. 1999;18:7328–33. doi: 10.1038/sj.onc.1203159. [DOI] [PubMed] [Google Scholar]
- 46.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–6. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 47.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 48.Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–9. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 49.Mazure NM, Pouysségur J. Atypical BH3-domains of BNIP3 and BNIP3L lead to autophagy in hypoxia. Autophagy. 2009;5:868–9. doi: 10.4161/auto.9042. [DOI] [PubMed] [Google Scholar]
- 50.Vande Velde C, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, et al. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol. 2000;20:5454–68. doi: 10.1128/MCB.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–93. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- 52.Mammucari C, Schiaffino S, Sandri M. Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy. 2008;4:524–6. doi: 10.4161/auto.5905. [DOI] [PubMed] [Google Scholar]
- 53.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez AM, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, et al. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2012;113:695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 55.Abdelmohsen K, Pullmann R, Jr., Lal A, Kim HH, Galban S, Yang X, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–57. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–8. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, et al. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–77. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim KM, Park SJ, Jung SH, Kim EJ, Jogeswar G, Ajita J, et al. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J Bone Miner Res. 2012;27:1669–79. doi: 10.1002/jbmr.1604. [DOI] [PubMed] [Google Scholar]
- 59.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 60.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–7. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 61.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villén J, Becker EB, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 62.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 63.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 64.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 65.Samuels Y, Diaz LA, Jr., Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–73. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 66.Hui RC, Gomes AR, Constantinidou D, Costa JR, Karadedou CT, Fernandez de Mattos S, et al. The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol Cell Biol. 2008;28:5886–98. doi: 10.1128/MCB.01265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ni YG, Wang N, Cao DJ, Sachan N, Morris DJ, Gerard RD, et al. FoxO transcription factors activate Akt and attenuate insulin signaling in heart by inhibiting protein phosphatases. Proc Natl Acad Sci U S A. 2007;104:20517–22. doi: 10.1073/pnas.0610290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–20. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.