Abstract
Granular corneal dystrophy type 2 (GCD2) is an autosomal dominant disease characterized by a progressive age-dependent extracellular accumulation of transforming growth factor β-induced protein (TGFBI). Corneal fibroblasts from GCD2 patients also have progressive degenerative features, but the mechanism underlying this degeneration remains unknown. Here we observed that TGFBI was degraded by autophagy, but not by the ubiquitin/proteasome-dependent pathway. We also found that GCD2 homozygous corneal fibroblasts displayed a greater number of fragmented mitochondria. Most notably, mutant TGFBI (mut-TGFBI) extensively colocalized with microtubule-associated protein 1 light chain 3β (MAP1LC3B, hereafter referred to as LC3)-enriched cytosolic vesicles and CTSD in primary cultured GCD2 corneal fibroblasts. Levels of LC3-II, a marker of autophagy activation, were significantly increased in GCD2 corneal fibroblasts. Nevertheless, levels of SQSTM1/p62 and of polyubiquitinated protein were also significantly increased in GCD2 corneal fibroblasts compared with wild-type (WT) cells. However, LC3-II levels did not differ significantly between WT and GCD2 cells, as assessed by the presence of bafilomycin A1, the fusion blocker of autophagosomes and lysosomes. Likewise, bafilomycin A1 caused a similar change in levels of SQSTM1. Thus, the increase in autophagosomes containing mut-TGFBI may be due to inefficient fusion between autophagosomes and lysosomes. Rapamycin, an autophagy activator, decreased mut-TGFBI, whereas inhibition of autophagy increased active caspase-3, poly (ADP-ribose) polymerase 1 (PARP1) and reduced the viability of GCD2 corneal fibroblasts compared with WT controls. These data suggest that defective autophagy may play a critical role in the pathogenesis of GCD2.
Keywords: LC3/MAP1LC3B, SQSTM1/p62, TGFBI, defective autophagy, rapamycin, ubiquitination
Introduction
Degenerative disorders are often characterized by the accumulation of intracellular or extracellular protein aggregates that form toxic multimeric complexes that eventually promote cell death. Cells have special surveillance systems to control the quality of proteins and organelles. Thus, WT cells effectively clear misfolded, aged, malfunctioning and damaged intracellular components.1,2 Two important proteolytic systems, the ubiquitin/proteasome pathway and the autophagy pathway, are responsible for the removal of abnormal proteins.1 The ubiquitin/proteasome pathway selectively degrades proteins, whereas autophagy is largely responsible for the degradation of several classes of macromolecules and for organelle turnover. Any alterations in components of these proteolytic systems may result in the intracellular accumulation and retention of misfolded proteins or defective organelles, causing pathological processes and disease.2 Similarly, an increase in intracellular aggresome levels due to functional alterations in one or both systems can occur with age.3,4
Autophagy occurs at a basal rate in all cells5 and is important for maintaining normal cellular homeostasis. This degradation system is induced through either decreased mechanistic target of rapamycin (MTOR) and/or activation by the BECN1 signaling pathway.6 BECN1 is a component of the class III phosphatidylinositol 3-kinase (PtdIns 3-kinase) complex that regulates autophagosome biogenesis. Although autophagy is primarily a survival process for the cell, it can also be a part of a cell death process. Growing evidence suggests that alterations in autophagy are involved in the pathogenesis of many diseases.7 Recent studies have determined that accumulation of disease-related proteins is highly dependent on both autophagy and the ubiquitin/proteasome pathway.
GCD2, also called Avellino corneal dystrophy, is an autosomal dominant disorder caused by an arginine-to-histidine substitution at codon 124 (R124H) in the transforming growth factor β-induced gene (TGFBI) on chromosome 5q31.8 TGFBI encodes a protein of 683 amino acid residues that contain a secretory signal peptide and an Arg-Gly-Asp (RGD) integrin recognition sequence.9 TGFBI is a component of the extracellular matrix (ECM), where it mediates migration and cell adhesion by interacting with integrins.10-12 Age-dependent progressive accumulation of hyaline and amyloid containing TGFBI in the corneal stroma is a hallmark of GCD2, which is characterized by the production of TGFBI in the corneal epithelia and stroma, interfering with corneal transparency.9,13,14 Moreover, the homozygous R124H mutations are the cause of the severe phenotype of GCD2 characterized by early-onset and confluent superficial opacity.15
Corneal fibroblasts, also known as keratocytes, have typical dendritic morphology and express keratan sulfate proteoglycans that are necessary for corneal structure, physiology and particularly for the maintenance of corneal transparency.16,17 Under pathological conditions, corneal transparency often cannot be maintained due to the degeneration of corneal fibroblasts.18,19 Consistent with this condition, corneal fibroblasts from GCD2 patients show degenerative features, including degenerated organelles such as mitochondria and rough endoplasmic reticulum.20 Recently, we have reported that the R124H mutation causes aberrant redistribution of mut-TGFBI into lysosomes.21 However, the precise reason for this redistribution is currently unclear. Furthermore, we have also shown that oxidative stress can induce apoptotic cell death of primary cultured GCD2 corneal fibroblasts through decreased catalase levels.22 Reactive oxygen species (ROS), especially H2O2, produced by starved cells, function as signaling molecules in starvation-activated autophagy and are essential for autophagosome formation.23,24 Generation of ROS through oxidative stress can also cause autophagic cell death, which can be regulated by the selective degradation of catalase.25 We have recently reported the presence of abnormal mitochondrial function in GCD2 corneal fibroblasts.26 These studies led us to hypothesize that GCD2 is associated with impaired autophagy, which ultimately leads to cell death. To examine this hypothesis, we investigated the physiological roles of autophagy in the pathogenesis of GCD2 by measuring the extent of autophagosome formation and the status of autophagy in primary cultured corneal fibroblasts from heterozygous (HE) or homozygous (HO) GCD2 patients and WT subjects.
Results
Characterization of GCD2 corneal fibroblasts
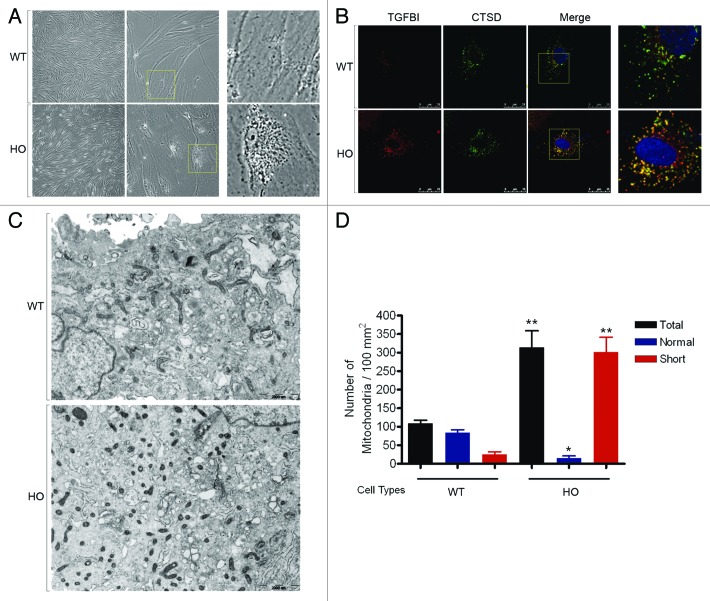
Phase contrast microscopic analysis was used to measure the morphological changes between WT and GCD2 corneal fibroblasts. GCD2 homozygous corneal fibroblasts displayed a number of morphological abnormalities, including increased size, intracellular deposits and senescence-like morphology, which were not seen in control cells (Fig. 1A). Results suggested that mut-TGFBI may accumulate intracellularly in GCD2 corneal fibroblasts. We then examined the subcellular localization of WT- and mut-TGFBI. Double immunostaining showed colocalization of the punctate TGFBI with CTSD, a lysosomal enzyme. Colocalization of TGFBI with CTSD indicates a lysosomal location of this protein, and GCD2 homozygous corneal fibroblasts showed much more extensive pattern of colocalized mut-TGFBI with CTSD than observed in WT corneal fibroblasts (Fig. 1B). Ultrastructural analyses using electron microscopy revealed that WT corneal fibroblasts had mitochondria with normal morphology (Fig. 1C). Ultrastructural analysis also indicated the presence of short, fragmented mitochondria with abnormal morphology in GCD2 corneal fibroblasts. Furthermore, GCD2 corneal fibroblasts had a higher number (2.86 ± 0.19 fold) of mitochondria compared with WT corneal fibroblasts (Fig. 1D). As shown in Figure 1D, WT corneal fibroblasts had 23.27 ± 5.78% proximal cells with fragmented or short mitochondrial form, which increased to 96.03 ± 1.23% in GCD2 corneal fibroblasts. Degradation of mitochondria by autophagy is a known process, and autophagy selectively degrades dysfunctional mitochondria.27 Mitochondrial fragmentation also occurs in response to apoptotic stimulation and this process requires fission.28,29 Dysfunctional mitochondria have been reported to accumulate in corneal fibroblasts isolated from GCD2 patients.26 These results indicated that accumulated mut-TGFBI and abnormal mitochondria in GCD2 corneal fibroblasts may be associated with defective autophagy, which could act as a protector or activate cell death processes such as apoptotic and/or autophagic cell death.
Figure 1. Morphological, histological and ultrastructural characterization of primary cultured GCD2 corneal fibroblasts. (A) Phase-contrast micrograph of WT and GCD2 HO corneal fibroblasts. Cells were analyzed and photographed using an inverted microscope. The GCD2 HO corneal fibroblasts showed a number of morphological abnormalities that are different from WT corneal fibroblasts, and the GCD2 cells contain different-sized intracellular deposits. The increased granularity within the cells was not seen in WT cells. (B) WT and GCD2 HO corneal fibroblasts were costained with anti-TGFBI and anti-CTSD antibodies. GCD2 homozygous corneal fibroblasts showed much more extensive pattern of colocalized mut-TGFBI with CTSD than observed in WT corneal fibroblasts. (C) Electron microscopy (EM) of WT and GCD corneal fibroblasts. Cells were fixed and processed for EM. Many abnormal mitochondria (short or fragmented) were observed in GCD2 cells compared with WT cells. Bars: 2000 nm. (D) Quantitation of mitochondrial numbers. Mitochondrial numbers were measured in six WT cells and six GCD2 cells. Results represent the mean ± SD *p < 0.05; **p < 0.01.
Mut-TGFBI accumulated intracellularly in GCD2 corneal fibroblasts
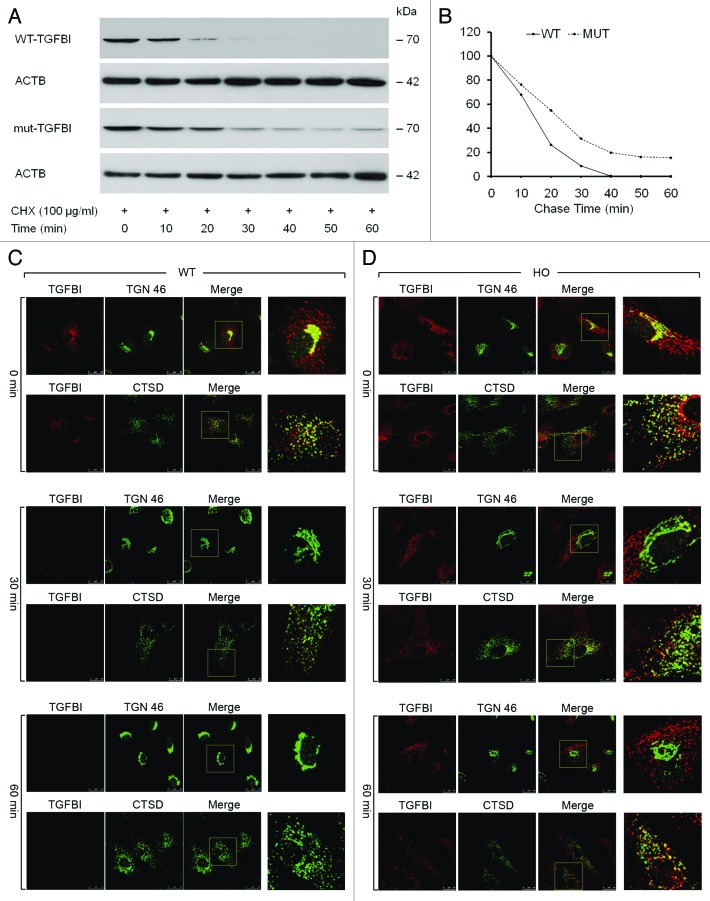
To measure the secretory status of intracellular TGFBI in WT and GCD2 HO corneal fibroblasts, cells were incubated in medium containing a protein synthesis inhibitor, cycloheximide (CHX, 100 µg/ml), for various time periods, and cell lysates were subjected to immunoblotting with anti-TGFBI antibody. As shown in Figure 2A and B, TGFBI was completely undetectable 40 min after CHX addition in WT corneal fibroblasts. In contrast, the level of mut-TGFBI remained constant in GCD2 HO corneal fibroblasts up to 60 min after CHX addition, despite the lack of ongoing protein synthesis (Fig. 2A and B). These results suggested that mut-TGFBI may accumulate in GCD2 corneal fibroblasts as a result of either impaired degradation or delayed extracellular secretion. To further investigate these possibilities, we examined the status of intracellular TGFBI using immunofluorescence analysis after exposure to CHX. The cells were again incubated for various time periods in medium containing CHX (100 µg/ml). WT and GCD2 HO corneal fibroblasts were stained with anti-TGFBI, anti-TGN 46 and anti-CTSD antibodies at various time intervals after CHX treatment. At 0 min after CHX treatment, both mut- and WT-TGFBI were found predominantly in the trans-Golgi network (TGN), as shown in Figure 2C and D. Thirty minutes after CHX treatment, WT-TGFBI could not be detected in the TGN, and only a small amount colocalized with CTSD in cytosolic vesicles (Fig. 2C). However, in GCD2 HO corneal fibroblasts, less mut-TGFBI was observed in the TGN after 30 min of CHX treatment (Fig. 2D). Instead, mut-TGFBI remained in cytosolic vesicles where it was colocalized with CTSD (Fig. 2D), and this colocalization persisted for up to 60 min (Fig. 2D). These cytosolic, CTSD-positive perinuclear vesicles could represent lysosomes or small bodies containing lysosomal enzymes (engulfed endosomes or autophagolysosomes). In summary, the massive colocalization of mut-TGFBI in vesicles containing lysosomal enzyme indicated that mut-TGFBI may be insufficiently degraded in GCD2 corneal fibroblasts.
Figure 2. Kinetics of TGFBI synthesis and secretion in WT and GCD2 corneal fibroblasts. (A) Corneal fibroblasts were incubated with 100 μg/ml CHX for the indicated time periods, and the cells were then harvested and subjected to immunoblot analysis using an anti-TGFBI antibody. WT-TGFBI was not detected in corneal fibroblasts after treatment for 50 min; in contrast, mut-TGFBI remained in corneal fibroblasts for up to 60 min after treatment. (B) Most TGFBI was secreted by WT corneal fibroblasts after 40 min. However, secretion of mut-TGFBI from GCD2 homozygous corneal fibroblasts was incomplete after 60 min. (C and D) mut-TGFBI accumulates intracellularly. WT and GCD2 HO corneal fibroblasts were incubated with 100 μg/ml CHX for the indicated times and then fixed and double stained with anti-TGFBI, anti-TGN 46 and anti-CTSD. Visualization was performed with FITC-conjugated goat anti-rat IgG for TGN 46 and CTSD and with rhodamine-conjugated goat anti-rabbit IgG for TGFBI. Images were obtained with a scanning laser confocal microscope separately for TGFBI, TGN 46 and CTSD. In the combined FITC and rhodamine images, the yellow color indicates overlap between the red and green fluorescent secondary antibodies. Both WT- and mut-TGFBI colocalized with the TGN and lysosomal compartments. (C) WT-TGFBI completely disappeared after 60 min, and it could not be observed inside the cell. (D) In contrast, mut-TGFBI was colocalized with lysosomes and was maintained even after 60 min.
TGFBI was degraded by autophagy
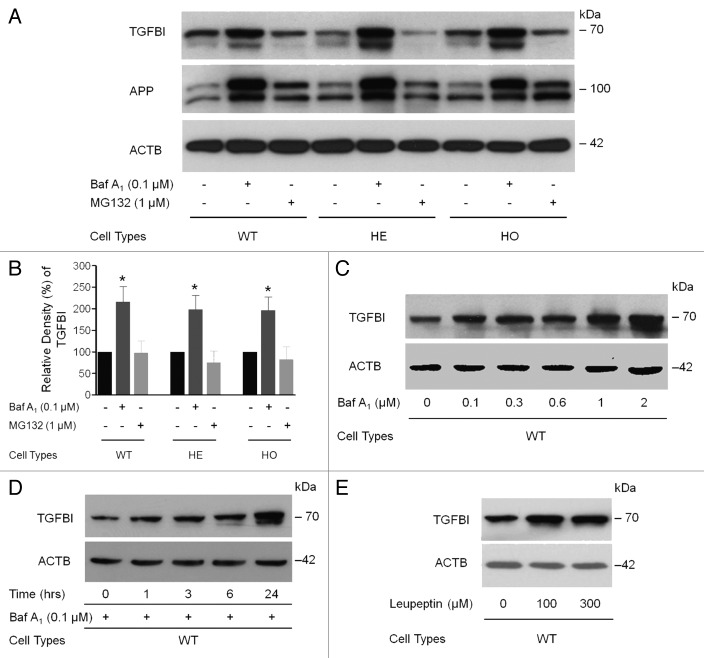
To determine if insufficient degradation of mut-TGFBI and the accumulation of abnormal mitochondria in GCD2 corneal fibroblasts were due to defects in any major degradation system, we focused on the two main cellular protein degradation pathways, the autophagy and the ubiquitin/proteasome pathways, by using specific inhibitors for each pathway. Corneal fibroblasts were incubated in complete medium for 12 h with or without 0.1 μM bafilomycin A1 an inhibitor of autophagy, and then TGFBI was detected using an anti-TGFBI antibody. Treatment with bafilomycin A1 significantly increased WT-TGFBI levels by 215.8 ± 35.5% relative to controls (Fig. 3A and B). TGFBI degradation was also inhibited in HE and HO corneal fibroblasts by 198.9 ± 75.7% and 196.8 ± 83.0%, respectively, vs. control levels (Fig. 3B). Furthermore, the effect of bafilomycin A1 on TGFBI levels was both dose-dependent (Fig. 3C) and time-dependent (Fig. 3D). We also analyzed the effects of MG132, a proteasome inhibitor, on the degradation of TGFBI and the amyloid β (A4) precursor protein (APP). Blockage of the ubiquitin/proteasome pathway by MG132 did not alter TGFBI levels (Fig. 3A and B). In further studies, WT corneal fibroblast cells were incubated with leupeptin, a moderate cell-permeable inhibitor of lysosomal cysteine proteases. Leupeptin treatment (100 or 200 µM) for 12 h increased the levels of intracellular TGFBI in WT (Fig. 3E), HE and HO cells (Fig. S1), demonstrating the importance of lysosomal enzyme activity. None of the inhibitors altered β-actin (ACTB) levels, indicating the specificity of this pathway for TGFBI. In addition, reverse transcription polymerase chain reaction (RT-PCR) analyses showed that these inhibitors did not interfere with TGFBI transcription as measured by mRNA expression (data not shown). These results demonstrated that autophagy was mainly responsible for TGFBI degradation.
Figure 3. TGFBI was degraded by autophagy. (A) The levels of intracellular TGFBI and APP [amyloid β (A4) precursor protein] in primary WT, HE and HO corneal fibroblasts were examined by immunoblot analysis following bafilomycin A1 (Baf A1) and MG132 treatment. At approximately 60–80% confluence, cells were treated with inhibitors for 12 h. (B) Quantification of the TGFBI levels in (A) from three independent experiments using image analysis software. Intracellular TGFBI levels were highly increased after Baf A1 (0.1 μM) treatment, whereas the levels were decreased by MG132 treatment (0.1 μM). (C) Primary corneal fibroblasts were incubated with Baf A1 (0.1, 0.3, 0.6, 1 or 2 μM) for 12 h and harvested. TGFBI levels were determined by immunoblot analysis with an anti-TGFBI antibody. The levels of intracellular TGFBI increased in a dose-dependent manner. (D) Primary corneal fibroblasts were incubated with Baf A1 (0.1 μM) for 1, 3, 6 and 12 h, and the TGFBI levels were determined by immunoblot as shown in (C). Intracellular TGFBI increased in a time-dependent manner. (E) Primary corneal fibroblasts were incubated with different doses of leupeptin (100 and 300 μg/ml) for 12 h and TGFBI levels were determined by immunoblot analysis with an anti-TGFBI antibody. Vertical bars: SD *p < 0.05; NS, not significant.
Autophagy markers were increased in cultured corneal fibroblasts from GCD2 patients
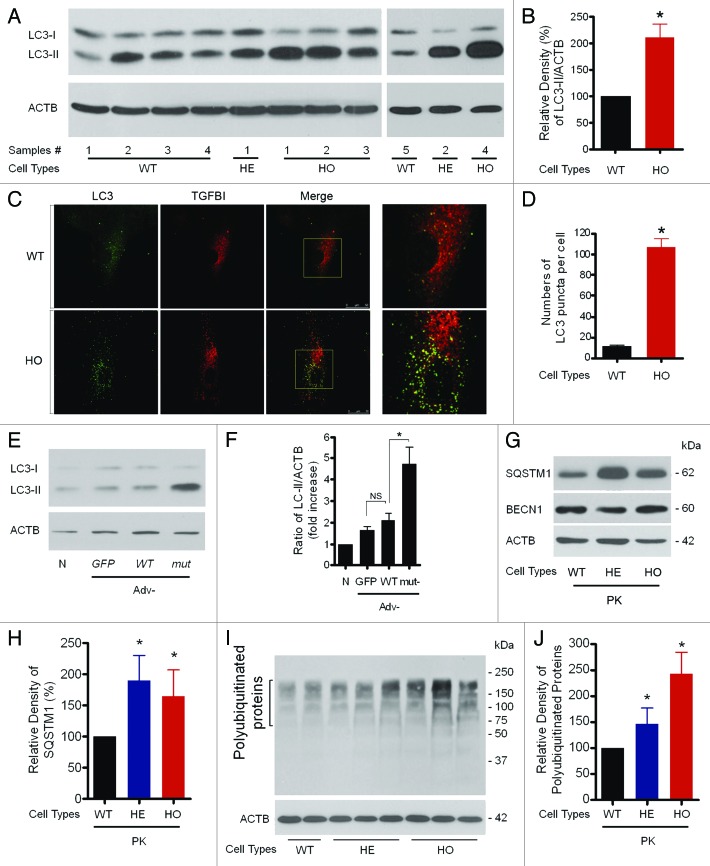
Next, we determined whether the massive colocalization of mut-TGFBI in vesicles containing lysosomal enzyme (Fig. 1B) or the presence of fragmented, abnormal mitochondria in GCD2 corneal fibroblasts (Fig. 1C and D) was involved in activation of, or defects in, the autophagic machinery. The autophagic state of both WT and GCD2 corneal fibroblasts were compared, using LC3 as a marker. As shown in Figure 4A, levels of LC3-II were significantly increased in GCD2 corneal fibroblasts under normal culture conditions. Quantification of three independent experiments showed an increase of approximately 2.1-fold (210.6 ± 53.32%) in the LC3-II/ACTB ratio in GCD2 corneal fibroblasts relative to WT controls (Fig. 4A and B). Because the levels of LC3-II correlate with the number of autophagosomes,30 the increased LC3-II/ACTB ratio may result from increased formation of autophagosomes. To test this, a detailed quantitative analysis was conducted by counting the number of endogenous LC3-labeled puncta in both WT and GCD2 HO corneal fibroblasts. As expected, GCD2 HO corneal fibroblasts contained numerous LC3-positive structures, whereas few LC3-labeled structures were detected in WT corneal fibroblasts. Quantitative analyses showed that the average number of LC3-reactive dots in WT corneal fibroblasts was 11.2 (SD ± 5.3, n = 10), whereas in GCD2 HO corneal fibroblasts, the average number of LC3 puncta increased to 84.2 (SD ± 27.2, n = 10) (Fig. 4C and D). Of note, most of the endogenous LC3 label colocalized with mut-TGFBI (Fig. 4C). To determine whether expression of exogenous mut-TGFBI could increase LC3-II levels in WT corneal fibroblasts, we assayed LC3 in WT corneal fibroblasts infected with either Adv-TGFBI and Adv-mut-TGFBI or the control Adv-GFP, by immunoblotting with an anti-LC3 antibody. LC3-II levels increased in WT fibroblasts infected with Adv-GFP (1.670 ± 0.154), Adv-WT-TGFBI (2.093 ± 0.333) and Adv-mut-TGFBI (4.739 ± 0.762) compared with noninfected cells (1.000). However, as shown in Figure 4E and F, LC3-II levels were increased more (about 2.2-fold) in WT corneal fibroblasts infected with Adv-mut-TGFBI than in cells infected with the Adv-WT-TGFBI (Fig. 4E and F).
Figure 4. Assessment of levels of autophagic vacuoles in GCD2 corneal fibroblasts. (A and B) At 60–80% confluence, cells were used for LC3 western blotting analysis. Representative LC3 blots and densitometric analysis of the LC3-II/ACTB ratio using samples from wild-type (WT), heterozygous (HE) and homozygous (HO) corneal fibroblasts (three independent experiments). (C) At 60–80% confluence, cells were used for LC3 staining. Representative pictures of WT and HO corneal fibroblasts expressing endogenous LC3. WT and homozygous corneal fibroblasts grown in complete media were fixed, permeabilized and immunostained with a monoclonal antibody to endogenous TGFBI (red) and a polyclonal antibody to endogenous LC3 (green). LC3 showed vesicular staining, and the number of LC3-positive puncta was higher in GCD2 HO corneal fibroblasts compared with control cells. Insets show a 3.5-fold magnification of the indicated region. A merged image of the red and green channels is shown in the third picture in each row; yellow indicates overlapping localization. (D) Endogenous LC3 puncta per cell were quantified from WT and GCD2 corneal fibroblasts. (E) Higher LC3-II levels in corneal fibroblasts overexpressing mut-TGFBI via adenovirus-mediated gene transfer. (F) Representative LC3 blots and densitometric analysis of the LC3-II/ACTB ratio in WT, Adv-GFP, Adv-WT-TGFBI and Adv-mut-TGFBI transfected corneal fibroblasts (three independent experiments). (G) SQSTM1 and BECN1 expression levels were analyzed using anti-SQSTM1 antibody and anti-BECN1 antibody in primary cultured corneal fibroblasts (PK) from patient corneas and age-matched WT corneas. (H) Levels of total polyubiquitinated proteins were analyzed using the anti-polyubiquitinated protein polyclonal antibody in primary cultured corneal fibroblasts from patient corneas and age-matched WT corneas. (I and J) Representative western blots of polyubiquitinated proteins and SQSTM1 protein were quantified from WT and GCD2 corneal fibroblasts. Polyubiquitinated proteins, SQSTM1 and BECN1, were normalized to ACTB. Error bars: SD from three independent experiments. *p < 0.05.
Increased levels of LC3-II and increased numbers of autophagosomes in GCD2 corneal fibroblasts can be due either to increased formation of autophagosomes or to inhibited degradation of autophagosomes. Defective autophagy causes accumulation of damaged mitochondria, polyubiquitin-containing protein aggregation and accumulation of SQSTM1 protein.31,32 Therefore, we measured levels of polyubiquitinated proteins and found that the levels were higher in GCD2 corneal fibroblasts compared with WT cells (Fig. 4H and J). Next, to examine whether TGFBI is ubiquitinated, we performed coimmunoprecipitation with anti-TGFBI antibody using lysate of WT and HO corneal fibroblasts treated with or without MG132, a ubiquitin proteasome inhibitor. The TGFBI immune complex was analyzed by western blot with the anti-ubiquitin antibody. When the lysates were immunoprecipitated with the anti-TGFBI antibody, ubiquitin was not detected in the TGFBI immune complex (Fig. S2). These results indicated that ubiquitination was not involved in targeting TGFBI to autophagosomes. Thus, TGFBI might be incorporated into autophagosomes in a ubiquitin-independent manner.
It is well known that SQSTM1 protein is involved in the targeting of polyubiquitinated proteins to autophagosomes and that SQSTM1 is selectively degraded via autophagy.33 Therefore, we measured the levels of SQSTM1 protein by western blotting, which showed an increase of SQSTM1 in GCD2 corneal fibroblasts (Fig. 4G and I). However, expression of BECN1 that is involved in the early stage of autophagosome formation34 was unchanged in GCD2 fibroblasts (Fig. 4G). The unchanged BECN1 level in GCD2 may explain the increased level of LC3-II in GCD2, and thus it may not be involved in increased autophagosome formation in GCD2 cells.
Fusion between autophagosomes and lysosomes was impaired in GCD2 corneal fibroblasts
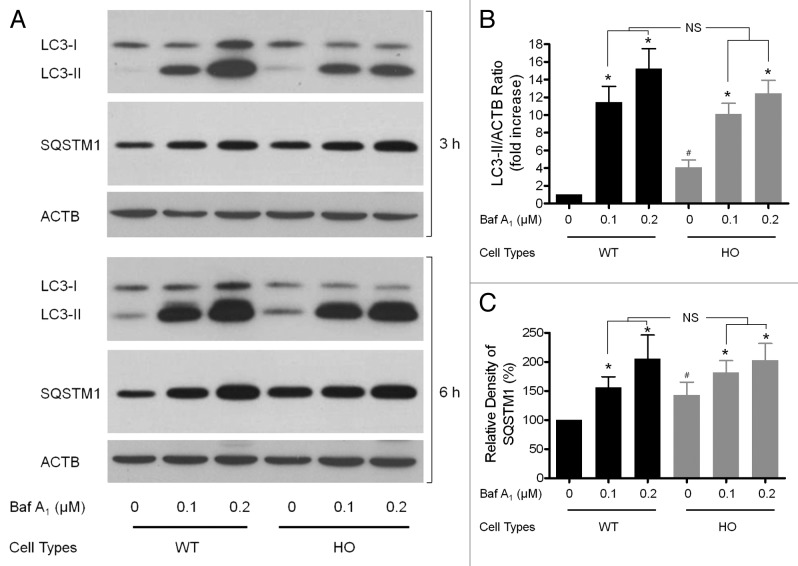
These findings raised the possibility that autophagy was defective in GCD2 corneal fibroblasts. Increased levels of LC3-II and number of autophagosomes in GCD2 corneal fibroblasts can be due either to increased formation of autophagosomes or to inhibited degradation of autophagosomes. To distinguish between these possibilities, we tested whether inhibition of fusion between autophagosomes and lysosomes could alter LC3-II levels in GCD2 corneal fibroblasts. If an inhibition of fusion between autophagosomes and lysosomes did not increase LC3-II levels in GCD2, compared with WT corneal fibroblasts, the results would suggest that there was already a block in autophagosome-lysosomal fusion.35Figure 5A and B show that bafilomycin A1 increased LC3-II levels in WT and GCD2 corneal fibroblasts. However, in GCD2 corneal fibroblasts, bafilomycin A1 did not increase LC3-II levels compared with WT corneal fibroblasts. Likewise, effects of bafilomycin A1 showed a similar trend in levels of SQSTM1 in both cell types (Fig. 5A and C). These results indicated that the increased LC3-II in GCD2 cells could be due to delayed autophagic flux.
Figure 5. Evaluation of LC3-II and SQSTM1 in the presence of Baf A1 after induction and/or inhibition of autophagy. (A) Corneal fibroblasts were cultured in the presence of Baf A1 (0.1–0.2 μM) for the indicated times and then subjected to immunoblot analysis using anti-LC3, anti-SQSTM1, anti-TGFBI and anti-ACTB antibody. (B and C) Quantification of the LC3-II, SQSTM1 and TGFBI levels in (A) from three independent experiments using image analysis software. Error bars: SD from three independent experiments. *p < 0.05. Baf A1 (0.1–0.2 μM) were added to the medium where indicated.
Suppression of autophagy increased susceptibility to cell death in GCD2
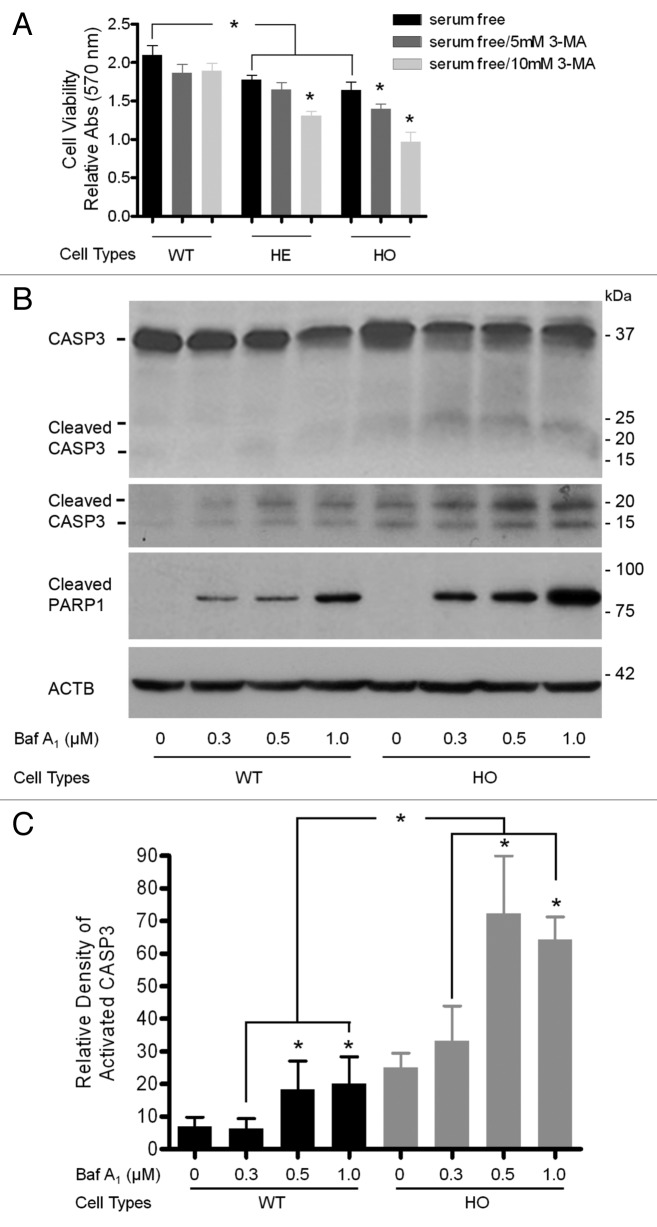
Autophagy can serve to protect cells,36,37 but may also paradoxically contribute to cell death.38,39 To examine whether defective autophagy promotes the survival or death of corneal fibroblasts, we assayed cell viability with or without 3-MA treatment for 12 h. In a dose-response manner, treatment with 3-MA resulted in a more significantly decreased cell proliferation of HE and HO corneal fibroblasts than of WT corneal fibroblasts (Fig. 6A). To further confirm the roles of defective autophagy in GCD2 corneal fibroblasts, we determined cell death by measuring caspase activation, because activation of caspases plays a central role in the execution of apoptosis. The cleavage of procaspase-3 (32 kDa) and the formation of corresponding active forms (17/19 kDa) were observed in both WT and GCD2 HO corneal fibroblasts when treated with bafilomycin A1 (ranging from 0.3 to 1.0 nM) for 36 h (Fig. 6B). However, different concentrations of bafilomycin A1 treatments revealed significantly more activated CASP3 and PARP1 in HO than in WT corneal fibroblasts (Fig. 6B). These results suggested that defective autophagy may contribute to cell death resulting from putative mut-TGFBI cytotoxicity.
Figure 6. Inhibition of autophagy led to reduced cell viability and CASP3 activation. At 60–80% confluence, cells were used for this study. (A) WT and GCD2 corneal fibroblasts were exposed to 3-MA for 14 h, and the number of viable cells was determined using an MTS assay. Cell viability was calculated by dividing the MTS OD value of an experimental sample by the MTS OD value of the control (cells treated with medium). (B) WT and GCD2 HO corneal fibroblasts treated with 0, 0.3, 0.5 or 1.0 μM bafilomycin A1 (Baf A1) for 12 h. Procaspase-3, cleaved CASP3, cleaved PARP1 and ACTB were determined by western blot. Cleaved CASP3 had two major fragments, migrating at 17 kDa and 19 kDa. Western blot analysis of extracts from WT and HO corneal fibroblasts, untreated or Baf A1-treated (36 h), showing CASP3 and PARP1 activation as evidence of induction of apoptosis. (C) Quantification of cleaved CASP3 levels in (B) from three independent experiments using image analysis software. Similar results were obtained from at least three independent experiments. Significantly more cell death occurred in GCD2 corneal fibroblasts relative to WT cells, indicating that GCD2 corneal fibroblasts were much more vulnerable to 3-MA‒treatment. Error bars: SD from three independent experiments. *p < 0.05.
Mut-TGFBI was cleared by autophagy inducer
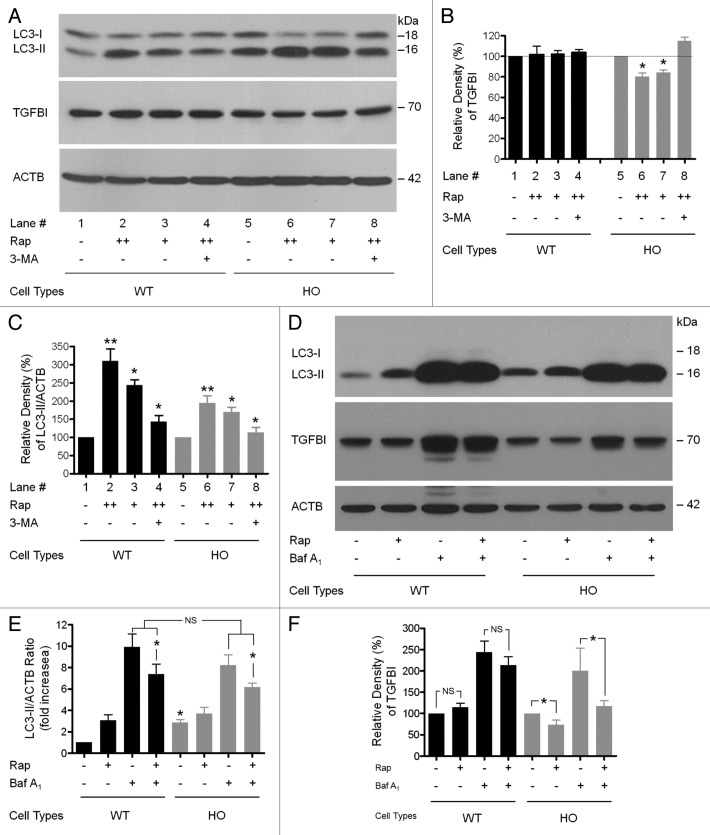
To test whether activation of autophagy could reduce accumulated mut-TGFBI in GCD2 corneal fibroblasts, both WT and GCD2 HO corneal fibroblasts were treated with either rapamycin (an autophagy activator) or 3-MA (an autophagy inhibitor), and the levels of TGFBI were examined by immunoblot analysis. Rapamycin treatment led to an increased ratio of LC3-II/ACTB in both WT and GCD2 HO corneal fibroblasts (Fig. 7A, lanes 2, 3, 6 and 7; Fig. 7B, lanes 2, 3, 6 and 7) and decreased levels (80.33 ± 5.85% with 200 nM treatment and 84.00 ± 4.58% with 100 nM treatment normalized to 100% without rapamycin) of mut-TGFBI in GCD2 HO corneal fibroblasts (Fig. 7A, lanes 6 and 7; Fig. 7C, lanes 6 and 7). However, rapamycin treatment did not alter the levels of WT-TGFBI in WT corneal fibroblasts (Fig. 7A, lanes 2 and 3; Fig. 7C, lanes 2 and 3). The addition of 3-MA to rapamycin-treated corneal fibroblasts abolished the effect of rapamycin on mut-TGFBI levels (Fig. 7A, lane 8 compared with lanes 6 and 7), but did not change WT-TGFBI levels (Fig. 7A, lane 4; Fig. 7C, lane 4). Together, our data indicated that the degradation of mut-TGFBI can be enhanced by activation of autophagy, but that mut-TGFBI may be incompletely degraded by autophagy under basal levels (nonrapamycin conditions) in GCD2 corneal fibroblasts.
Figure 7. Rapamycin-mediated activation of autophagy enhanced the clearance of Mut-TGFBI. At 60–80% confluence, cells were treated with inhibitors for 14 h. (A) Cells were treated with either rapamycin or DMSO, and extracts were probed with anti-TGFBI and anti-LC3 antibodies. Rapamycin (Rap) decreased the levels of mut-TGFBI in HO corneal fibroblasts. Representative blots are shown of samples from three independent experiments. (B) Autophagy was measured as the ratio of LC3-II/ACTB from immunoblots shown in (A) following normalizing of protein levels to ACTB using densitometry. (C) Quantification of the TGFBI levels in (A). Baf A1 (0.1–0.2 μM) and Rap (100 nM) were added to the medium where indicated. (D) WT and GCD2 HO corneal fibroblasts were treated with Rap (100 nM) and Baf A1 (0.1 μM) individually and in combination for 14 h. Cells were lysed and immunoblotted with anti-LC3 (first panel), anti-TGFBI (second panel) or anti-ACTB (third panel) antibodies. Autophagy was measured as the ratio of LC3-II/ACTB and is represented as the fold increase in this ratio relative to WT untreated corneal fibroblasts. (E) The ratio of LC3-II/ACTB shown in (D). (F) Quantification of the TGFBI levels shown in (D). Results represent the mean ± SD (n = 3). *p < 0.05; **p < 0.01; NS, not significant.
Autophagy inducers can further increase LC3-II levels in bafilomycin A1-treated cells.40,41 In this study, rapamycin treatment increased LC3-II in both WT and GCD2 corneal fibroblasts (Fig. 7D). However, rapamycin significantly decreased LC3-II levels in the presence of bafilomycin A1 in both cell types (Fig. 7D; Fig. S3), suggesting an increase in degradation of the autophagosome and/or induction of autophagosome-lysosome fusion. Furthermore, although WT-TGFBI did not significantly change with rapamycin-only treatment, levels of mut-TGFBI were significantly reduced by the treatments (Fig. 7D and F). These data indicated that rapamycin reduced accumulated autophagosomes by bafilomycin A1, but had additional effects on autophagy. Thus, the autophagy-inhibitory effect of bafilomycin A1 was rescued by rapamycin. Rapamycin treatments also eliminated the mut-TGFBI accumulated by blockage of autophagosome-lysosomal fusion. Notably, in the presence of rapamycin, mut-TGFBI was significantly decreased in GCD2 HO corneal fibroblasts compared with WT cells (Fig. 7D and F). Together, these findings provided the possibility for new therapeutic approaches for GCD2 based on pharmacological activation of autophagy. In addition, to assess whether increased LC3-II in GCD2 cells resulted from defective or protective autophagy, the changes of GCD2 cell death were examined after treatment with or without bafilomycin A1 and/or rapamycin. Cells were divided into four groups according to treatment: control (no treatment), bafilomycin A1 (treated with 0.1 μM bafilomycin A1), rapamycin (treated with 100 nM rapamycin) and combination (treated with both bafilomycin A1 and rapamycin). Levels of cell death were measured by fluorescence-activated cell sorting (FACS) analysis.22 Cell death was detected in both types of cells after bafilomycin A1 treatment, but levels of cell death were higher in HO GCD2 cells than WT cells (Fig. S4). Interestingly, rapamycin treatment only induced cell death of HO GCD2 cells. Furthermore, the combined treatment of bafilomycin A1 and rapamycin greatly increased cell death of HO GCD2 cells, but cell death of WT cells was only caused by treatment with bafilomycin A1 (Fig. S4). Thus, levels of cell death in WT cells showed no difference between cells treated with bafilomycin A1 alone and bafilomycin A1 with rapamycin (combination). Consequently, these results also indicated that increased LC3-II and autophagosomes may be due to defective autophagy, as opposed to being some type of protective mechanism in GCD2 corneal fibroblasts.
Discussion
In this study, we investigated the role of autophagy in degeneration of GCD2 corneal fibroblasts. Our results revealed striking differences in autophagy between GCD2 and WT corneal fibroblasts, which led us to conclude that autophagy is defective in this disease. Thus, defective autophagy in this disease causes dysfunction of corneal fibroblasts. This was demonstrated by the lack of autophagic clearance of mut-TGFBI, accumulation of dysfunctional mitochondria, increased ubiquitinated proteins and autophagosomes, and by increased levels of SQSTM1 and LC3-II in GCD corneal fibroblasts. Not only was accumulated mut-TGFBI toxic to GCD2 cells, but autophagy inducers were also capable of causing elimination of accumulated mut-TGFBI.
Our data demonstrated that autophagy was the main intracellular degradation mechanism for TGFBI, because TGFBI accumulated in cells treated with bafilomycin A1, an autophagy inhibitor. Furthermore, inhibitors of lysosomal cathepsins, such as leupeptin, substantially reduced the rate of TGFBI degradation. TGFBI is found within lysosomal compartments of corneal fibroblasts. Most notably, the levels of mut-TGFBI in lysosomes appeared to be much greater than the levels of WT-TGFBI. However, it remains to be determined which mechanisms are responsible for lysosomal targeting of TGFBI as a secretory protein. Most proteins are sorted in the TGN into different Golgi-derived secretory vesicles, which translocate their contents to different intracellular destinations. Some proteins processed in TGN are delivered to lysosomes, and others are delivered to secretory vesicles in a constitutive or regulated fashion.42 In this study, we found that both mut- and WT-TGFBI were localized to TGN, and later appeared in lysosomes, then in the cell media.
Translated TGFBI may be translocated into the endoplasmic reticulum via its signal peptide and then matured during passage through the Golgi. Therefore, TGFBI molecules may be transported to the ECM via conventional secretory vesicles. However, we cannot rule out the possibility that TGFBI might also be secreted via the unconventional secretory pathway, which is involved in autophagy and endocytosis. Although autophagy is considered as degradative machinery, it also participates in protein secretion. For example, the autophagy-based unconventional secretory route for extracellular delivery of interleukin-1 β (IL1B) has recently been reported.43-45 Here, we proposed the possibility that accumulated mut-TGFBI in GCD2 cells might be secreted into the ECM via the unconventional secretory pathway. TGFBI may undergo reinternalization through either the formation of clathrin-coated vesicles or caveolae-mediated endocytosis and then be selectively sorted into multivesicular bodies (MVBs), which accumulate small membrane vesicles termed exosomes. Exosomes, small 40- to 100-nm cup-shaped vesicles surrounded by a lipid bilayer,43 are generated as intraluminal vesicles within MVBs. Fusion of MVBs with the plasma membrane results in the release of exosomes into the ECM.43 Furthermore, MVBs can also fuse with autophagosomes for degradation. Autophagy induction diverts MVBs toward the autophagic pathway with the consequent inhibition of exosome release and leads to increased associations between MVBs and autophagosomes, resulting in the generation of big, enlarged organelles (amphisomes). These studies suggested that MVBs, which contain mut-TGFBI, might be accumulated due to decreased or incomplete fusion between MVBs and either autophagosomes or lysosomes in GCD2 corneal fibroblasts. In addition, exosome-mediated secretion of aggregation-prone proteins such as β-amyloid, α-synuclein and prion protein,44-46 also reveals the possibility that TGFBI might also be secreted and ultimately deposited in the corneal matrix via this mechanism. However, whether or not extracellular deposition of mut-TGFBI occurs through exocytosis in corneas of GCD2 patients remains to be investigated.
After inhibition of protein synthesis by CHX, mut-TGFBI remained inside the GCD2 corneal fibroblasts. Immunofluorescence staining of GCD2 corneal fibroblasts also showed that mut-TGFBI was primarily retained in lysosomes and persisted for long periods. It is likely that trafficking defects, problems in autophagy, and/or impaired lysosomal functions were responsible for the incomplete, slower degradation of mut-TGFBI. Among several possibilities, we determined that defective autophagy could be involved in accumulated mut-TGFBI in GCD2 corneal fibroblasts. This idea was subsequently confirmed by the following experimental data. First, although the levels of LC3-II and SQSTM1 were more increased in GCD2 corneal fibroblasts compared with WT corneal fibroblasts, no differences in LC3-II and SQSTM1 levels were found between GCD2 and WT cells after blocking autophagosome-lysosome fusion. Second, the number of endogenous LC3 puncta was significantly increased in GCD2 corneal fibroblasts and these puncta colocalized with mut-TGFBI. Third, levels of LC3-II and SQSTM1 were significantly increased in GCD2 compared with WT corneal fibroblasts. Finally, GCD2 corneal fibroblasts contained dysfunctional mitochondria (short or fragmented mitochondrial shapes) and polyubiquitinated proteins. It is well known, in contrast to the ubiquitin/proteasome system, that the autophagy/lysosome system not only degrades proteins, but also intracellular organelles such as mitochondria.47-49 In particular, damaged mitochondria are preferentially targeted for autophagy. In addition, accumulation of ubiquitinated proteins has been observed in a defective autophagy model and likely results from increased polyubiquitinated substrates with longer half-lives.50 Therefore, these results also indicated that autophagy was defective in GCD2 corneal fibroblasts.
However, the above results did not resolve whether defective autophagy in GCD2 corneal fibroblasts is a mechanism for degeneration of GCD2 corneal fibroblasts. This question is particularly important, as defective or excessive activation of autophagy is also associated with cell death that can occur either in the absence of detectable signs of apoptosis or concomitantly with apoptosis. Furthermore, according to several studies, lysosomal accumulation of undegraded substrates results in impaired autophagosome-lysosome fusion. As a consequence of this block, toxic proteins and dysfunctional mitochondria accumulate, ultimately leading to cell death.51 It is likely that fragmented mitochondria may release proapoptotic factors such as cytochrome c, which can activate the intrinsic apoptotic pathway.52,53 Thus, GCD2 corneal fibroblasts with impaired autophagy may have an increased susceptibility to cell death. Our data also supported this idea. Treatment of cells with a well-known inhibitor of the initiation stage of autophagy (3-MA)54 significantly reduced cell viability of GCD2 corneal fibroblasts compared with WT corneal fibroblasts. In addition, blocking autophagosome-lysosome fusion with bafilomycin A1, an end stage inhibitor of autophagy, activated CASP3 and PARP1 in GCD2 corneal fibroblasts more than in WT corneal fibroblasts. This disease specifically causes severe corneal fibroblast degeneration compared with other TGFBI-expressing cell types such as skin fibroblasts and epithelial cells. Thus, corneal fibroblasts may be more susceptible to defective autophagy than other cell types. This may be because other cells undergo more rapid division than corneal fibroblasts, which may be helpful in preventing the accumulation of misfolded or aggregated proteins by cell division-mediated dilution. Therefore, corneal fibroblasts may be more susceptible to cell death from defective autophagy because they are quiescent, remain in resting phase (G0), and have a longer life span.55 This may explain the prevalence of corneal fibroblast degeneration in GCD2.
Further points to be emphasized from this study were not only that autophagy was defective in GCD2, but also that it could be cytotoxic to corneal fibroblasts. Autophagy inducers are indeed capable of providing therapeutic treatment options for GCD2. Induction of autophagy has also been reported for therapeutic treatments in age-related diseases.56 In one study, it is proposed that aggregation‐prone proteins could be cleared by induction of autophagy.57 Thus, the autophagy inducer, rapamycin, has been used to modify Huntington’s disease pathogenesis. Huntington’s disease is caused by an abnormal polyglutamine expansion (polyQ) repeat in the disease protein huntingtin, and is characterized by inclusion bodies of aggregated huntingtin protein and by neuronal cell death. In cultured cells and in Drosophila and mouse models, rapamycin reduces levels of mutant huntingtin and attenuates toxicity through activation of autophagy.58 Berger et al. report that rapamycin reduces both the toxicity of mutant microtubule-associated protein tau in fronto-temporal dementia/tauopathy and also the polyglutamine-containing neurodegenerative protein ATXN3 in a Drosophila disease model.59 We also observed a reduction in the level of mut-TGFBI in rapamycin-treated GCD2 corneal fibroblasts. However, no change in TGFBI levels was detected in WT corneal fibroblasts. These results indicated that treatment with rapamycin had a significant effect only in cells with defective autophagy. The above results strongly suggested that the activation of autophagy is important for the clearance of accumulated mut-TGFBI in GCD2 patients. Taken together, these data revealed that the therapeutic potential of MTOR inhibition deserves important consideration. Furthermore, because the mechanism of action of rapamycin is known and rapamycin is already used clinically,60 this drug or related analogs may be suitable candidates for therapeutic use in GCD2.
The molecular mechanisms underlying the defective autophagy in GCD2 corneal fibroblasts are still unknown. Wen et al.,61 have reported that PI3K/AKT/MTOR signaling is activated in TGFBI-deficient cell lines. Furthermore, it has been demonstrated that constitutive activation of the MTOR signaling pathway induces defective autophagy.62 We therefore suggest that, although TGFBI mutations associated with GCD2 pathogenesis are still not identified, these point mutations could result in either a loss of function or gain of function. Loss of function of TGFBI may induce defective autophagy through activation of the PI3K/AKT/MTOR signaling pathway.
In conclusion, we propose that the lack of autophagic clearance of proteins such as mut-TGFBI and of organelles caused by impairment of the autophagy/lysosomal degradation pathway, may contribute to GCD2 pathogenesis. Although the molecular mechanisms leading to the defective autophagy are still not known, our studies suggested that activation of autophagy may be a therapeutic strategy for patients with GCD2.
Materials and Methods
Materials
The following compounds were obtained from Sigma-Aldrich: Proteasomal inhibitor MG132 (C2211), lysosomal inhibitor bafilomycin A1 (B1793), leupeptin (L2884), rapamycin (R117), cycloheximide (CHX) (C4859), dimethylsulfoxide (DMSO) (D8418) and 3-methyladenine (3-MA) (M9281). The SuperSignal West Pico Chemiluminescent Substrate was obtained from Pierce (Pierce, NC14080KR).
Isolation and culture of primary corneal fibroblasts
WT (n = 5), HE (n = 3) and HO (n = 4) primary corneal fibroblasts were prepared using previously described methods.22 The age, gender and diagnosis for all GCD2 patients from which cells were generated for this study are listed in Table 1. Donor confidentiality was maintained in accordance with the Declaration of Helsinki and was approved by the Severance Hospital IRB Committee (CR04124), Yonsei University, Seoul, Korea. GCD2 was diagnosed by DNA sequence analysis for TGFBI mutations. After removal of the corneal button for penetrating keratoplasty, the remaining corneal rims were harvested for WT corneal fibroblast culture. The medical records of the control donors from the eye bank of Yonsei University Severance Hospital did not show any history of genetic or systemic metabolic disease. The fibroblasts grown from the pieces of corneal rims were treated as WT controls, and DNA sequencing analysis was performed to verify genetic normality of TGFBI. Table 1 presents information regarding the corneal fibroblasts used in this study.
Table 1. Subject data for GCD2 and wild-type samples.
| Wild type | Heterozygous | Homozygous | |||
|---|---|---|---|---|---|
|
Sex/Age |
Mean Age |
Sex/Age |
Mean Age |
Sex Age |
Mean Age |
| F/20 |
|
F/37 |
|
F/13 |
|
| M/10 |
|
F/20 |
|
M/6 |
|
| M/45 |
26.4 |
F/49 |
33 |
F/27 |
20.5 |
| M/22 |
|
|
|
M/36 |
|
| M/35 | |||||
F, female; M, male
Transmission electron microscopic examination of corneal fibroblasts
Primary cultured corneal fibroblasts were fixed overnight, dehydrated and processed for electron microscopy as described previously.26 Evaluation was performed using a transmission electron microscope (JEM1200 EX2; JEOL Ltd.).
Treatment of cells with 3-MA, bafilomycin A1, leupeptin and rapamycin
The autophagy inhibitor 3-MA was freshly dissolved in culture medium at 37°C for 1 h before use. To analyze the effects of 3-MA on TGFBI accumulation, corneal fibroblasts were incubated with 3-MA and analyzed for TGFBI levels by immunoblot analysis. Bafilomycin A1, which inhibits fusion between autophagosomes and lysosomes by blocking the vacuolar adenosine triphosphate (ATP) pump, was used at 100 nM. Leupeptin, which inhibits cathepsins, was dissolved in dimethylsulfoxide (DMSO). Rapamycin was dissolved in DMSO immediately before use and added to the culture medium at concentrations of 100–200 nM.
Western blot analysis
Cell lysates from corneal fibroblasts were prepared using a radioimmunoprecipitation assay buffer (RIPA buffer; 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM TRIS-HCl, pH 7.4) containing protease inhibitors (Complete Mini Protease Inhibitor Cocktail Tablet, 1836170) and phosphatase inhibitors (PhosSTOP, Roche, 04906845001). Crude cell lysates were centrifuged at 10,000 × g for 10 min at 4°C to remove nuclear fragments and tissue debris. A portion of the supernatant was used to determine the total protein concentration using a bicinchoninic acid kit (Pierce, 23228). Total cellular protein was electrophoresed on TRIS-glycine SDS polyacrylamide gels. Proteins were transferred onto polyvinylidene difluoride membranes (Millipore Corp., IPVH00010) and then blocked in 5% nonfat dry milk (Santa Cruz, sc-2325) in TRIS-buffered saline containing Tween-20 (TBS-T) [20 mM TRIS (pH 7.5), 150 mM NaCl, 0.1% Tween 20] at room temperature for 1 h. After washing three times with TBS-T, blots were incubated with primary antibodies to TGFBI (0.2 µg/ml; R&D Systems, AF2935), ubiquitin (1:100 dilution; Sigma-Aldrich, U5379), ACTB (1:5000 dilution; Sigma-Aldrich, A-5441) and, LC3 (Cell Signaling Technology, 3868), BECN1 (Cell Signaling Technology, 3738), CASP3 (Cell Signaling Technology, 9662), cleaved CASP3 (Cell Signaling Technology, 9661), cleaved PARP1 (Cell Signaling Technology, 5625) (1:1000 dilution each) and SQSTM1 antibody (1:1000 dilution; BD Transduction Laboratories, 610833) overnight at 4°C. After three washes with TBS-T, blots were incubated with secondary antibodies conjugated to horseradish peroxidase at room temperature for 1 h. Horseradish peroxidase-linked anti-mouse IgG (1:5000 dilution; Amersham Pharmacia Biotechnology, NA931V) or anti-rabbit IgG (1:5000 dilution; Amersham Pharmacia Biotechnology, NA934V) was used as a secondary antibody. Western blots were visualized using the enhanced chemiluminescence system (Pierce, NCI4080KR). Immunoreactive protein bands were scanned at two intensities, and the optical densities of the bands were quantified using ImageJ software, version 1.37 (Wayne Rasband, National Institutes of Health), corrected by background subtraction, and normalized to the intensity of the corresponding ACTB protein bands.
Kinetics of TGFBI secretion
WT and GCD2 HO corneal fibroblasts were grown to 70–80% confluency on culture slides (BD Falcon Labware, REF 354108) and then were incubated for various time periods (0, 30 or 60 min) with 100 µg/ml CHX. Cells were then processed for immunocytochemical staining.
Immunocytochemical staining
WT and HO corneal fibroblasts grown on culture slides (BD Falcon Labware, REF 354108) were permeabilized and fixed in methanol at −20°C for 3 min. Cells were washed with phosphate-buffered saline (PBS), blocked with 10% bovine serum albumin (Sigma-Aldrich, A3311) with PBS for 10 min, and incubated with primary antibody in blocking buffer for 1 h at room temperature (RT). Cells were hybridized with secondary antibodies for 1 h at RT. The coverslips were mounted on glass slides using Vectashield mounting medium (Vector Labs Inc., H-1200). Cells were viewed under a Leica TCS SP5 confocal microscope (Leica, Microsystems CMS GmbH, Germany). The following primary antibodies were used: monoclonal anti-TGFBI (1:300 dilution; kindly provided by Dr. IS, Kim, Kyungpook National University, Korea), polyclonal anti-trans golgi network (TGN) 46 (5 μg/ml; Abcam, ab50595), anti-CTSD rabbit polyclonal antibody (5 μg/ml; Calbiochem, IM16) and anti-LC3 (1:100 dilution; Cell Signaling Technology, 3868). The following secondary antibodies were used: Alexa 594 (red)-conjugated anti-rabbit IgG (1:200 dilution; Vector Laboratories Inc., DI-1488) and fluorescein isothiocyanate (green)-labeled anti-mouse IgG (1:200 dilution; Jackson ImmunoResearch Laboratories, 315-097-003).
Adenoviral vectors
Adenoviruses expressing green fluorescent protein (GFP) or WT- and mut-TGFBI were generated using the AdEasy system (developed by Seolin Bioscience Co., Ltd.). The open reading frame of TGFBI was subcloned into a transfer vector. The resultant plasmids were linearized with Pme I and cotransformed with the pAdEasy into Escherichia coli BJ5183 cells. Recombination was confirmed by restriction analysis and sequencing. High titer stocks were generated by transfecting linearized recombinant plasmid with Pac I into 293 cells. The effective titer was determined by the frequency of GFP-positive 293 cells 30 h after infection. For infection, primary corneal fibroblasts were plated in 30 mm 6-well dish, and approximately 1–3 × 107 infectious units per dish were added. Cells were infected in the presence of polybrene (4 μg/ml) (Sigma-Aldrich, 107689).
Cell viability
Corneal fibroblasts were plated in 96-well plates at a density of 10,000 cells/well and incubated overnight. Cell proliferation was determined using the CellTiter 96 Aqueous One Solution Reagent Cell Proliferation Assay (MTS assay) (Promega, G3580). Briefly, the culture medium was removed, and 20 µl MTS solution was added to each well in 100 μl culture medium. Cultures were incubated at 37°C for 2 to 4 h under 95% humidity and 5% CO2. Optical density was measured at 490 nm using a microplate reader (VERSAmax, Molecular Devices).
Statistical analysis
The results were statistically evaluated for significance (p < 0.05) using one-way ANOVA. Results are expressed as the mean ± SD. All data were processed using GraphPad Prism software version 4.0 (GraphPad Software Inc.).
Supplementary Material
Acknowledgments
This work was supported by the Converging Research Center Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2012K001354) and by a Midcareer Researcher Program grant through the NRF funded by the MEST (No. 2011-0028699).
Glossary
Abbreviations:
- GCD2
granular corneal dystrophy type 2
- TGFBI
transforming growth factor β-induced protein
- mut-TGFBI
mutant TGFBI
- LC3/MAP1LC3B
microtubule-associated protein 1 light chain 3β
- WT
wild type
- HE
heterozygous
- HO
homozygous
- PARP1
poly (ADP-ribose) polymerase 1
- MTOR
mechanistic target of rapamycin
- R124H
arginine-to-histidine substitution at codon 124
- TGFBI
transforming growth factor β-induced gene
- ECM
extracellular matrix
- ROS
reactive oxygen species
- CHX
cycloheximide
- TGN
trans-Golgi network
- APP
amyloid β (A4) precursor protein
- Adv
adenoviral vectors
- GFP
green fluorescent protein
- FACS
fluorescence-activated cell sorting
- MVBs
multivesicular bodies
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/22067
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/22067
References
- 1.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Finkbeiner S, Cuervo AM, Morimoto RI, Muchowski PJ. Disease-modifying pathways in neurodegeneration. J Neurosci. 2006;26:10349–57. doi: 10.1523/JNEUROSCI.3829-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward WF. Protein degradation in the aging organism. Prog Mol Subcell Biol. 2002;29:35–42. doi: 10.1007/978-3-642-56373-7_3. [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Vicente M, Sovak G, Cuervo AM. Protein degradation and aging. Exp Gerontol. 2005;40:622–33. doi: 10.1016/j.exger.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim Biophys Acta. 2009;1792:3–13. doi: 10.1016/j.bbadis.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–23. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munier FL, Korvatska E, Djemaï A, Le Paslier D, Zografos L, Pescia G, et al. Kerato-epithelin mutations in four 5q31-linked corneal dystrophies. Nat Genet. 1997;15:247–51. doi: 10.1038/ng0397-247. [DOI] [PubMed] [Google Scholar]
- 9.Skonier J, Neubauer M, Madisen L, Bennett K, Plowman GD, Purchio AF. cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA Cell Biol. 1992;11:511–22. doi: 10.1089/dna.1992.11.511. [DOI] [PubMed] [Google Scholar]
- 10.LeBaron RG, Bezverkov KI, Zimber MP, Pavelec R, Skonier J, Purchio AF. Beta IG-H3, a novel secretory protein inducible by transforming growth factor-beta, is present in normal skin and promotes the adhesion and spreading of dermal fibroblasts in vitro. J Invest Dermatol. 1995;104:844–9. doi: 10.1111/1523-1747.ep12607024. [DOI] [PubMed] [Google Scholar]
- 11.Billings PC, Herrick DJ, Kucich U, Engelsberg BN, Abrams WR, Macarak EJ, et al. Extracellular matrix and nuclear localization of beta ig-h3 in human bladder smooth muscle and fibroblast cells. J Cell Biochem. 2000;79:261–73. doi: 10.1002/1097-4644(20001101)79:2<261::AID-JCB90>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 12.Kim JE, Jeong HW, Nam JO, Lee BH, Choi JY, Park RW, et al. Identification of motifs in the fasciclin domains of the transforming growth factor-beta-induced matrix protein betaig-h3 that interact with the alphavbeta5 integrin. J Biol Chem. 2002;277:46159–65. doi: 10.1074/jbc.M207055200. [DOI] [PubMed] [Google Scholar]
- 13.Klintworth GK. Advances in the molecular genetics of corneal dystrophies. Am J Ophthalmol. 1999;128:747–54. doi: 10.1016/S0002-9394(99)00358-X. [DOI] [PubMed] [Google Scholar]
- 14.Korvatska E, Henry H, Mashima Y, Yamada M, Bachmann C, Munier FL, et al. Amyloid and non-amyloid forms of 5q31-linked corneal dystrophy resulting from kerato-epithelin mutations at Arg-124 are associated with abnormal turnover of the protein. J Biol Chem. 2000;275:11465–9. doi: 10.1074/jbc.275.15.11465. [DOI] [PubMed] [Google Scholar]
- 15.Okada M, Yamamoto S, Inoue Y, Watanabe H, Maeda N, Shimomura Y, et al. Severe corneal dystrophy phenotype caused by homozygous R124H keratoepithelin mutations. Invest Ophthalmol Vis Sci. 1998;39:1947–53. [PubMed] [Google Scholar]
- 16.Cintron C, Schneider H, Kublin C. Corneal scar formation. Exp Eye Res. 1973;17:251–9. doi: 10.1016/0014-4835(73)90176-0. [DOI] [PubMed] [Google Scholar]
- 17.Cintron C, Kublin CL. Regeneration of corneal tissue. Dev Biol. 1977;61:346–57. doi: 10.1016/0012-1606(77)90304-9. [DOI] [PubMed] [Google Scholar]
- 18.Jester JV, Moller-Pedersen T, Huang JY, Sax CM, Kays WT, Cavangh HD, et al. The cellular basis of corneal transparency: evidence for ‘corneal crystallins’. J Cell Sci. 1999;112:613–22. doi: 10.1242/jcs.112.5.613. [DOI] [PubMed] [Google Scholar]
- 19.Qazi Y, Wong G, Monson B, Stringham J, Ambati BK. Corneal transparency: genesis, maintenance and dysfunction. Brain Res Bull. 2010;81:198–210. doi: 10.1016/j.brainresbull.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akhtar S, Meek KM, Ridgway AE, Bonshek RE, Bron AJ. Deposits and proteoglycan changes in primary and recurrent granular dystrophy of the cornea. Arch Ophthalmol. 1999;117:310–21. doi: 10.1001/archopht.117.3.310. [DOI] [PubMed] [Google Scholar]
- 21.Kim BY, Olzmann JA, Choi SI, Ahn SY, Kim TI, Cho HS, et al. Corneal dystrophy-associated R124H mutation disrupts TGFBI interaction with Periostin and causes mislocalization to the lysosome. J Biol Chem. 2009;284:19580–91. doi: 10.1074/jbc.M109.013607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi SI, Kim TI, Kim KS, Kim BY, Ahn SY, Cho HJ, et al. Decreased catalase expression and increased susceptibility to oxidative stress in primary cultured corneal fibroblasts from patients with granular corneal dystrophy type II. Am J Pathol. 2009;175:248–61. doi: 10.2353/ajpath.2009.081001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–7. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–60. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–7. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim TI, Kim H, Lee DJ, Choi SI, Kang SW, Kim EK. Altered mitochondrial function in type 2 granular corneal dystrophy. Am J Pathol. 2011;179:684–92. doi: 10.1016/j.ajpath.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–53. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–25. doi: 10.1016/S1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 29.Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, et al. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002;159:931–8. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 32.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 33.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–5. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 36.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue L, Fletcher GC, Tolkovsky AM. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol Cell Neurosci. 1999;14:180–98. doi: 10.1006/mcne.1999.0780. [DOI] [PubMed] [Google Scholar]
- 39.Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–93. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3:331–8. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarkar S, Davies JE, Huang ZB, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–52. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 42.Blázquez M, Shennan KI. Basic mechanisms of secretion: sorting into the regulated secretory pathway. Biochem Cell Biol. 2000;78:181–91. doi: 10.1139/o00-010. [DOI] [PubMed] [Google Scholar]
- 43.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, et al. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A. 2006;103:11172–7. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, et al. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30:6838–51. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, et al. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004;101:9683–8. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kissová I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–74. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–11. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–29. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 51.Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–40. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Settembre C, Annunziata I, Spampanato C, Zarcone D, Cobellis G, Nusco E, et al. Systemic inflammation and neurodegeneration in a mouse model of multiple sulfatase deficiency. Proc Natl Acad Sci U S A. 2007;104:4506–11. doi: 10.1073/pnas.0700382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ravikumar B, Berger Z, Vacher C, O’Kane CJ, Rubinsztein DC. Rapamycin pre-treatment protects against apoptosis. Hum Mol Genet. 2006;15:1209–16. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 54.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 55.Francesconi CM, Hutcheon AE, Chung EH, Dalbone AC, Joyce NC, Zieske JD. Expression patterns of retinoblastoma and E2F family proteins during corneal development. Invest Ophthalmol Vis Sci. 2000;41:1054–62. [PubMed] [Google Scholar]
- 56.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 57.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 58.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 59.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–42. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 60.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 61.Wen GY, Hong M, Li BY, Liao WP, Cheng SK, Hu BR, et al. Transforming growth factor-β-induced protein (TGFBI) suppresses mesothelioma progression through the Akt/mTOR pathway. Int J Oncol. 2011;39:1001–9. doi: 10.3892/ijo.2011.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ng S, Wu YT, Chen B, Zhou J, Shen HM. Impaired autophagy due to constitutive mTOR activation sensitizes TSC2-null cells to cell death under stress. Autophagy. 2011;7:1173–86. doi: 10.4161/auto.7.10.16681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.