Abstract
Autophagy is a vital process through which cellular material and dysfunctional organelles are degraded and recycled, and it is inhibited by the metabolic checkpoint kinase MTOR. Autophagy also targets intracellular bacteria (a process termed xenophagy) for lysosomal degradation, thereby playing a key role in innate immunity. In the past few years, the identification of molecules, such as CALCOCO2/NDP52, SQSTM1/p62 and ubiquitin, implicated in the specific targeting of intracellular bacteria, received considerable attention. However, it remains unclear how xenophagy is initiated, since this process commonly occurs in metabolically replete cells. In a recent study, we demonstrated that infection with Shigella and Salmonella triggered an early state of intracellular amino acid (AA) starvation causing MTOR dissociation from endomembranes, downregulation of MTOR activity and activation of the EIF2AK4/GCN2-EIF2S1/eIF2α/ATF3 signaling axis. We also observed that AA starvation was caused by host membrane damage, which appeared to be transient in the case of Salmonella and sustained in Shigella-infected cells, thus highlighting the existence of key timing disparities in xenophagy triggering, depending on the bacterial pathogen. Together, our findings demonstrate that xenophagy is only one arm of a more general metabolic switch geared toward AA starvation in bacteria-infected cells.
Keywords: amino acid starvation, mTOR, Salmonella, Shigella, bacteria
Macroautophagy (hereafter termed autophagy) is a central metabolic process, conserved in all eukaryotic cells, which mediates the targeting, lysosomal degradation and recycling of cellular contents, such as damaged organelles or large molecular complexes. Autophagy regulation is tightly associated with the metabolic status of cells, allowing this process to provide a transient source of recycled nutrients during starvation. At the molecular level, the kinase target of rapamycin (TOR; MTOR in mammals) serves as a central metabolic sensor, and MTOR inhibition by rapamycin provides a starvation signal that is sufficient to turn on autophagy.
The autophagy system was recently found to target intracellular bacteria, thus providing an efficient cell-autonomous mechanism to target and send these pathogens to lysosomes for degradation. Although elegant, this bacterial killing mechanism remains puzzling, in particular because its ignition mechanism implies the existence of critical and mainly uncharacterized links between innate immunity and metabolic regulation. Another conundrum in the use of autophagy as an intrinsic microbial killing system relies on the fact that this mechanism must display some level of specificity toward bacterial targeting, in order to avoid that cells undergoing xenophagy would die from excessive “self-eating.” This issue of the specificity in xenophagy targeting has been a central question in the past few years, and an ever-expanding arsenal of host proteins appears to contribute to the specific targeting of intracellular bacteria in infected cells. These molecules include ubiquitinated proteins, CALCOCO2, SQSTM1, OPTN/optineurin, LGALS8/galectin 8, SEPT/septin and TECPR1. Although important to address the issue of bacterial targeting, these studies have to assume the existence of an ongoing “autophagic potential” that would allow rapid formation of autophagic structures following specific detection of the intruders. Since bacterial infection typically occurs in metabolically replete cells (at least in most experimental systems used to study xenophagy), this suggests that infection likely changes the metabolic status of cells, thus affecting the triggering and (as discussed below) the timing of xenophagy induction.
In epithelial cells infected with Shigella flexneri, a human enteric pathogen that invades host cells and rapidly gets access to the cytosol, we first noticed that MTOR signaling (in particular the levels of EIF4EBP1/4EBP1 and RPS6KB1/S6K1 phosphorylation) is significantly blunted, and immunofluorescence analysis revealed that this inhibition correlates with the dissociation of MTOR from host LAMP2+ late endosome/lysosome (LE/Ly) membranes. Because recent evidence has demonstrated that AA starvation, and not the lack of other nutrients, specifically provokes dispersion of MTOR complex 1 (MTORC1) from LE/Ly, we speculated that Shigella-induced MTOR inhibition is caused by a state of host AA starvation. Direct measurement of l-leucine/l-isoleucine pools in Shigella-infected cells confirmed this hypothesis (Fig. 1). Moreover, we noticed that Shigella infection also triggers a second crucial host pathway that depends on AA starvation, the EIF2AK4→EIF2S1→ATF3 integrated stress signaling cascade. In agreement, Shigella infection triggered the accumulation of EIF2S1-dependent mRNA stress granules, and microarray analyses revealed the existence of a critical ATF3-driven transcriptional signature in Shigella-infected cells.
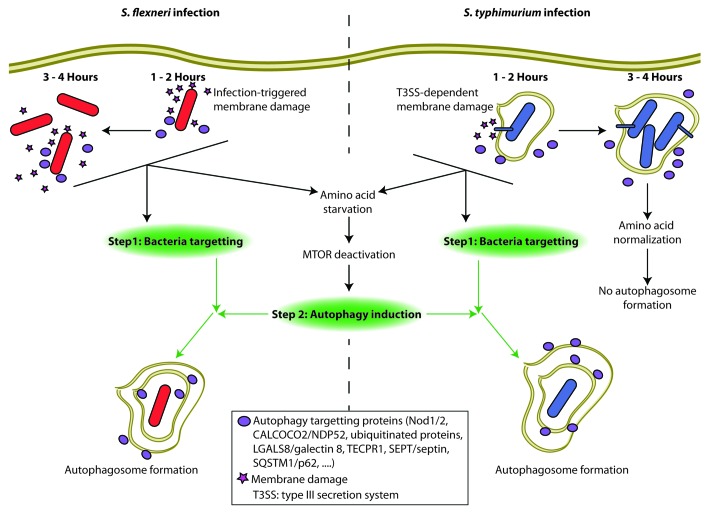
Figure 1. Clearance of intracellular bacteria by autophagy requires both targeting and autophagy induction. (1–2 h) Infection with either S. flexneri or S. typhimurium triggers membrane damage and amino acid starvation responses that lead to the deactivation of MTOR and the induction of autophagy. Simultaneously, intracellular bacteria are recognized by several autophagy targeting proteins that direct the activated autophagy machinery, and autophagosomes are formed. (3–4 h) A subset of autophagy targeting proteins continue to target both S. flexneri and S. typhimurium for degradation in autophagosomes. During S. flexneri infection, membrane damage and amino acid stress responses persist and drive prolonged MTOR deactivation allowing for continued autophagosome formation. In contrast, the transient nature of the membrane damage and amino acid stress responses triggered by S. typhimurium lead to MTOR reactivation and no autophagosome formation at late timepoints despite the presence of autophagy targeting proteins on the SCV.
We next analyzed how these pathways were modulated by Salmonella enterica serovar Typhimurium, another human enteric pathogen. Contrary to Shigella, Salmonella remains confined in an intracellular compartment known as the Salmonella-containing vacuole (SCV), which is progressively remodeled and matured through the action of specific bacterial effectors. Interestingly, the effect of Salmonella infection on MTOR signaling and AA starvation responses appears biphasic: the first phase (1–2 h post-infection) is characterized by MTOR inhibition, AA starvation and ATF3 induction. In a second phase (3–4 h post-infection), these host responses rapidly normalize and MTORC1 is found to accumulate strongly at the surface of the maturing SCVs. The accumulation of MTOR at the SCV is explained by the fact that the RAG GTPase complex, which serves as a membrane anchor for MTORC1, is strongly enriched at the surface of these vacuoles. However, it remained to be understood why AA starvation responses were shut off at later times post-infection. Analysis of cytosolic AA levels in Salmonella-infected cells reveals that the AA pools are progressively replenished, most likely through the import of AA from the extracellular compartment. This observation also suggested that the Salmonella-dependent event that had caused the initial AA starvation is no longer active at 3–4 h post-infection.
In searching for the mechanistic events causing AA starvation in infected cells, we noticed that aseptic damage to host membranes with digitonin or glycyl-l-phenylalanine 2-naphthylamide is sufficient to cause induction of AA stress responses, suggesting that the induction of these pathways in infected cells could be caused by host membrane damage. In agreement, and using CALCOCO2 accumulation in immunofluorescence as a marker of membrane damage, we noticed that while Salmonella causes only transient membrane damage to the SCV (as previously reported by Brumell’s group), Shigella triggers a prolonged accumulation of CALCOCO2 in the cytosol of infected cells. Therefore, these data provide strong correlative evidence that host AA starvation responses in infected cells are caused by membrane damage (Fig. 1), although the exact mechanism linking these events remains unclear.
Finally, we analyzed the impact of the induction of these pathways on host autophagy responses. Interestingly, we observed that autophagy induction in Salmonella-infected cells is only transient and peaks at 1–2 h post-infection, in agreement with previous studies, and this corresponds with a time when AA starvation responses are maximal. Our results also suggest that the normalization of MTOR signaling at 3–4 h post-infection (at a time when MTORC1 is mainly associated with the SCV membrane) is the prime reason for the decrease in autophagy targeting of Salmonella at later time points, because stimulation of Salmonella-infected cells with rapamycin is sufficient to restore long-lasting xenophagy responses to this pathogen.
In summary, our results provide evidence that the same cellular event, that is, membrane damage, provides two distinct yet interdependent signals in bacteria-infected cells: it drives the peri-bacterial accumulation of proteins critical for autophagic targeting (such as CALCOCO2, SQSTM1 or ubiquitinated proteins) and simultaneously generates a cellular stress response (identified as AA starvation) that inhibits MTOR, thereby favoring the metabolic switch required for full-blown induction of autophagic responses. Moreover, our studies also highlighted the fact that different bacterial pathogens can cause AA starvation responses with distinct kinetic profiles. Intriguingly, progressive downregulation of autophagic responses at late time points of infection has been previously reported for bacterial pathogens such as Salmonella, Listeria, or Mycobacterium tuberculosis. Our results suggest that this host response profile is not necessarily the result of active bacterial escape from xenophagy targeting, but could be caused by kinetic differences in the modulation of AA starvation responses, MTOR signaling and cellular metabolism.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/21863