Abstract
The key autophagic lipid sensors are Atg18 in yeast and the WIPI proteins in mammals. Atg18 and the WIPIs belong to the PROPPIN family of proteins. PROPPINs are seven- bladed β-propellers that bind to phosphatidylinositol 3-phosphate (PtdIns3P) and phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2]. In order to understand how PROPPINs bind phosphoinositides, we have determined the crystal structure of a representative, biochemically tractable PROPPIN, Hsv2 of Kluveromyces lactis. The structure revealed that PROPPINs contain two phosphoinositide binding sites which cooperate with a hydrophobic anchoring loop in membrane binding. These three binding elements cooperate in function, as demonstrated by the incremental loss of function in Atg18 mutants impaired in combinations of the two phosphoinositide binding sites and the hydrophobic loop.
Keywords: Atg18, Atg21, WIPI, autophagy, membrane binding
Macroautophagy is a cellular process that is conserved from yeast to mammals in which cytoplasmic constituents are engulfed by a double-membrane vesicle termed the autophagosome, and targeted to the vacuole or lysosome for degradation. One of the major signaling events in autophagy is when the autophagy-specific class III PtdIns 3-kinase complex I synthesizes PtdIns3P at the nascent phagophore, the precursor structure to the autophagosome. Various effector proteins recognize PtdIns3P, but the most important for autophagy are Atg18 in yeast and WIPI1 to WIPI4 in mammals. All of these proteins are biochemically competent to bind both PtdIns3P and PtdIns(3,5)P2, however, the latter lipid is not known to have a direct role in autophagy in yeast. The ability of Atg18 to bind PtdIns(3,5)P2 is important for a second biological function of this protein in vacuole homeostasis.
Atg18, its two fungal homologs Atg21 and Hsv2, and the mammalian WIPI proteins are all predicted seven-bladed β-propellers that bind PtdIns3P, and are collectively termed PROPPINS. PROPPINS share a conserved FRRG motif which is involved in PtdInsP binding and therefore membrane targeting. We sought to explore the molecular mechanism of membrane binding by PROPPINS, and solved the crystal structure of Hsv2 at 3.0-Å resolution. The crystal structure confirms that Hsv2 folds into a seven-bladed β-propeller. Surprisingly, the two Arginine residues of the conserved FRRG motif are directed toward different blades of the propeller and coordinate two distinct sulfate ions. One sulfate is bound by blade 5 and the other by blade 6. The sulfate sites were interpreted as potential markers for two distinct lipid binding sites, termed site 1 and site 2. A comparison of blades 5 and 6 reveals a conserved pattern of charged and polar residues and most of these residues, especially a key His and Arg, are conserved in all PROPPINs. Mutation of residues in either binding site disrupts the lipid binding of Hsv2 as analyzed by liposome affinity-isolation studies. Despite their similarities, the PtdInsP binding sites are not identical and have different affinities for PtdIns3P- and PtdIns(3,5)P2-containing liposomes. Site 2 appears to be the more important of the two for binding to PtdIns(3,5)P2, while both sites are equally important for PtdIns3P binding.
Another interesting feature of the PROPPIN family revealed by the Hsv2 crystal structure is the presence of a flexible loop located between β strands 3 and 4 in blade 6. Sequence analysis of different family members demonstrates that this region is variable in both length and amino acid composition but shows a significant presence of aromatic and hydrophobic residues. Indeed, most PROPPIN family members except WIPI4 contain at least 3 aromatic residues in this region. Consistent with the possibility that this loop is involved in membrane binding, WIPI4 binds more weakly to PtdIns3P-containing liposomes as compared with the other yeast and human PROPPINs.
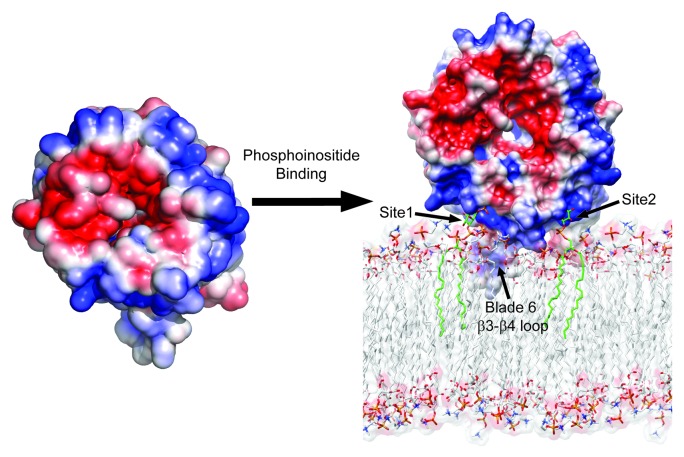
When the blade 6 β3-β4 loop is either deleted, replaced with a GlySerGlySerGlySer linker, or the three aromatic residues are mutated to Ala, there is a complete loss in the liposome binding of Hsv2. These data led us to propose an edge-on model (Fig. 1) for membrane binding wherein two PtdIns3P molecules are bound by sites 1 and 2, and the hydrophobic β3-β4 loop of blade 6 anchors itself in the membrane interface.
Figure 1. Structural model for phosphatidylinositol phosphate binding and membrane targeting by Atg18 family proteins. Electrostatic potential surface of free and membrane bound forms of Kluyveromyces lactis Hsv2 with saturating blue and red at ± 3 kT/e. The lipid molecules bound at site 1 and site 2 are shown as green stick models.
To test the predictions of our model in yeast, we studied the effect of various Atg18 lipid-binding mutants on autophagy. These experiments were performed in atg18Δ S. cerevisiae using three standard yeast autophagy assays: GFP-Atg8 processing, GFP-Atg8 microscopy and Pho8Δ60 activity. Consistent with the Hsv2 liposome-binding studies, the His and Arg double mutants within sites 1 and 2 of Atg18 show a decrease in autophagy. Furthermore, a combination of site 1 and 2 mutations with the blade 6 β3-β4 triple aromatic residue mutation result in an essentially complete loss of function, with a phenotype identical to that of the atg18Δ mutant.
Generally, the congruence between the structural analysis, in vitro biochemistry, and cell function is gratifying. One discrepancy that has yet to be explained is that recombinant Atg18 has a strong preference to bind PtdIns(3,5)P2-containing liposomes in vitro, even though its autophagic function is to recognize PtdIns3P. These in vitro studies need to be viewed with caution, however, because recombinant Atg18 forms a large aggregate of ill-defined stoichiometry and is probably not representative of its functional form in cells. In autophagy, Atg18 functions as part of a complex with the much larger Atg2 protein. We predict that in the context of the Atg2-Atg18 complex, the in vitro lipid-binding specificity would reveal a higher affinity for PtdIns3P, more consistent with its biological function. Another minor discrepancy is that the site 1 mutant of Atg18 seems to eliminate vacuolar localization, which is postulated to depend on PtdIns(3,5)P2, yet the site 1 mutant of Hsv2 retains strong in vitro binding to PtdIns(3,5)P2. While the Hsv2 structure and in vitro biochemistry has generally been an excellent predictor of the cellular properties of Atg18 mutants, there is clearly at least one significant difference in detail.
Acknowledgment
This research was supported by the Intramural Research Program of the NIH, NIDDK, and by a fellowship from the NIGMS to M.J.R.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/22077