Abstract
RBX1/ROC1 is an essential subunit of the largest multiunit Cullin-RING E3 ligase (CRL), which controls the degradation of diverse substrates, thereby regulating numerous cellular processes. Recently, we reported that RBX1 is overexpressed in hepatocellular carcinomas (HCC) and its expression is negatively correlated with patient survival. Moreover, siRNA silencing of RBX1 inhibits the proliferation of liver cancer cells both in vitro and in vivo by inducing CDKN1A/p21-dependent cell senescence. Interestingly, independent of senescence, RBX1 knockdown also triggers an autophagy response, due, at least in part, to the accumulation of the MTOR-inhibitory protein DEPTOR, a recently identified CRL substrate. Biologically, blockage of autophagy significantly enhances the growth-suppressive effect of RBX1 knockdown by triggering massive apoptosis, indicating that the autophagy response upon RBX1 knockdown serves as a survival signal in liver cells. Similar observations were also made in many types of human cancer cells upon inhibition of CRL by MLN4924. These findings suggest that RBX1-CRL is a promising anti-cancer drug target and provide proof-of-concept evidence for a novel drug combination of RBX1-CRL inhibitor and autophagy inhibitor for effective treatment of human cancer.
Keywords: ROC1, RBX1, Cullin-RING E3 ligase, autophagy, senescence, DEPTOR, MTOR, neddylation, MLN4924
RBX1 is a RING subunit of Cullin-RING ligase and is required for CRL to direct a timely degradation of diverse substrates, thereby regulating numerous cellular processes under both physiological and pathological conditions. As an essential subunit of the CRL complex, RBX1 binds to cullins at its N terminus and ubiquitin-loaded E2 at its RING-H2 finger-containing C terminus, and effectively catalyzes the transfer of ubiquitin from E2 to specific substrates for proteasome-targeted degradation (Fig. 1). Physiologically, RBX1 is required for embryonic development in Caenorhabditis elegans, Drosophila and mouse by regulating cell proliferation and differentiation.
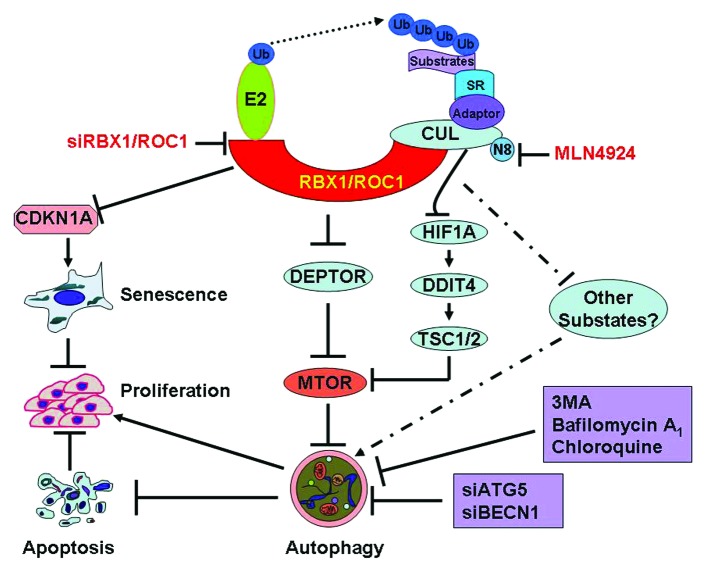
Figure 1. Inactivation of RBX1-CRL E3 ligase triggers protective autophagy. RBX1 is an essential subunit of CRL, which binds to CUL and substrate receptors to assemble functional CRLs. As an appealing anticancer target, RBX1 knockdown induces CDKN1A/p21-dependent senescence to suppress the growth of liver cancer cells. Interestingly, RBX1 knockdown also triggers an autophagy response by inducing the accumulation of the MTOR-inhibitory protein DEPTOR, resulting in the inactivation of MTOR. Similarly, CRL inhibitor MLN4924 also triggers autophagy by blockage of MTOR signals via DEPTOR as well as the HIF1A-DDIT4/REDD1-TSC1/2 axis. The autophagy response serves as a prosurvival signal since blockage of the autophagy pathway via siRNA silencing of essential autophagy genes (ATG5 or BECN1) or treatment with autophagy inhibitors significantly enhances the growth-suppressive effect of RBX1-CRL inactivation by triggering massive apoptosis in cancer cells. SR, substrate receptor, 3MA, 3-methyladenine, N8, NEDD8.
Our recent work showed that RBX1 is overexpressed in a number of human cancers, including breast, liver and lung. More importantly, we found that RBX1 overexpression in hepatocellular carcinomas directly correlates with poor patient survival. Moreover, RBX1 knockdown via siRNA silencing suppresses the growth of liver cancer cells both in vitro and in vivo. Further mechanism analysis revealed that RBX1 knockdown induces the accumulation of the CRL substrate CDKN1A/p21, resulting in cell senescence to suppress cell proliferation (Fig. 1). These findings suggest that RBX1 is an appealing drug target for liver cancers, and its overexpression may serve as a prognosis biomarker.
More interestingly, we found that RBX1 knockdown triggers a senescence-independent autophagic response in liver cancer cells, given the fact that RBX1 knockdown effectively induces (1) the conversion of LC3-I (cytosolic form) to LC3-II (membrane-bound lipidated form); (2) the degradation of the autophagy-selective substrate SQSTM1/p62; (3) the formation of EGFP-LC3 puncta; (4) the formation of acidic vesicular organelles (AVOs); (5) the formation of double-membrane autophagosomes; and (6) an intact autophagic flux. In addition, we found that RBX1 silencing also induces autophagy in other cancer cells (such as HeLa cervical cancer cells and H1299 lung carcinoma cells). Similarly, we found that CRL inactivation by MLN4924, a specific inhibitor of the NEDD8-activating enzyme (NAE1), which causes cullin deneddylation and subsequent inactivation of CRL activity, triggers autophagy in many human cancer lines derived from carcinomas of brain, breast, cervix, colon, and liver. These findings indicate that autophagy is a universal cellular response to CRL inactivation.
To address how RBX1 knockdown induces autophagy, we then screened for potential CRL substrates, which are accumulated and trigger autophagy upon CRL inactivation by RBX1 silencing. We found that the MTOR-inhibitory protein DEPTOR, a recently well-characterized CRL substrate, is accumulated upon RBX1 knockdown. The accumulation of DEPTOR inhibits MTOR activity and thus induces autophagy, whereas DEPTOR knockdown largely abrogates RBX1 knockdown-triggered autophagy. Similarly, we found that inactivation of CRL by MLN4924 also induces autophagy through modulating the DEPTOR-MTOR axis as well as the HIF1A-DDIT4/REDD1-TSC1/2-MTOR axis (Fig. 1).
Since we found that RBX1 knockdown induces both DEPTOR-dependent autophagy and CDKN1A-dependent senescence in liver cancer cells, we further determined the potential relevance of these cellular responses. We found that RBX1 knockdown causes a sequential induction of autophagy and senescence, with autophagy occurring at the early stage, suggesting that the autophagy response may play a role in facilitating senescence, a phenomenon seen in other cell models. However, we found that blockage of the autophagy pathway by knockdown of autophagy essential genes BECN1 and ATG5 has no obvious effect on senescence induction upon RBX1 knockdown in liver cells. Thus, RBX1 knockdown triggers autophagy and senescence independently, probably resulting from the accumulation of different sets of CRL substrates.
Autophagy may play a dual role, prosurvival or prodeath, dependent on different cellular stresses. To address the biological significance of the autophagy response in RBX1-silenced cells, we blocked the autophagy-signaling pathway via siRNA silencing of ATG5 and BECN1 in RBX1-silenced cells and evaluated the effects on RBX1 knockdown-induced growth suppression. We found that blockage of the autophagy pathway significantly enhances the growth-suppressive effect of RBX1 knockdown by triggering massive cell apoptosis (Fig. 1). Similarly, we found that pharmaceutical inactivation of CRL by MLN4924 also triggers a protective autophagy in many types of human cancer cells and combination of MLN4924 with an autophagy inhibitor or ATG5 knockdown enhances apoptosis. These findings demonstrate that the autophagy response, induced by CRL inactivation through either RBX1 knockdown or small molecule inhibitors, serves as a survival signal to protect cancer cells. Thus, strategic combination of inhibitors of RBX1-CRL and autophagy may significantly increase the therapeutic efficacy against many types of human cancer.
Acknowledgments
This work is supported by National Natural Science Foundation Grant of China (31071204, 81172092), National Basic Research Program of China (973 program, 2012CB910302) and Key Project of Shanghai Municipal Health Bureau (2010012) to L.J., National Natural Science Foundation Grant of China to D.Y. (81101811) and J.L. (91129702, 81125001), as well as Postdoctoral Science Foundation of China to D.Y. (KLF101054). The work is also partly supported by the NCI grants (CA118762 and CA156744) to Y.S.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/22024