Abstract
Interferon Regulatory Factor 6 (IRF6) is a transcription factor that, in mammals, is required for the differentiation of skin, breast epithelium, and oral epithelium. However, the transcriptional targets that mediate these effects are currently unknown. In zebrafish and frog embryos Irf6 is necessary for differentiation of the embryonic superficial epithelium, or periderm. Here we use microarrays to identify genes that are expressed in the zebrafish periderm and whose expression is inhibited by a dominant-negative variant of Irf6 (dnIrf6). These methods identify Grhl3, an ancient regulator of the epidermal permeability barrier, as acting downstream of Irf6. In human keratinocytes, IRF6 binds conserved elements near the GHRL3 promoter. We show that one of these elements has enhancer activity in human keratinocytes and zebrafish periderm, suggesting that Irf6 directly stimulates Grhl3 expression in these tissues. Simultaneous inhibition of grhl1 and grhl3 disrupts periderm differentiation in zebrafish, and, intriguingly, forced grhl3 expression restores periderm markers in both zebrafish injected with dnIrf6 and frog embryos depleted of Irf6. Finally, in Irf6 deficient mouse embryos, Grhl3 expression in the periderm and oral epithelium is virtually absent. These results indicate that Grhl3 is a key effector of Irf6 in periderm differentiation.
Introduction
Interferon Regulatory Factor 6 (IRF6) drives the differentiation of epithelia with barrier function via unknown mechanisms. In mice deficient for Irf6, epidermal keratinocytes fail to differentiate terminally, resulting in the absence of an epidermal permeability barrier (Ingraham et al. 2006; Richardson et al. 2006; Biggs et al. 2012). Moreover, the superficial layer of the oral epithelium, the oral periderm is missing, resulting in adhesion between the palatal shelves and the tongue (Richardson et al. 2009). Mutations in IRF6 cause two syndromic forms of cleft lip and palate (CL/P): Van der Woude and Popliteal Pterygium syndromes (Kondo et al. 2002). They are also associated with non-syndromic CL/P (Zucchero et al. 2004; Koillinen et al. 2005; Srichomthong et al. 2005; Pegelow et al. 2008; Desmyter et al. 2010). In patients with such mutations, abnormal differentiation of the epidermis or oral periderm may contribute to CL/P pathogenesis of CL/P. In addition, IRF6 appears to be necessary for normal wound healing (Jones et al. 2010), promotes differentiation of the breast epithelium (Bailey et al. 2008), and functions as a tumor suppressor in squamous cell carcinoma (Botti et al. 2011). A recent study identified hundreds of genomic loci that are bound by IRF6 in human keratinocytes (Botti et al. 2011), but it is unclear which of these apparent transcriptional targets effect epithelial differentiation.
The aim of this study was to characterize the position of Irf6 within the gene regulatory network that controls differentiation of the periderm in zebrafish, as many aspects of mammalian skin biology are conserved in fish (Li et al. 2011). The periderm is a simple squamous epithelium that is the most superficial layer of an embryo and forms the first permeability barrier to dehydration in mouse embryos (M’Boneko and Merker 1988). In both fish and amphibian embryos, the periderm forms in early gastrula stage and is necessary for structural integrity of the embryo during this period (Fukazawa et al. 2010). We previously showed that the periderm was disrupted in both zebrafish embryos injected with RNA encoding a dominant-negative Irf6 variant (dnIrf6) and in frog embryos depleted of maternal Irf6: the expression of various periderm markers was lost, and the periderm itself ruptured (Sabel et al. 2009). Here, through microarray analysis, in vivo reporter studies, and epistasis experiments, we identify the gene encoding Grainyhead-like 3 (Grhl3), an ancient mediator of epithelial integrity, as a direct effector of Irf6 in periderm differentiation.
Results
Potential regulators of periderm differentiation
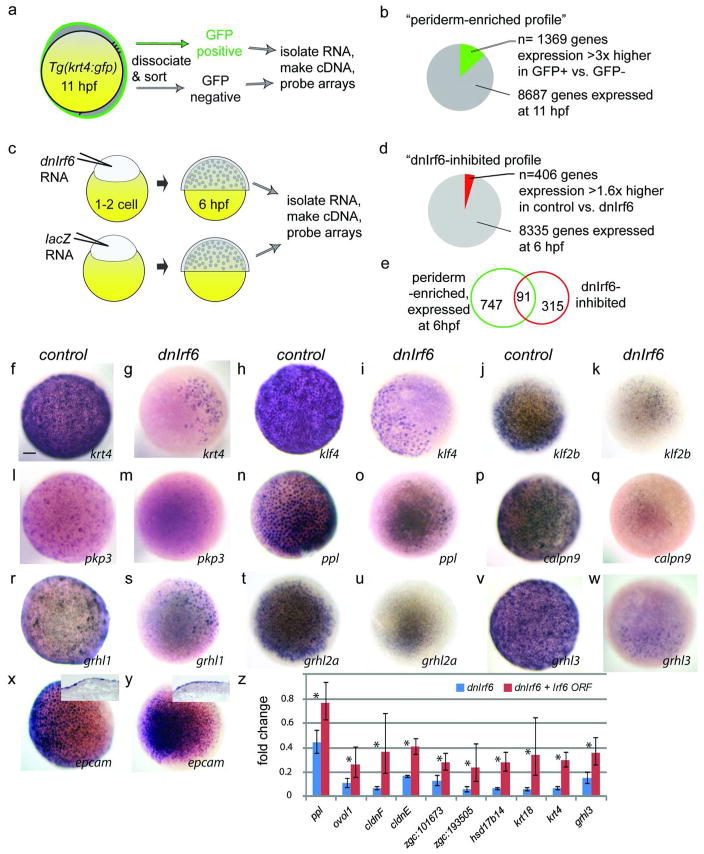
We assessed the gene expression profile of isolated zebrafish periderm cells using the Tg(krt4:gfp) transgenic zebrafish line, in which GFP is robustly expressed in simple epithelia of the embryo (Gong et al. 2002). Through cell sorting and microarray analyses, we identified a subset of genes expressed in GFP+ cells at levels at least three-fold higher than in GFP− cells (Fig.1a,b). This “periderm-enriched profile” (Supplemental Table S1) is significantly enriched for genes with ontology terms typical of those present in other epithelia (See Methods). The seventy transcription factor-encoding genes represented in this profile (Table S1) potentially participate in the periderm gene regulatory network, either upstream or downstream of Irf6.
Figure 1. Microarray analysis of zebrafish periderm.
a,c Schematic representation of methods used to create the periderm-enriched (a) and the dnIrf6-inhibited (c) profile. b,d, Pie charts showing genes whose expression is b 3× higher in FACS-sorted GFP-positive versus GFP-negative cells (c) or significantly reduced (>1.7 fold) in dnIrf6-injected compared to lacZ-injected embryos (d). e Venn diagram portraying the overlap of sets in b and d. f–y Animal-pole view of mid-gastrula stage embryos, uninjected or injected with dnIrf6, and processed to reveal expression of the indicated gene. All are expressed in periderm and all except y (epcam) are strongly reduced in dnIrf6-injected embryos. Scale bar: f, 100 μM. z qRT-PCR for genes whose expression is reduced by dnIrf6 and significantly elevated (with respect to dnIrf6 alone) by co-injection with Irf6 (p <0.05).
To learn where Irf6 fits into this network, we created a profile of genes whose expression is altered in the presence of dnIrf6. We did not use antisense morpholino oligonucleotides (MO), the standard tools for reducing gene expression in zebrafish embryos, because they are commonly ineffective against maternally-encoded transcripts (Draper et al. 2001) and this proved to be the case for Irf6 (Fig. S1). Instead we modified the dnIrf6 generated for our earlier study(Sabel et al. 2009) by fusing the Irf6 DNA-binding domain to the Engrailed repressor domain. We injected embryos with an RNA encoding either dnIrf6 or β-galactosidase (i.e. lacZ RNA), harvested RNA at 6 hours post fertilization (hpf; such embryos rupture by 7 hpf), and probed microarrays as described above. We identified a subset of genes that are expressed at significantly lower levels in dnIrf6-injected than in lacZ-injected embryos, and refer to this as the “dnIrf6-inhibited profile” (Fig.1c,d, Table S2); it contains many genes that encode adhesion molecules, consistent with rupture of the periderm in dnIrf6-injected embryos.
Of the genes present in both the periderm-enriched and dnIrf6-inhibited profiles (Fig. 1E) (Table S3), ten are classified by the gene ontology term, “transcription factor activity,” and therefore may participate in the periderm gene regulatory network that acts downstream of Irf6 (Table 1). Among these, grhl1 and klf2b are of special interest because homologues of these genes, Grhl3 and Klf4, respectively, are required to generate the epidermal permeability barrier in mice (Ting et al. 2005) (Segre et al. 1999). However, probes for the zebrafish orthologs of neither Grhl3 nor Klf4 are present in the microarray we deployed. Therefore we assessed the expression of these genes, and that of several others common to the two profiles, by in situ hybridization. We found that all are indeed down-regulated in dnIrf6-injected embryos vs. control-injected embryos (Fig. 1f-w), and that epcam, which is present in only the former profile, is indeed expressed in dnIrf6-injected embryos (Fig. 1x,y). Moreover, we found that co-injecting an RNA encoding full-length Irf6 partially reversed the effects of dnIrf6 (Fig. 1z), supporting the idea that those effects result from interference with Irf6 and/or closely related proteins. Genes in the periderm-enriched profile whose expression was significantly elevated in the presence of dnIrf6 are candidates for repression by Irf6, although most are likely affected indirectly because dnIrf6 is a constitutive repressor. Interestingly, p63, which regulates epidermal differentiation in mice and zebrafish, and directly regulates Irf6 expression, ranks among these genes (Bakkers et al. 2002; Lee and Kimelman 2002; Moretti et al. 2010; Thomason et al. 2010). In summary, profling experiments revealed several genes that are likely to be members of the periderm gene regulatory network that act downstream of Irf6.
Table 1.
Genes encoding transcription factors in both the periderm-enriched and dnIrf6-inhbited profiles
| Gene ID | Gene name |
|---|---|
| ENSDART00000082425 | GATA-binding protein 2a |
| ENSDART00000021159 | LIM homeobox 1b |
| ENSDART00000080693 | LIM homeobox 5 |
| ENSDART00000060861 | forkhead box P2 |
| ENSDART00000025153 | GATA-binding protein 3 |
| ENSDART00000052521 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| ENSDART00000052570 | paired-like homeodomain transcription factor 2 |
| ENSDART00000017456 | ventral expressed homeobox |
| ENSDART00000087454 | Grainyhead-like 1 |
| ENSDART00000059188 | klf2b, Kruppel-like factor 2b |
Irf6 activates epithelial enhancers adjacent to Grhl3
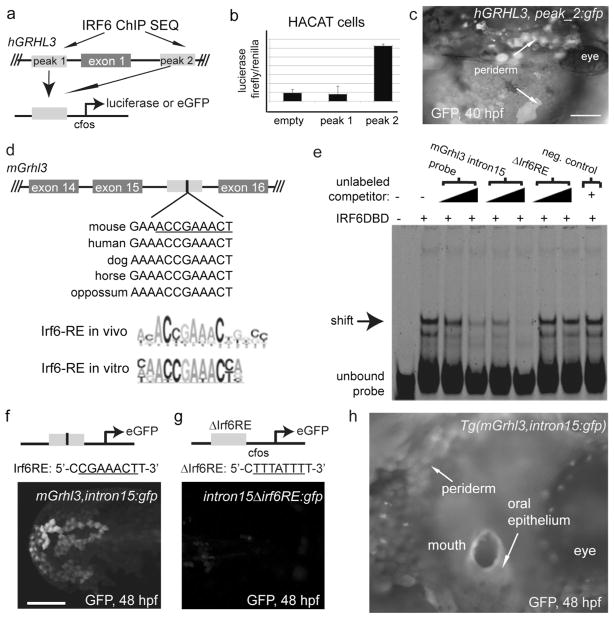
We next sought to test the possibility that Irf6 directly activates grhl3 expression in the periderm. In a recent ChIP experiment conducted in human keratinocytes, IRF6 was found to be bound at two locations near the first exon of GRHL3 – about 2 kb upstream, and about 3 kb downstream (intronic) (Fig. 2a) (Botti et al. 2011). Both peaks are within chromatin elements that are conserved among mammals and are sites of DNAse hypersensitivity in human keratinocytes (ENCODE project). Thus, these elements may have cis-regulatory function. In addition, microarray analysis revealed that GRHL3 expression is reduced in cells depleted of IRF6, suggesting that IRF6 directly activates GRHL3 expression in keratinocytes (Botti et al. 2011). To test this possibility, we amplified the conserved elements containing these peaks and cloned them into a firefly luciferase reporter vector containing a minimal promoter (Fig. 2a). In a human keratinocyte cell line transfected with the construct containing the intron-1 element, but not in one tranfsected with the construct containing the the upstream element, luciferase levels were significantly higher than in a line transfected with a control-construct (Fig. 2b). Although neither IRF6-bound element is detectably conserved in zebrafish, the functions of mammalian enhancer elements without detectable counterparts in zebrafish often nonetheless are conserved in zebrafish embryos (Fisher et al. 2006; McGaughey et al. 2008; McGaughey et al. 2009). We thus cloned the elements into a reporter vector that contains a minimal promoter and the gene encoding green fluorescent protein (GFP)(Fig. 2a) (vector described in Fisher et al. 2006), and injected these constructs into wild-type zebrafish embryos. About 20% of the embryos injected with hGRHL3_peak2:gfp exhibited GFP expression in contiguous patches of periderm cells (recognizable due to their characteristic polygonal morphology), starting at high stage and continuing to at least 72 hpf (shown on the yolk at 48 hpf, Fig. 2c) (numbers of injected embryos for reporter experiments are presented in Methods). Embryos injected with hGRHL3_peak1:gfp or the vector lacking either element did not exhibit these patches. These studies support the idea that Irf6 directly activates Grhl3 expression in both human keratinocytes and the zebrafish periderm.
Figure 2. Enhancer activity of DNA elements near GRHL3.
a Positions of Irf6 binding peaks in human keratinocytes from a ChIP-SEQ experiment (Botti et al. 2011), and construction of luciferase and GFP reporter vectors with these peaks. b Results of luciferase reporter assay. c,f,g,h Transient (c,f,g) and stable (h) transgenic zebrafish embryos of the indicated constructs with GFP expression in the periderm. d Schematic representation of a conserved element within GRHL3 intron 15 (gray box), which contains a conserved Irf6 response element (IRF6-RE). Position-weighted matrix for IRF6-RE defined in vitro (Little et al. 2009) and in vivo (Botti et al. 2011). e Electromobility shift assay with human IRF6DBD and a labeled probe that matches 25 base pairs of the hGrhl3 intron 15 that encompass the putative Irf6RE. Scale bars: c, 100 μm; f, 200 μm.
As an alternative approach for identifying Irf6-dependent enhancers, we searched mammalian and zebrafish genomes for the presence of conserved, non-coding elements that possess the consensus Irf6 response element (Little et al. 2009). Although we did not detect any that are conserved between mammals and zebrafish, we found one (within intron 15 of GRHL3) that is conserved among mammals (Fig. 2d). Although IRF6 binding of this site was not reported in keratinocytes (Botti et al. 2011), it could potentially occur in other cell types including those of the periderm. Indeed, we found that an oligonucleotide that matches Grhl3 intron sequence encompassing the apparent Irf6 binding site competes with a control probe for the binding of Irf6DBD in vitro, and that a version of the oligo in which this site has been altered lacks this ability (Fig. 2e). To test the potential enhancer activity of the conserved element within GRHL3 intron 15, we amplified it (972 bp) from the murine genome and cloned it into the GFP reporter vector (creating mGrhl3intron15:gfp). In both transient and stable transgenic embryos harboring this construct, GFP is expressed in the periderm (Fig 2f,h). In addition, we generated a construct in which six bases within the Irf6RE are mutated (as in the in vitro binding experiment described above), creating intron15ΔIrf6RE:gfp (Fig. 2g). In embryos injected with this construct, GFP expression was rarely detected in periderm cells, and was completely extinguished by 48 hpf (Fig. 2g). In summary a conserved element within intron 15 of mammalian Grhl3 has periderm enhancer activity that depends on the presence of a consensus Irf6 binding site.
Inhibition of grhl1 and grhl3 disrupts periderm development
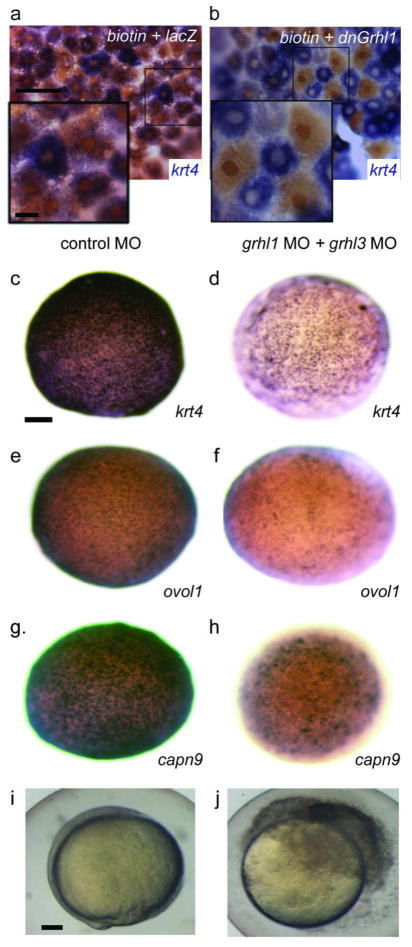
Three members of the Grhl family are expressed in the zebrafish periderm (Janicke et al. 2010). Because Grhl family members share a core DNA consensus binding site (Ting et al. 2005; Wilanowski et al. 2008; Rifat et al. 2010) (Boglev et al. 2011), we reasoned that overexpressing the DNA binding domain (DBD) of any is likely to displace them all from DNA. We thus injected zebrafish embryos with an RNA encoding the Xenopus Xgrhl1 DBD (dnXgrhl1). This resulted in stalled epiboly and embryonic rupture, much as in dnIrf6-injected embryos (not shown, 29 of 30 injected embryos)(Tao et al. 2005). We found that mosaic injection of the dnXgrhl1 blocked krt4 expression in cell-autonomous fashion (Fig. 3a,b). To determine which family members are essential for development of the zebrafish periderm, we injected MOs known to target the grhl1 start codon (Janicke et al. 2010) or to block the splicing of grhl3 (confirmed in Fig. S2). Epiboly was not affected when either was injected alone, but when both were injected simultaneously, the expression of krt4, capn9, and ovol1b was significantly reduced, epiboly was delayed, and the embryos ruptured at the animal pole (Fig. 3c–j). RT-PCR confirmed that the combination of MOs targeting grhl1 and grhl3 reduced expression of krt4 (Fig. S2). Thus, Grhl1 and Grhl3 function redundantly to promote periderm differentiation in zebrafish.
Figure 3. Knockdown of grhl family members results in loss of periderm markers.
a,b Animal-pole view of 8hpf embryos injected mosaically with a, biotinylated dextran mixed with lacZ or b, dnXgrhl1 mRNA. Embryos were fixed at shield stage and processed to reveal krt4 expression (purple) and biotin (brown). Periderm cells inheriting lacZ (brown nuclei) express krt4 variably, like periderm cells in uninjected embryos, while those inheriting dnXGrhl1 (brown nuclei) lack krt4. c–j Embryos injected with control MO (c,e,g,i), or grhl1 and grhl3 MOs (d,f,h,j), fixed at 8 hpf, and processed to reveal expression of the indicated gene, or allowed to develop until 10 hpf (i–j), at which time control MO-injected embryo have normal morphology (i), and grhl1/grhl3 MO-injected embryos have ruptured (j). Scale bars: a, 50 μm; a inset 10 μm; c, 100 μm; i, 100 μm.
Forced expression of grhl3 partially restores epiboly in dnIrf6-injected embryos
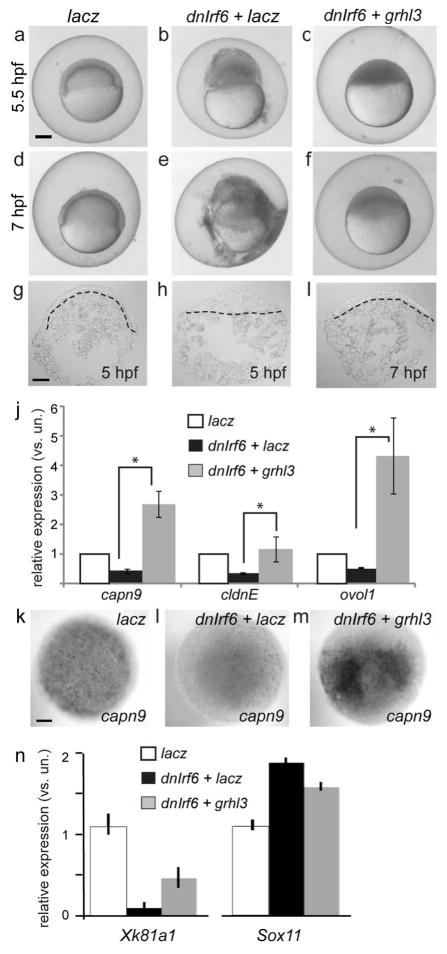
During epiboly, blastomeres in control embryos migrate towards the vegetal pole, producing doming of the yolk towards the animal pole (Fig. 4b,h). In the large majority of embyros injected with dnIrf6 mRNA and subsequently injected with a control mRNA (i.e., lacZ), these movements do not appear to occur (Fig. 4g and 4h), and virtually all have ruptured through the animal pole by the time control embryos reach 6.5 hpf (Fig. 4b) (94%, n=114 injected embryos). By contrast, in the majority of dnIrf6 -injected embryos that are instead injected with the grhl3 mRNA, blastomeres move towards the vegetal pole, the yolk undergoes doming, and embryos are still alive at 6.5 hpf (Fig. 4c,f,i) (70%, n=119). Moreover they express the periderm markers claudinE, capn9, and ovol1 at higher levels, as demonstrated by qRT-PCR analysis (Fig. 4j) and confirmed by in situ hybridization for capn9 (Fig. 4k–m). Most such embryos nonetheless ultimately rupture by the time control embryos reach 7.5 hpf (81%, n=119). Thus grhl3 mRNA improved survival during epiboly of dnIrf6-injected embryos without obviously rescuing the overall morphology of embryos.
Figure 4. Injection of grhl3 restores periderm markers in dnIrf6-injected embryos.
A–I Images of live (a–f) and DAPI-stained sections of (g–i) embryos injected with the indicated RNA. Insets, magnified images of the boxed area. g–i The doming of the yolk apparent in lacz-injected embyros (g), is lost in dnIrf6-injected embryos (h), and partially restored in dnIrf6-and-grhl3 -injected embryos (i). j qRT-PCR analysis of mRNA levels. capn9, p=0.03, cldnE, p=0.003, ovol1, p=0.048.k–m 5-hpf embryos processed for capn9 expression. n qRT-PCR analysis of levels of the periderm marker xk81a1, and the deep blastomere marker sox11, in Xenopus embryos derived from oocytes injected with control MO or irf6 MO and injected as zygotes with the XGrhl3 mRNA, as indicated. By ANOVA analysis, xk81a1, p< 0.0002, sox11, p< 0.03. Error bars, standard deviation. Scale bars: a, 200 μm; g, 100 μm.
We also tested the ability of Grhl3 to reverse the effects of MO-mediated knockdown of the irf6 mRNA in frog embryos. Using the oocyte-transfer method to inhibit maternal irf6, we confirmed the earlier report (Sabel et al. 2009) of a strong reduction of the superficial epithelium marker Xk81a1 and up-regulation of the deep-cell marker, sox11 (Fig. 4n). In embryos that were additionally injected with the Xenopus grhl3 mRNA, xk81a1 levels were significantly closer to those in control embryos (Fig. 4n). Thus, Irf6-mediated activation of grhl3 expression appears to be a critical component in the periderm gene regulatory network in fish and in frogs.
Expression of Grhl3 in murine Irf6-deficient embryos
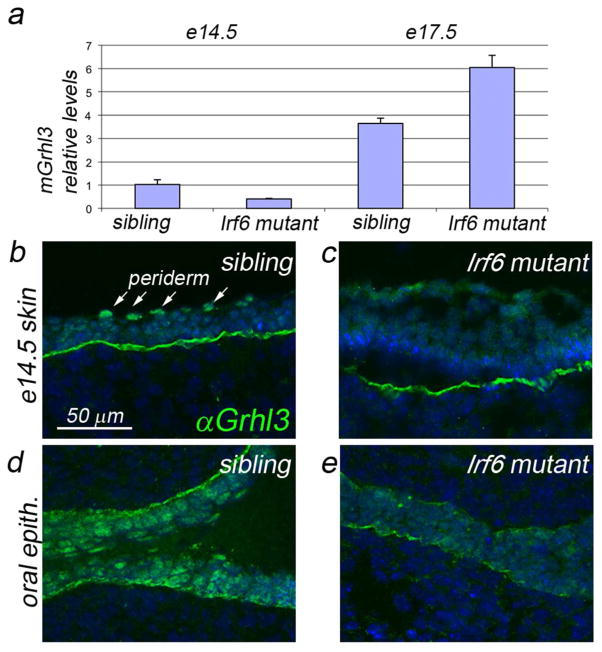
We harvested epidermis from mouse embryos that expressed wild-type and/or a null allele of Irf6 (i.e., homozygous control, heterozgous mutant or homozygous mutant) at E14.5 and E17.5, and measured Grhl3 levels by qRT-PCR. In homozygous mutants, Grhl3 was reduced at E14.5 but elevated at E17.5 with respect to the levels in siblings (wild-type or heterozygous mutant) (Fig. 5a). The elevation at the later time point is consistent with the observation that epidermal stratification is abnormal (the basal and spinous layers are expanded and exhibit inappropriate gene expression, and the granulosum and cornified layers are absent) (Ingraham et al. 2006; Richardson et al. 2006). To determine the reason for the reduction of Grhl3 expression at E14.5, we evaluated anti-Grhl3 immunoreactivity (Grhl3 IR) in coronal head sections from homozgyous Irf6 mutants and their siblings at E14.5. In the sibling embryos, nuclear Grhl3 IR was detected in all layers of the epidermis, with expression highest in periderm-cell nuclei (Fig. 5b). In Irf6 mutants, Grhl3 IR was generally lower and non-nuclear, and the characteristic periderm expression was absent (Fig. 5c). Similar changes in the gene expression pattern and the epidermal structure were observed in the oral epithelium (Fig. 5d,e). Together these findings suggest that Irf6 directly activates Grhl3 expression in the periderm and oral epithelium in mice, but that in keratinocytes, Irf6 contributes to the activation of Grhl3 expression, but is not essential for it.
Figure 5. Expression of Grhl3 in Irf6-deficient mice.
a qRT-PCR analysis of Grhl3 levels in epidermis harvested at the indicated stage: e14.5, 2 replicates; e17.5, 3 replicates; p<0.03. b–e Anti-Grhl3 immunofluorescence on the indicated tissue at the indcates stages. In the epidermis of sibling control embryos (b), Grhl3 immunoreactivity (IR) is prominent in all epidermal and peridermal nucleii. In mutant embyros (c), Grhl3 IR is diffuse, non-nuclear, and weak to undetectable in the periderm. In the oral epithelium of control siblings (d), Grhl3 IR is strong in in all nuclei, and strongest in the oral periderm. In mutant embryos (e), Grhl3 IR is highly reduced. Scale bar: b, 50 μm.
Finally, we generated mutants doubly heterozygous for Irf6 and Grhl3. In newborn Irf6gt/+; Grhl3del/+ pups, we detected neither gross morphological defects in the face, ears, limbs or oral cavity, nor obvious histological defects in skin or oral epithelium (n=6) (not shown). Thus, if Irf6 and Grhl3 function in the same pathway during formation of the permeability barrier, the dosage of neither gene is limiting in double heterozygotes.
Discussion
Irf6 and the periderm gene regulatory network
Here we have conducted profiling experiments that have shed light on the role of Irf6 in zebrafish periderm development. We used conservative criteria in identifying genes in the in the periderm-enriched and dnIrf6-inhibited profiles, and probes for some of the genes that are important for epithelial development, including grhl3 itself, were not represented on the microarray we deployed. Nonetheless, the periderm-enriched profile includes at least seventy genes encoding transcription factors, all of which are candidate participants in the periderm transcriptional gene regulatory network. Those that are also present in the dnIrf6-inhibited profile include grhl1, tfap2c, klf2b, and c/ebp beta. Each of these genes or a homologue has previously been implicated in epithelial development. Notably, many of the regulatory genes in the periderm profile are not in the dnIrf6-sensitive profile, and these could play roles upstream of, or parallel to, Irf6 during epithelial development. Candidates for regulatory proteins acting upstream of Irf6 include aPKC (Chalmers et al. 2003), Ikk1 (Fukazawa et al. 2010), and p63 (Moretti et al. 2010; Thomason et al. 2010). Thus, our work has helped to place Irf6 within the gene regulatory network that governs differentiation of the zebrafish periderm, a tissue that serves as a model for other simple squamous epithelia, including mammalian oral periderm.
Grhl3 appears to be a direct effector of Irf6 in the periderm
Our results suggest that Irf6 directly regulates grhl3 expression, and that in some tissues it may do so in conjunction with other Irf family members. Specifically, we show that Grhl3 expression is strongly down regulated in the periderm of zebrafish embryos injected with dnIrf6, consistent with previous findings from Xenopus embryos derived from oocytes injected with irf6 MO (Sabel et al. 2009), Irf6 mutant mouse embryos (Ingraham et al. 2006; Richardson et al. 2006), and human keratinocytes transfected with an shRNA targeting human IRF6 (Botti et al. 2011). Moreover, we have identified two conserved non-coding elements close to GRHL3 – one known to be bound by IRF6 in keratinocytes, and another strongly conserved Irf6 consensus binding site – that both have zebrafish periderm enhancer activity. We have also found that Grhl3 expression is not completely dependent on Irf6 in all tissues, for instance in mouse keratinoctyes; it is possible that in such contexts Irf6 homologues cooperate with Irf6 to regulate target-gene expression. We note that the periderm-enriched profile includes two irf6 homologues (i.e., irf4a and irf11). The presence of these genes, together with maternally encoded irf6, may explain why dnIrf6 effectively reduces grhl3 expression upon injection into zebrafish eggs, whereas the irf6 MO does not (our unpublished finding). We note also, that a ChIP-SEQ experiment found that IRF4 binds intron 15 of GRHL3 in a lymphoblast cell line (ENCODE project, Rick Myers laboratory, HudsonAlpha Institute). In summary, the available genetic and biochemical evidence indicates that Irf6, possibly in conjunction with its homologs, directly activates grhl3 expression in epithelial structures.
Clinical and evolutionary implications
Because Grhl3 appears to act in concert with Irf6 in the oral periderm gene regulatory network, mutations in GRHL3, and in genes encoding other members of this network, are candidates to contribute risk for CL/P. Whereas mutations in IRF6 have been estimated to confer 12% of inherited risk for non-syndromic CL/P (Zucchero et al. 2004), the majority of such risk has not been ascribed to any gene (Yuan et al. 2011). In mice, Irf6 is necessary for differentiation of the oral epithelium, particularly that of the oral periderm (Richardson et al. 2009). Grhl3 is expressed in the zebrafish oral epithelium (our unpublished findings) and in mouse embryos (Auden et al. 2006). Furthermore, as recently reported, mouse Grhl2 mutants exhibit orofacial clefts (Pyrgaki et al. 2011). Although our macroscopic analyses of Irf6gt/+;Grhl3del/+ double heterozygous mice at birth did not detect gross anomalies in skin development, it will be important to assess embryos for the presence of oral adhesions. Importantly, the absence of a phenotype in double heterozygotes is not strong evidence against two genes acting in the same pathway. Indeed, the related defects in barrier function in Grhl3 and Irf6 mutants suggests that the two genes promote skin differentiation through the same pathway (Ting et al. 2005; Ingraham et al. 2006; Richardson et al. 2006). Also, the ability of Grhl3 to rescue the expression of periderm markers in dnIrf6-injected embryos, as shown here, is strong evidence that they act in the pathway that promotes periderm differentiation. The data presented here motivate an assessment of over-transmission of specific alleles of GRHL3 (or other GRHL family members) in patients with non-syndromic CL/P, and of mutations in GRHL3 in patients with syndromic forms of CL/P. In this context it is intriguing that three linkage studies have detected a risk locus at 1p36, where GRHL3 resides (Prescott et al. 2000; Martinelli et al. 2001; Moreno et al. 2004), and Van der Woude Syndrome 2 (VWS2), which includes oro-facial clefting, maps to an interval that contains GRHL3 (Koillinen et al. 2001).
Finally, our data lead us to speculate how the role of IRF6 in epithelial development evolved. The IRFs are ancient, having arisen alongside muticellurity in animals (Nehyba et al. 2009), and the GRHL family has an ancient role in promoting barrier formation (Ting et al. 2005). Most members of the IRF family regulate the expression of genes that encode interferons and proteins that promote the differentiation of dendritic cells (Gabriele and Ozato 2007); a unifying theme of targets of other IRFs is an involvement in innate immunity. However, given that the most important aspect of innate immunity is the epidermal permeability barrier, it is plausible that GRHL3 activation is not a novel function of IRF6 activity, but instead reflects an ancient interaction that evolved as one feature of the innate immune response.
Methods
Dissociation of zebrafish embryos, FACS and RNA preparation
For production of the periderm-enriched profile, about 200 heterozygous transgenic tg(krt4:gfp)(Gong et al. 2002) embryos were reared to 11 hpf, pestle homogenized, and dissociated with phosphate buffered saline (PBS) containing 0.25% trypsin and 1 mM EDTA (Gibco # 25200, Invitrogen, Carlsbad, CA) for 30 minutes at 33°C. Cells were resuspended in PBS plus 3% fetal bovine serum (FBS) and sorted on a FACS DiVa (Becton Dickinson, Franklin Lakes, NJ), directly into a buffer containing guanidinium thiocyanate for subsequent RNA isolation using the RNEasy Plus Mini Kit (i.e., Buffer RLT, QIAGEN, Valencia, CA). RNA was subjected to two rounds of amplification using the Message Amp II aRNA Amplification Kit (Invitrogen). For production of the dnIrf6-inhibited profile, embryos were injected with dnIrf6 or the lacZ mRNA, raised to 6 hpf and homogenized. RNA was extracted from the dnIrf6- or control-RNA injected embryos as described above, without an amplification step. RNA was pooled from a total of approximately 100 embryos of each type; both types were generated on two separate injection days.
cDNA preparation and probe hybridization
For production of the periderm-enriched profile, we synthesized and labeled cDNA a single time, and hybridized 6 arrays each with the two classes of probes, using cDNA microarrays in the 12-plex format (Design number 090505_Zv7_EXPR_HX12, Roche NimbleGen, Madison, WI). The periderm-enriched profile was thus generated from a single biological replicate (of about 200 animals) and 6 technical replicates. Based on technical replicates, the FDR-adjusted p values for all genes in the periderm profile were lower than 10−8. For the dnIrf6-inhibited profile, three cDNA synthesis reactions were performed on the pooled RNA, probes were generated from each cDNA preparation, and three arrays (technical replicates) were hybridized with each probe reaction. Differentially expressed genes were selected based on an FDR-adjusted p-value cutoff of 0.05, and a fold-change cutoff of 1.7. Because the periderm-enriched and dnIrf6-inhibited profiles were created from embryos at different ages, the periderm-enriched profile was filtered of all genes whose expression at 6 hpf in lacz-injected embryos was not above threshold (defined in Supplememental Methods) before overlap between the two profiles was evaluated.
Reporter experiments
Construction of reporters to test enhancer function of the human and mouse genomic elements is described in Supplemental Methods. At 24 hpf, 20% (20 of 97 injected embryos) of zebrafish embryos injected with hGRHL3_peak2:gfp exhibited patches of 10 or more GFP-positive periderm cells, whereas none of those injected with hGRHL3_peak1:gfp (30 injected embryos), or with the pGW_cfosEGFP vector lacking an insert (28 injected embryos), did so.
For both the wild-type Irf6RE and the Irf6RE variant, embryos were injected with 5 nl of plasmid (0.05 ng/μl) plus tol2 mRNA (25ng/μl) at the single-cell stage. Embryos at shield stage (6 hpf) were scored, using a Leica epifluorescence compound microscope, for GFP expression in greater than 10 contiguous periderm cells (mGrhl3intron15:gfp, 40%, n=79 embryos; 0%, intron15 ΔIrf6RE, n=91 embryos). A 5 μm step z-stack was taken and the “auto-blend” feature of Photoshop CS3 was used to merge it into a single image (Fig. 2F). We raised injected founders and identified two that transmitted GFP in the F1 generation. One exhibited high-level GFP expression throughout the periderm, like the transient transgenic embryos (shown in Fig 2H); the other exhibited GFP expression in facial periderm, in hatching gland cells, and in unidentified oral structures.
Supplementary Material
Figure S1 Western blot analysis of lysates of zebrafish eggs and embryos. Western blot analysis of zebrafish embryo lysates, gel loaded with lysates of embryos at the indicated stage, from uninjected or irf6 5′UTR MO-injected embryos as indicated. Anti-Irf6 immunoreactivity detected in lysates of unfertilized eggs and 2hpf (128-cell stage) embryos supports the presence of maternally-encoded Irf6 at these stages. Irf6 immunoreactivity in lysates of irf6 5′UTR MO-injected embryos is slightly reduced, supporting the specificity of the antisera. Blot was reprobed with anti-alpha-Tubulin to confirm similar levels of protein were loaded in each lane.
Figure S2 A morpholino targeting E4I4of grhl3 prevents proper splicing A. An antisense morpholino targeting E4I4of grhl3 (red) prevents proper splicing. Primers (green) were designed to lie in exon 4 and exon 6 of grhl3 (black). The larger splicing product (arrow) was cloned and sequenced to reveal that intron 4 (lower case) was not spliced from the transcript and contained a premature stop codon. cDNA synthesis was also performed with nuclease free water as a negative control (-RT). This cDNA template yielded no PCR products (not shown).
Supplemental Table S1 Periderm-enriched profile. These are all genes whose average expression (across technical replicates) in GFP-expressing cells was greater than 300 units (average expression among all probes was about 1500 units) and whose expression in GFP-expressing cells was at least 3 fold greater than in GFP-non-expressing cells. Genes that match the gene ontology (GO) term, “regulation of transcription, DNA dependent” (www.http://david.abcc.ncifcrf.gov/) are in bold. Note this GO term may not capture all transcription-factor-encoding genes.
Supplemental Table S2 dnIrf6-inhibited profile. These are genes whose average expression (across biological replicates) in control (lacZ-injected) embryos was significantly higher (FDR adj. p value < 0.05) than in dnIrf6-injected embryos.
Supplemental Table S3 The intersection of periderm-enriched and dnIrf6-inhibited profiles.
Acknowledgments
Supported by grants from March of Dimes (1-F08-412 to RAC) and the NIH (GM067841 to R.A.C., GM08399904 to D.W.H., R01GM77429 to D.S.W, T32 GM008629 and T32 GM082729 to J.L.W., and DE13513 to B.C.S). G.dlG and R.S. were supported by grants 5T32DC000040-17(PI: Bruce Gantz), and 5R37DE008559-21 (PI: Jeffery Murray). YAK was supported by 1F31DE022696-01. We are grateful to Adam Dupuy, Andrew Chalmers, Matthias Hammerschidt and Boggi Anderson for reagents.
Footnotes
The authors declare no conflicts of interest.
References
- Auden A, Caddy J, Wilanowski T, et al. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr Patterns. 2006;6(8):964–970. doi: 10.1016/j.modgep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Bailey CM, Abbott DE, Margaryan NV, et al. Interferon regulatory factor 6 promotes cell cycle arrest and is regulated by the proteasome in a cell cycle-dependent manner. Mol Cell Biol. 2008;28(7):2235–2243. doi: 10.1128/MCB.01866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkers J, Hild M, Kramer C, et al. Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev Cell. 2002;2(5):617–627. doi: 10.1016/s1534-5807(02)00163-6. [DOI] [PubMed] [Google Scholar]
- Biggs LC, Rhea L, Schutte BC, et al. Interferon regulatory factor 6 is necessary, but not sufficient, for keratinocyte differentiation. J Invest Dermatol. 2012;132(1):50–58. doi: 10.1038/jid.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boglev Y, Wilanowski T, Caddy J, et al. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Developmental biology. 2011;349(2):512–522. doi: 10.1016/j.ydbio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Botti E, Spallone G, Moretti F, et al. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1110931108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers AD, Strauss B, Papalopulu N. Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development. 2003;130(12):2657–2668. doi: 10.1242/dev.00490. [DOI] [PubMed] [Google Scholar]
- Desmyter L, Ghassibe M, Revencu N, et al. IRF6 Screening of Syndromic and a priori Non-Syndromic Cleft Lip and Palate Patients: Identification of a New Type of Minor VWS Sign. Mol Syndromol. 2010;1(2):67–74. doi: 10.1159/000313786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30(3):154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, et al. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312(5771):276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Fukazawa C, Santiago C, Park KM, et al. poky/chuk/ikk1 is required for differentiation of the zebrafish embryonic epidermis. Dev Biol. 2010 doi: 10.1016/j.ydbio.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele L, Ozato K. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 2007;18(5–6):503–510. doi: 10.1016/j.cytogfr.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Gong Z, Ju B, Wang X, et al. Green fluorescent protein expression in germ-line transmitted transgenic zebrafish under a stratified epithelial promoter from keratin8. Dev Dyn. 2002;223(2):204–215. doi: 10.1002/dvdy.10051. [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat Genet. 2006;38(11):1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke M, Renisch B, Hammerschmidt M. Zebrafish grainyhead-like1 is a common marker of different non-keratinocyte epidermal cell lineages, which segregate from each other in a Foxi3-dependent manner. Int J Dev Biol. 2010;54(5):837–850. doi: 10.1387/ijdb.092877mj. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JL, Canady JW, Brookes JT, et al. Wound complications after cleft repair in children with Van der Woude syndrome. J Craniofac Surg. 2010;21(5):1350–1353. doi: 10.1097/SCS.0b013e3181ec6aad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koillinen H, Lahermo P, Rautio J, et al. A genome-wide scan of non-syndromic cleft palate only (CPO) in Finnish multiplex families. J Med Genet. 2005;42(2):177–184. doi: 10.1136/jmg.2004.019646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koillinen H, Wong FK, Rautio J, et al. Mapping of the second locus for the Van der Woude syndrome to chromosome 1p34. Eur J Hum Genet. 2001;9(10):747–752. doi: 10.1038/sj.ejhg.5200713. [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32(2):285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev Cell. 2002;2(5):607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- Li Q, Frank M, Thisse CI, et al. Zebrafish: a model system to study heritable skin diseases. The Journal of investigative dermatology. 2011;131(3):565–571. doi: 10.1038/jid.2010.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little HJ, Rorick NK, Su LI, et al. Missense mutations that cause Van der Woude syndrome and popliteal pterygium syndrome affect the DNA-binding and transcriptional activation functions of IRF6. Hum Mol Genet. 2009;18(3):535–545. doi: 10.1093/hmg/ddn381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M’Boneko V, Merker HJ. Development and morphology of the periderm of mouse embryos (days 9–12 of gestation) Acta Anat (Basel) 1988;133(4):325–336. doi: 10.1159/000146662. [DOI] [PubMed] [Google Scholar]
- Martinelli M, Scapoli L, Pezzetti F, et al. Linkage analysis of three candidate regions of chromosome 1 in nonsyndromic familial orofacial cleft. Ann Hum Genet. 2001;65(Pt 5):465–471. doi: 10.1017/s000348000100882x. [DOI] [PubMed] [Google Scholar]
- McGaughey DM, Stine ZE, Huynh JL, et al. Asymmetrical distribution of nonconserved regulatory sequences at PHOX2B is reflected at the ENCODE loci and illuminates a possible genome-wide trend. BMC Genomics. 2009;10:8. doi: 10.1186/1471-2164-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey DM, Vinton RM, Huynh J, et al. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18(2):252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno LM, Arcos-Burgos M, Marazita ML, et al. Genetic analysis of candidate loci in non-syndromic cleft lip families from Antioquia-Colombia and Ohio. Am J Med Genet A. 2004;125A(2):135–144. doi: 10.1002/ajmg.a.20425. [DOI] [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo Iacono N, et al. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest. 2010;120(5):1570–1577. doi: 10.1172/JCI40267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehyba J, Hrdlickova R, Bose HR. Dynamic evolution of immune system regulators: the history of the interferon regulatory factor family. Molecular biology and evolution. 2009;26(11):2539–2550. doi: 10.1093/molbev/msp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegelow M, Peyrard-Janvid M, Zucchelli M, et al. Familial non-syndromic cleft lip and palate--analysis of the IRF6 gene and clinical phenotypes. Eur J Orthod. 2008;30(2):169–175. doi: 10.1093/ejo/cjm097. [DOI] [PubMed] [Google Scholar]
- Prescott NJ, Lees MM, Winter RM, et al. Identification of susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a two stage genome scan of affected sib-pairs. Hum Genet. 2000;106(3):345–350. doi: 10.1007/s004390051048. [DOI] [PubMed] [Google Scholar]
- Pyrgaki C, Liu A, Niswander L. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Developmental biology. 2011;353(1):38–49. doi: 10.1016/j.ydbio.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Jiang R, et al. Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum Mol Genet. 2009;18(14):2632–2642. doi: 10.1093/hmg/ddp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, et al. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006;38(11):1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- Rifat Y, Parekh V, Wilanowski T, et al. Regional neural tube closure defined by the Grainy head-like transcription factors. Developmental biology. 2010;345(2):237–245. doi: 10.1016/j.ydbio.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Sabel JL, d’Alencon C, O’Brien EK, et al. Maternal Interferon Regulatory Factor 6 is required for the differentiation of primary superficial epithelia in Danio and Xenopus embryos. Dev Biol. 2009;325(1):249–262. doi: 10.1016/j.ydbio.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nature genetics. 1999;22(4):356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Srichomthong C, Siriwan P, Shotelersuk V. Significant association between IRF6 820G->A and non-syndromic cleft lip with or without cleft palate in the Thai population. J Med Genet. 2005;42(7):e46. doi: 10.1136/jmg.2005.032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Kuliyev E, Wang X, et al. BMP4-dependent expression of Xenopus Grainyhead-like 1 is essential for epidermal differentiation. Development. 2005;132(5):1021–1034. doi: 10.1242/dev.01641. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, et al. Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J Clin Invest. 2010;120(5):1561–1569. doi: 10.1172/JCI40266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308(5720):411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- Wilanowski T, Caddy J, Ting SB, et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. The EMBO journal. 2008;27(6):886–897. doi: 10.1038/emboj.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Blanton SH, Hecht JT. Genetic causes of nonsyndromic cleft lip with or without cleft palate. Advances in oto-rhino-laryngology. 2011;70:107–113. doi: 10.1159/000322486. [DOI] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351(8):769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Western blot analysis of lysates of zebrafish eggs and embryos. Western blot analysis of zebrafish embryo lysates, gel loaded with lysates of embryos at the indicated stage, from uninjected or irf6 5′UTR MO-injected embryos as indicated. Anti-Irf6 immunoreactivity detected in lysates of unfertilized eggs and 2hpf (128-cell stage) embryos supports the presence of maternally-encoded Irf6 at these stages. Irf6 immunoreactivity in lysates of irf6 5′UTR MO-injected embryos is slightly reduced, supporting the specificity of the antisera. Blot was reprobed with anti-alpha-Tubulin to confirm similar levels of protein were loaded in each lane.
Figure S2 A morpholino targeting E4I4of grhl3 prevents proper splicing A. An antisense morpholino targeting E4I4of grhl3 (red) prevents proper splicing. Primers (green) were designed to lie in exon 4 and exon 6 of grhl3 (black). The larger splicing product (arrow) was cloned and sequenced to reveal that intron 4 (lower case) was not spliced from the transcript and contained a premature stop codon. cDNA synthesis was also performed with nuclease free water as a negative control (-RT). This cDNA template yielded no PCR products (not shown).
Supplemental Table S1 Periderm-enriched profile. These are all genes whose average expression (across technical replicates) in GFP-expressing cells was greater than 300 units (average expression among all probes was about 1500 units) and whose expression in GFP-expressing cells was at least 3 fold greater than in GFP-non-expressing cells. Genes that match the gene ontology (GO) term, “regulation of transcription, DNA dependent” (www.http://david.abcc.ncifcrf.gov/) are in bold. Note this GO term may not capture all transcription-factor-encoding genes.
Supplemental Table S2 dnIrf6-inhibited profile. These are genes whose average expression (across biological replicates) in control (lacZ-injected) embryos was significantly higher (FDR adj. p value < 0.05) than in dnIrf6-injected embryos.
Supplemental Table S3 The intersection of periderm-enriched and dnIrf6-inhibited profiles.