Abstract
Developing a detailed understanding of enzyme function in the context of an intracellular signal transduction pathway requires minimally invasive methods for probing enzyme activity in situ. Here, we describe a new method for monitoring enzyme activity in living cells by sandwiching live cells between two vertical silicon nanowire (NW) arrays. Specifically, we use the first NW array to immobilize the cells, and then present enzymatic substrates intracellularly via the second NW array by utilizing the NWs’ ability to penetrate cellular membranes without affecting cells’ viability or function. This strategy, when coupled with fluorescence microscopy and mass spectrometry, enables intracellular examination of protease, phosphatase and protein kinase activities, demonstrating the assay’s potential in uncovering the physiological roles of various enzymes.
Keywords: Silicon nanowire, sandwich assembly, live cell assay, enzyme activity
Enzymes mediate a wide range of cellular processes, and dynamic control over their activities is crucial for proper signal transduction. Traditionally, enzyme activity has been studied in vitro in tissue homogenates or with purified proteins(1, 2) using spectrophotometric, calorimetric, chromatographic, radiometric, or mass spectroscopic detection (See(3-5) for review papers). While these methods provide powerful tools for characterizing enzymes, sometimes down to the single molecule level,(6-10) they do so by separating the enzymes from their actual biological contexts. In reality, enzymatic behavior may be different in the cellular context because multiple cellular components and extracellular stimuli, absent in vitro, may modulate endogenous enzyme activity.(11) Clearly, cell-based assays that can accurately monitor enzyme behavior would be highly beneficial in uncovering the true physiological functions of enzymes.
To date, most of cell-based assays have been based upon fluorescence measurements.(11-20) For instance, fluorogenesis from substrate-quencher conjugates(21-23) as well as fluorescence enhancement due to conformational changes(24) have been widely employed to visualize and quantify molecular processes. Employing strategies based on fluorescence resonance energy transfer (FRET), a variety of serine/threonine and tyrosine kinases and small GTPases have been monitored in living cells with high spatiotemporal resolution.(25-27) Unfortunately, cell-to-cell variations in the expression levels of genetically encoded probe molecules, as well as the background signal from the unpaired probes, have presented problems in these studies.(20, 28) Moreover, simultaneous monitoring of multiple enzyme activities in the same cell has been difficult, although recent studies demonstrated the use of two FRET pairs for observing two different enzyme activities at the same time.(29, 30)
Nanomaterials with well-defined morphologies are emerging as a promising platform for interfacing with biological systems.(31-42) In particular, vertical silicon nanowires (NWs), when used as a culture substrate, have been demonstrated to provide direct physical access to the cells’ interior without affecting their viability or function (Figure 1a). The same NW platform has also been utilized to efficiently introduce a variety of foreign biomolecules into living cells.(31, 36, 43) While our NW platform permits cell membrane penetration, thereby mediating delivery of foreign molecules, prolonged culturing of cells on nanopillars have resulted in cell membrane wrapping,(37, 40) implying the dependence of behaviors of cells interfaced with nanostructures on their physical properties as well as contact duration.
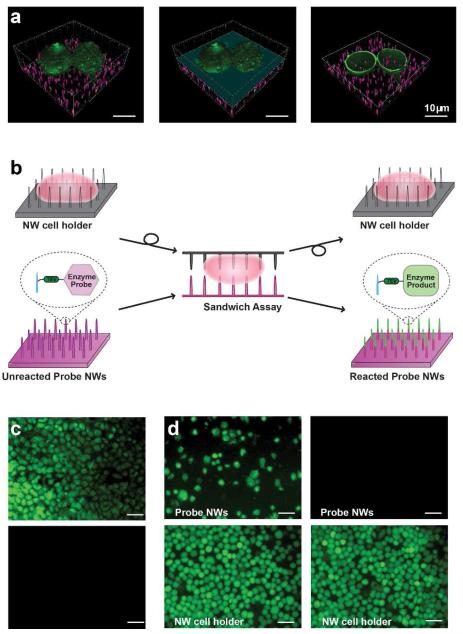
Figure 1.
(a) Confocal microscope images of membrane-labeled HeLa cells (green) on fluorescently labeled NWs (magenta). Removing the top half of the cell on NWs (left panel), as indicated by the light blue plane (middle panel), reveals NWs that reside inside the cell (right panel). Scale bar, 10 μm. (b) Schematic of the NW-cell sandwich assembly. (c) Covalent tethering of peptides to NWs prevents errant release. Fluorescein-labeled peptide molecules were either electrostatically tethered (top) or covalently attached (bottom) to probe NWs and the degree of their release into sandwiched cells was assessed after an overnight sandwich incubation. Electrostatically bound peptides were readily delivered into the cytoplasm of cells on the NW cell holder while covalently bound peptides were not transferred. Scale bar, 50 μm. (d) Validation of cell immobilization on a NW cell holder. HeLa cells, seeded in excess on the NW cell holder, were either untreated (left) or treated briefly with a TrypLE Express solution (right). After overnight sandwich incubation and disassembly, both samples were treated with CFDA for cell visualization. Weakly bound, multilayered cells were transferred from the NW cell holder to probe NWs (upper-left), whereas all cells stayed on the NW cell holder in the TrypLE Express-treated sample (note absence of cells, upper-right). Scale bar, 50 μm.
Here we report a new, minimally invasive method for monitoring endogenous enzyme activity in living cells by leveraging the ability of NWs to access the cell cytoplasm. The essence of our live-cell assay is to use NWs to present covalently attached enzymatic substrates directly inside living cells so that these molecules interact with cytoplasmic enzymes. To facilitate the cell detachment from these “probe NWs” without cell lysis after a well-defined interaction period, we use another set of NWs as a “NW cell holder” to immobilize the cells (Figure 1b). We expose the enzymatic substrates to living cells by “sandwiching” the cells between the probe NWs and the NW cell holder. Once the NW cell holder is withdrawn by disassembling this NW-cell sandwich, changes induced to the enzyme substrates are examined using conventional analysis tools, such as fluorescence microscopy and mass spectrometry. We show that this strategy enables activity measurements of several endogenous enzymes in living cells, demonstrating the assay’s potential for probing enzyme function in situ.
We first characterized the feasibility of this strategy and optimized its implementation. We used the sandwich assay format in which the NW cell holder was placed on top of the probe NWs (Figure 1b) because the inverse configuration was found to lead to inefficient cellular impalement by the substrate-presenting NWs. To confirm that the weight of the NW cell holder (22 mg, 5 × 5 mm, 8.6 Pa, 1.4 × 10−4 Pa/cell) did not destroy the cells underneath, we examined cell viability and membrane integrity using carboxyfluorescein diacetate (CFDA) and propidium iodide (PI). CFDA becomes fluorescent carboxylates inside cells through the action of active cellular esterase, and gets trapped within live cells.(44) The PI molecule, on the other hand, stains the cell nuclei only when the cell membranes are damaged.(44) HeLa cells on NWs, whether sandwiched or not, showed positive CFDA fluorescence with no nuclear PI staining (See Supporting Information (SI) Figure S1), indicating that sandwiching affected neither cell viability nor membrane integrity. In addition, we have performed the quantitative real-time polymerase chain reaction (PCR) analyses of for sandwiched HeLa cells for 4 hrs, and the transcript levels of five common housekeeping genes were similar to those in cells cultured on NWs without sandwich incubation (SI Figure S2). Moreover, when cells on NWs after the sandwich incubation for 4 hours and disassembly were cultured for three more days, they appeared to grow at a rate similar to non-sandwiched cells on NWs (data not shown), indicating that sandwiched cells can proliferate normally.
Next, we optimized the NW surface modification protocol to ensure that the enzyme substrates (typically peptides) tethered to the probe NWs were neither removed nor damaged during sandwiching. When we attached fluorescein-labeled peptides to the probe NWs via weak electrostatic binding, HeLa cells exhibited bright intracellular fluorescence after overnight incubation inside a NW-cell sandwich (Figure 1c, upper), indicating the intracellular release of these peptides, as previously observed.(36) To prevent such release, we covalently attached fluorescein-labeled peptides to probe NWs using cysteine-maleimide chemistry (see SI for details). In this case, even after overnight incubation, HeLa cells did not show any significant cellular fluorescence after sandwich disassembly (Figure 1c, bottom). This observation proves that covalently attached peptides were both firmly anchored during our sandwich incubation and not degraded randomly by cellular enzymes. The integrity of the peptide incubated in a sandwich was further confirmed by mass spectrometric analysis (data no shown). Intriguingly, when we explored surface modification more closely by employing gold nanoparticles (AuNPs) that can be visualized by SEM, we found that a majority of AuNPs bound to NWs, particularly to their upper parts (SI Figure S3), indicating preferential modification of the NW tops. This result implies that the top parts of the NWs might be more oxidized, leading to differential surface reactivity.
We also developed experimental protocols for preventing transfer of cells from our NW cell holder to probe NWs. First, we co-coated our NW cell holder with both 3-aminopropyltrimethoxysilane (APTMS) and fibronectin to improve cellular adhesion (see SI). Second, we increased the length of both of the cell-holding NWs and the probe NWs from 1-2 μm to 3-5 μm (typical height of HeLa cells on the NW surface is about 5-8 μm) to tightly attach the cells on the NW.(45) It should also be noted that utilized NWs were slightly tapered with diameters of 200-400 nm at their bottoms and ~100 nm at their tops, and arrayed at ~100 NWs per 400 μm2. In the case of a HeLa cell whose surface area is approximately 300-400 μm2, 70-100 NWs are expected to be in contact with each single cell. Finally, and most importantly, we covered NW cell holders with a densely packed monolayer of cells by plating 3-4 fold more cells than those required for 100% confluency in a given area and treated them with a TrypLE Expresssolution supplied as 1X to remove loosely adhered cells. After this treatment, no cell transfer was observed from the NW cell holder to probe NWs even after an overnight sandwich incubation (Figure 1d, upper-right column. The upper-left column in Figure 1d shows the control experiment without a TrypLE Express solution treatment for comparison).
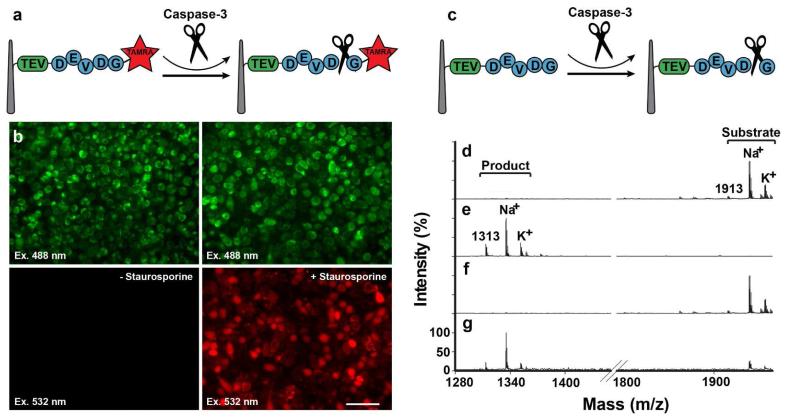
Having established the experimental protocols for the sandwich assay, we used it to measure the activity of endogenous caspase-3, an apoptotic protease that plays an essential role in both apoptosis and inflammation.(46) Since caspase-3 recognizes the peptide sequence, Asp-Glu-Val-Asp (DEVD) with cleavage occurring on the second D residue,(46, 47) we constructed a caspase-3 substrate peptide that contains the DEVD sequence with a 5-carboxytetramethylrhodamine (TAMRA: fluorescent label) linked at the N terminus (Figure 2, see Table S1 for the entire sequence). In this assay, active caspase-3 should result in red fluorescent TAMRA being released into HeLa cells sandwiched between NWs. To induce caspase-3 in our samples, we incubated the NW-cell sandwich in the presence of an apoptosis-inducing reagent staurosporine for 4 hours (see SI). As expected, when the NW cell was disassembled and the cells were examined by fluorescence microscopy, TAMRA fluorescence was only seen in HeLa cells cultured in the presence of staurosporine (Figure 2b, bottom-right column). It should be noted that not all assayed cells exhibited TAMRA fluorescence – although this could be the result of individual cells responding differently to staurosporin on this short time scale,(47) we cannot exclude the possibility that some cells were not penetrated by NWs. Importantly, the retention of TAMRA fluorescence indicates the maintenance of cell membrane integrity under our sandwich condition and proves that the sandwich assembly neither affected cell viability nor caused any significant apoptotic behavior.
Figure 2.
Assaying caspase-3 activity using a NW-cell sandwich assembly. (a-b) fluorescence-based caspase-3 assay. (a) Assay schematic. (b) TAMRA-labeled substrate peptides were covalently attached to the probe NWs and sandwiched onto HeLa cells immobilized on the NW cell holder. This assembly was then incubated either without (left) or with staurosporine (right), followed by examination for peptide release into the cell cytoplasm by fluorescence microscopy (top panels: all cells visualized by using autofluorescence due to laser excitation at 488 nm; bottom panels: TAMRA-peptide fragments visualized by excitation at 532 nm). (c-g) A mass spectrometry-based caspase-3 assay. (c) Assay schematic. Caspase-3 substrate peptide molecules were covalently attached to the probe NWs, and treated (d) without or (e) with purified caspase 3. Bound peptides were subsequently released with TEV and analyzed by MALDI-TOF (Voyager analyzer). Substrate-containing probe NWs were sandwich-assembled with HeLa cells on the NW cell holder, which was incubated either (f) without or (g) with staurosporine, and processed for TEV cleavage and MALDI-TOF analysis.
To complement the fluorescence experiments and extend the utility of our technique, we then developed an experimental protocol for mass spectrometry-based analyses. In these experiments, once the sandwich culture was completed, we disassembled the NW-cell sandwich and analyzed the enzyme substrates attached to probe NWs using a matrix-assisted laser desorption/ionization (MALDI)-time-of-flight (TOF) mass spectrometer. Specifically for the caspase-3 sandwich assay, we modified the previously reported caspase-3 substrate peptide,(47) by inserting a Tobacco etch virus (TEV) protease recognition site (Figure 2c, see Table S1 for the entire sequence) – this site is used to remove the covalent linkage between the probe NWs and enzyme substrates to facilitate the mass spectrometric analysis. Unreacted caspase-3 substrate, when examined by mass spectrometry after TEV cleavage, had a mass peak at 1913 Da, with two more peaks arising from cationic adducts at 1935 Da (Na+) and 1961 Da (K+) (Figure 2d). Treatment with purified caspase-3 in vitro caused a mass shift to 1313 Da with cationic adduct peaks at 1335 Da (Na+) and 1351 Da (K+) (Figure 2e). When we performed a sandwich assay with HeLa cells, we observed unmodified substrate mass peaks (Figure 2f) similar to those obtained in the unreacted sample (Figure 2d). After the staurosporine treatment for 4 hours to activate caspase-3, however, we detected decreased molecular mass peaks as expected (with a small amount of unreacted forms, Figure 2g). Collectively, these results not only confirmed our fluorescence studies but also validated the utility of combining mass-spectrometry analysis with our sandwich assay.
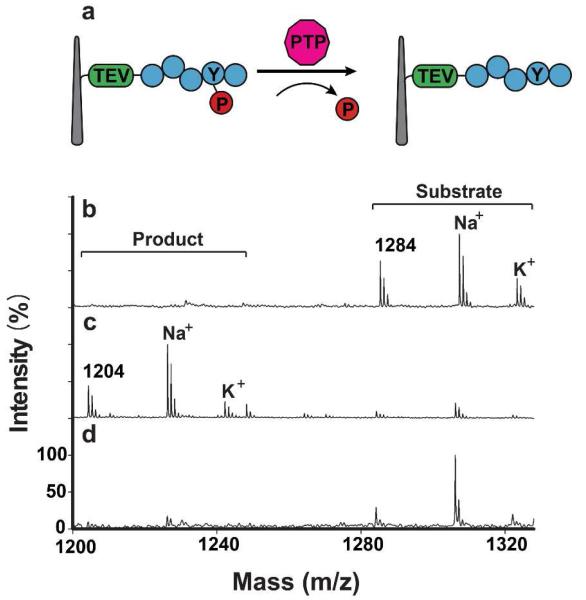
Next, we investigated the endogenous activity of protein tyrosine phosphatases (PTPs) that are always functional in HeLa cells even in the absence of external stimuli. PTPs remove phosphate groups from phosphorylated tyrosine residues on proteins, thereby controlling phosphorylated states of many important signaling proteins and modulating their activity.(28) Using a general peptide substrate that could be modified by any kind of cellular PTP (see Table S1 for the entire sequence), we performed the same mass spectrometry-based analysis after a one-hour-long sandwich assay (Figure 3a). Unreacted substrates exhibited mass peaks at 1284 Da with two cationic adduct peaks (1307 Da and 1323 Da for Na+ and K+ adducts, respectively) (Figure 3b). After the sandwich assay, however, the mass spectra exhibited three new major peaks corresponding to the de-phosphorylated form of the attached peptide and its two cationic adducts (1204 Da, 1227 Da and 1243 Da) (Figure 3c). We confirmed that this reaction was mediated by PTPs by pretreating a set of sandwiched cells with two PTP inhibitors, α-bromo-4-hydroxyacetophenone and sodium orthovanadate (see SI); in the presence of these inhibitors, we observed little dephosphorylated peaks (Figure 3d). We further performed 5 independent PTP sandwich assays with independently prepared NWs and HeLa cells (SI Figure S4). The results were highly reproducible, yielding the average conversion from the substrate to the dephosphorylated product to be 82.7 ± 1.9% (S.D.). In addition, when we attempted to measure endogenous protein kinase activity, PKCδ among various protein kinases was found to show a small but significant activity in unstimulated HeLa cells (SI Figure S5).
Figure 3.
(a) Schematic for a PTP activity assay using a NW-cell sandwich assembly. (b-d) MALDI-TOF spectra for: (b) substrate peptides cleaved by TEV; (c) peptides cleaved from NWs by TEV after an hour of sandwich assembly with immobilized HeLa cells; and, (d) peptides cleaved by TEV after an hour-long sandwich assay with the inclusion of the PTP inhibitors, α-bromo-4-hydroxyacetophenone (80 μM) and sodium orthovanadate (200 μM). Substrate and product peptides show three distinct mass peaks including two adducts (Na+ and K+), respectively.
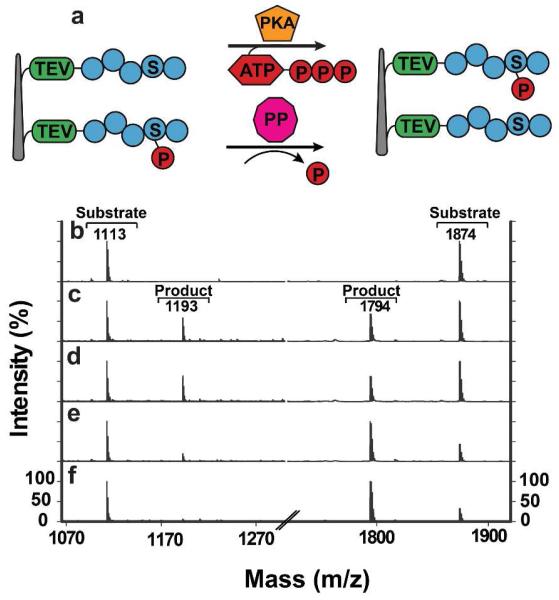
Finally, we used the sandwich assay to probe the time dependent activities of protein kinases that play critical roles in regulating cellular signal transduction pathways and are implicated in many human diseases.48 One particular kinase that we studied is protein kinase A (PKA) that is known to be activated by cAMP in a spatiotemporal manner.(11, 49) To study PKA activity, we attached two peptide substrates to our probe NWs: one containing a PKA-sensitive sequence (refer to Table S1 for the entire sequence) that we optimized for maximal activity in vitro using purified PKA (data not shown), and a second containing a pre-phosphorylated PKA-sensitive sequence (Table S1 for the entire sequence) that could be dephosphorylated by serine/threonine phosphatases (Figure 4a). We then applied our sandwich assay to PC3 cells by treating them with forskolin that raises intracellular cAMP levels. Upon addition of forskolin, sandwiched samples were incubated for various time periods (0, 15, 30 and 60 minutes) to monitor the enzyme’s kinetics. As shown in Figure 4c, after 15-minute incubation, the peptide substrates displayed a molecular mass increase or decrease of 80 Da, suggesting a phosphorylated or dephosphorylated peptide product, respectively (conversion percentages: 36.9% for the PKA substrate and 40.5% for the phosphatase substrate). Although the same pattern of activity was observed for a 30-minute treatment (Figure 4d), the phosphorylated peak for the PKA substrate almost vanished for the 60 minute time point (Figure 4e), concomitant with a dramatic increase of the dephosphorylated peak for the phosphatase substrate (to the level similar to the forskolin untreated sample, Figure 4f). Intriguingly, when stimulated by forskolin, PC3 cells showed elevated cAMP levels by 15 minutes that persisted through 60 minutes (SI Figure S6). Together, these data suggest that, despite continuously elevated cAMP levels, the interplay between PKA and phosphatases controls the duration and magnitude of PKA signaling.
Figure 4.
Assaying the time course of PKA activity. (a) Schematic for the PKA assay based on PKA substrate and phosphatase substrate peptides. (b-e) A sandwich assembly using PC3 cells was incubated in the presence of a PKA inducer forskolin, followed by MALDI-TOF analysis. The time dependence of forskolin’s effect on PKA activation was monitored at (b) 0, (c) 15, (d) 30 and (e) 60 minutes post forskolin addition. (f) A sandwich assembly using PC3 cells was incubated in the absence of forskolin for 60 minutes. A phosphorylated product peak appeared temporarily, but disappeared afterwards, suggesting transient kinase activation.
Here, we describe a novel strategy for detecting endogenous enzyme activity in live cells by using NW arrays to both immobilize cells and introduce exogenous enzymatic substrates. This sandwich assay is compatible with both fluorescence- and mass spectrometry-based readouts. In particular, the use of mass spectrometry offers many distinct advantages: it overcomes many of the limitations of fluorescence-based methods (e.g., high background, low dynamic range, transfection difficulties, etc.), and permits massively parallel monitoring of enzyme activities via the design of multiple peptide substrates linked to a single set of NWs. Since enzyme kinetics and its dependence on inducers/inhibitors can be readily probed using our methodology, we anticipate this sandwich assay to be a useful tool for screening the effects of drugs on living cells. Moreover, by integrating NW-assisted biomolecule delivery with our live-cell enzyme activity assay, perturbations and observation of cellular signal transduction could be achieved, enabling unprecedented studies of enzymatic function in situ.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Proteogenomics Research Program through the National Research Foundation of Korea funded by the Korean Ministry of Education, Science and Technology (EGY), the KIST grant (EGY), NIH Pioneer award (HP, 5DP1OD003893-03), and NIH CEGS Award (HP, 1P50HG006193-01).
Footnotes
Supporting Information
Experimental details including NW fabrication, cell culture methods, peptide substrate sequences, cell viability testing protocols, fluorescence imaging, and MALDI-TOF measurements. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1).Min DH, Su J, Mrksich M. Angewandte Chemie-International Edition. 2004;43(44):5973–5977. doi: 10.1002/anie.200461061. [DOI] [PubMed] [Google Scholar]
- 2).Parker L, Engel-Hall A, Drew K, Steinhardt G, Helseth DL, Jabon D, McMurry T, Angulo DS, Kron SJ. Journal of Mass Spectrometry. 2008;43(4):518–527. doi: 10.1002/jms.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Enzyme Assays: A Practical Approach. Oxford University Press, Inc.; New York: 2002. [Google Scholar]
- 4).Liesener A, Karst U. Anal Bioanal Chem. 2005;382(7):1451–1464. doi: 10.1007/s00216-005-3305-2. [DOI] [PubMed] [Google Scholar]
- 5).Greis KD. Mass Spectrom Rev. 2007;26(3):324–339. doi: 10.1002/mas.20127. [DOI] [PubMed] [Google Scholar]
- 6).Kim S, Blainey PC, Schroeder CM, Xie XS. Nat Methods. 2007;4(5):397–399. doi: 10.1038/nmeth1037. [DOI] [PubMed] [Google Scholar]
- 7).Gorris HH, Rissin DM, Walt DR. Proc Natl Acad Sci U S A. 2007;104(45):17680–17685. doi: 10.1073/pnas.0705411104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. Nature. 2011;473(7348):484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Lamboy JA, Kim H, Lee KS, Ha T, Komives EA. Proc Natl Acad Sci U S A. 2011;108(25):10178–10183. doi: 10.1073/pnas.1102226108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Lee G, Yoo J, Leslie BJ, Ha T. Nat Chem Biol. 2011;7(6):367–374. doi: 10.1038/nchembio.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Kovarik ML, Allbritton NL. Trends Biotechnol. 2011;29(5):222–230. doi: 10.1016/j.tibtech.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Hobson JP, Liu SH, Rono B, Leppla SH, Bugge TH. Nature Methods. 2006;3(4):259–261. doi: 10.1038/nmeth862. [DOI] [PubMed] [Google Scholar]
- 13).Zhang J, Allen MD. Molecular Biosystems. 2007;3(11):759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 14).Terai T, Nagano T. Curr Opin Chem Biol. 2008;12(5):515–521. doi: 10.1016/j.cbpa.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 15).Frommer WB, Davidson MW, Campbell RE. Chemical Society Reviews. 2009;38(10):2833–2841. doi: 10.1039/b907749a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Gershenson A. Curr Opin Chem Biol. 2009;13(4):436–442. doi: 10.1016/j.cbpa.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 17).Heller DA, Jin H, Martinez BM, Patel D, Miller BM, Yeung TK, Jena PV, Hobartner C, Ha T, Silverman SK, Strano MS. Nat Nanotechnol. 2009;4(2):114–120. doi: 10.1038/nnano.2008.369. [DOI] [PubMed] [Google Scholar]
- 18).Edgington LE, Verdoes M, Bogyo M. Current Opinion in Chemical Biology. 2011;15(6):798–805. doi: 10.1016/j.cbpa.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Kardash E, Bandemer J, Raz E. Nature Protocols. 2011;6(12):1835–1846. doi: 10.1038/nprot.2011.395. [DOI] [PubMed] [Google Scholar]
- 20).Okumoto S, Jones A, Frommer WB. Annu Rev Plant Biol. 2012;63(1):663–706. doi: 10.1146/annurev-arplant-042110-103745. [DOI] [PubMed] [Google Scholar]
- 21).Sharma V, Wang Q, Lawrence DS. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2008;1784(1):94–99. doi: 10.1016/j.bbapap.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Hu M, Li L, Wu H, Su Y, Yang PY, Uttamchandani M, Xu QH, Yao SQ. J Am Chem Soc. 2011;133(31):12009–12020. doi: 10.1021/ja200808y. [DOI] [PubMed] [Google Scholar]
- 23).Myochin T, Hanaoka K, Komatsu T, Terai T, Nagano T. J Am Chem Soc. 2012;134(33):13730–13737. doi: 10.1021/ja303931b. [DOI] [PubMed] [Google Scholar]
- 24).Lawrence DS, Wang QZ. Chembiochem. 2007;8(4):373–378. doi: 10.1002/cbic.200600473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Kiyokawa E, Hara S, Nakamura T, Matsuda M. Cancer Sci. 2006;97(1):8–15. doi: 10.1111/j.1349-7006.2006.00141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Aoki K, Kiyokawa E, Nakamura T, Matsuda M. Philos Trans R Soc Lond B Biol Sci. 2008;363(1500):2143–2151. doi: 10.1098/rstb.2008.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Seong JY, Ouyang MX, Kim T, Sun J, Wen PC, Lu SY, Zhuo Y, Llewellyn NM, Schlaepfer DD, Guan JL, Chien S, Wang YX. Nature Communications. 2011;2(406):1–9. doi: 10.1038/ncomms1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Zhang ZY. Annu Rev Pharmacol Toxicol. 2002;42:209–234. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 29).Wu XL, Simone J, Hewgill D, Siegel R, Lipsky PE, He LS. Cytometry Part A. 2006;69A(6):477–486. doi: 10.1002/cyto.a.20300. [DOI] [PubMed] [Google Scholar]
- 30).Ouyang M, Huang H, Shaner NC, Remacle AG, Shiryaev SA, Strongin AY, Tsien RY, Wang Y. Cancer Res. 2010;70(6):2204–2212. doi: 10.1158/0008-5472.CAN-09-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Kim W, Ng JK, Kunitake ME, Conklin BR, Yang P. J Am Chem Soc. 2007;129(23):7228–7229. doi: 10.1021/ja071456k. [DOI] [PubMed] [Google Scholar]
- 32).Hallstrom W, Martensson T, Prinz C, Gustavsson P, Montelius L, Samuelson L, Kanje M. Nano Letters. 2007;7(10):2960–2965. doi: 10.1021/nl070728e. [DOI] [PubMed] [Google Scholar]
- 33).Park J, Bauer S, von der Mark K, Schmuki P. Nano Letters. 2007;7(6):1686–1691. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 34).Cohen-Karni T, Timko BP, Weiss LE, Lieber CM. Proc Natl Acad Sci U S A. 2009;106(18):7309–7313. doi: 10.1073/pnas.0902752106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Liu YC, Rieben N, Iversen L, Sorensen BS, Park J, Nygard J, Martinez KL. Nanotechnology. 2010;21(24):245105. doi: 10.1088/0957-4484/21/24/245105. [DOI] [PubMed] [Google Scholar]
- 36).Shalek AK, Robinson JT, Karp ES, Lee JS, Ahn DR, Yoon MH, Sutton A, Jorgolli M, Gertner RS, Gujral TS, MacBeath G, Yang EG, Park H. Proc Natl Acad Sci U S A. 2010;107(5):1870–1875. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Xie C, Hanson L, Xie W, Lin Z, Cui B, Cui Y. Nano Lett. 2010;10(10):4020–4024. doi: 10.1021/nl101950x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Park SY, Park J, Sim SH, Sung MG, Kim KS, Hong BH, Hong S. Advanced Materials. 2011;23(36):H263–H267. doi: 10.1002/adma.201101503. [DOI] [PubMed] [Google Scholar]
- 39).Yu MR, Huang Y, Ballweg J, Shin H, Huang MH, Savage DE, Lagally MG, Dent EW, Blick RH, Williams JC. ACS Nano. 2011;5(4):2447–2457. doi: 10.1021/nn103618d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Hanson L, Lin ZC, Xie C, Cui Y, Cui B. Nano Lett. 2012;12(11):5815–5820. doi: 10.1021/nl303163y. [DOI] [PubMed] [Google Scholar]
- 41).Kim DJ, Seol JK, Lee G, Kim GS, Lee SK. Nanotechnology. 2012;23(39):395102. doi: 10.1088/0957-4484/23/39/395102. [DOI] [PubMed] [Google Scholar]
- 42).VanDersarl JJ, Xu AM, Melosh NA. Nano Letters. 2012;12(8):3881–3886. doi: 10.1021/nl204051v. [DOI] [PubMed] [Google Scholar]
- 43).Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, Gat-Viks I, Tonti E, DeGrace MM, Clauser KR, Garber M, Eisenhaure TM, Yosef N, Robinson J, Sutton A, Andersen MS, Root DE, von Andrian U, Jones RB, Park H, Carr SA, Regev A, Amit I, Hacohen N. Cell. 2011;147(4):853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Dive C, Watson JV, Workman P. Cytometry. 1990;11(2):244–252. doi: 10.1002/cyto.990110205. [DOI] [PubMed] [Google Scholar]
- 45).Shan Y, Hao X, Shang X, Cai M, Jiang J, Tang Z, Wang H. Chemical Communications. 2011;47(12):3377–3379. doi: 10.1039/c1cc00040c. [DOI] [PubMed] [Google Scholar]
- 46).Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Cell Death Differ. 2007;14(1):44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- 47).Su J, Rajapaksha TW, Peter ME, Mrksich M. Anal Chem. 2006;78(14):4945–4951. doi: 10.1021/ac051974i. [DOI] [PubMed] [Google Scholar]
- 48).Kaidanovich-Beilin O, Eldar-Finkelman H. Physiology (Bethesda) 2006;21:411–418. doi: 10.1152/physiol.00022.2006. [DOI] [PubMed] [Google Scholar]
- 49).Zhang J, Ma Y, Taylor SS, Tsien RY. Proc Natl Acad Sci U S A. 2001;98(26):14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.