Abstract
We have developed a multiplex reporter system to monitor multiple biological variables in real-time. The secreted Gaussia luciferase was fused to ten different epitope tags (Gluctag), each expressed in different tumor cells. By immunobinding of the tags followed by Gluctag detection, this system allowed the independent and real-time monitoring of mixed cell cultures in vitro and of mixed subcutaneous and intracranial tumor subpopulations in vivo.
To study complex processes in systems biology, tools are needed that give insight into multiple molecular and cellular interactions simultaneously and in real-time1,2. Several cellular reporter systems are in use to shed light on biological processes, many based on luciferase enzymes and their photon production upon luciferin substrate addition, including Firefly luciferase (Fluc)3, Renilla luciferase (Rluc)4, and Gaussia luciferase (Gluc)5,6. These bioluminescent reporters have been used to monitor biological processes such as cellular proliferation and differentiation7, transcription factor activity8, translational repression by miRNA9,10, and intercellular interactions11, reviewed in12. Other commonly used reporters are based on the green fluorescent protein (GFP), the red fluorescent protein (RFP) and, more recently, near infra-red fluorescent proteins13,14 which can be multiplexed together to monitor several processes simultaneously using spectral unmixing in conjunction with fluorescence molecular tomography15. These reporters have several disadvantages including low sensitivity due to autofluorescence, complexity, and the requirement of expensive instrumentation. Further, similar to bioluminescence, limited numbers of variables can be monitored simultaneously, limiting their use in multiplex screening assays and cellular heterogeneity studies. Here, we developed a multiplex bioluminescence assay that allows monitoring of many variables in real-time within the same biological system. This assay is based on Gluc since this reporter is highly sensitive, naturally secreted, and can be detected in the conditioned medium of cells in culture as well as in the blood of animals ex vivo, allowing real-time monitoring of cellular variables5,6.
Results
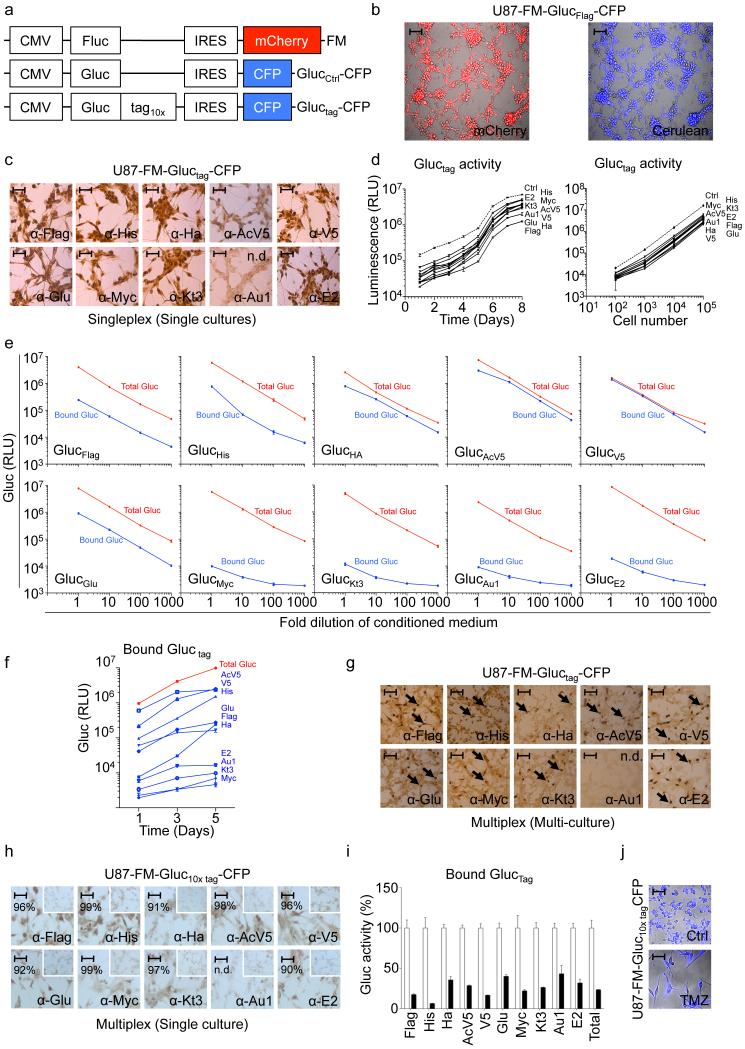
We constructed a library of lentiviral vectors consisting of the Gluc cDNA fused to ten different epitope tags at its C-terminus, under the constitutively active cytomegalovirus (CMV) promoter resulting in GlucFlag, GlucHis, GlucHA, GlucAcV5, GlucV5, GlucGlu, GlucMyc, GlucKt3, GlucAu1, GlucE2, and the control reporter construct without tag, GlucCtrl. These lentiviral vectors also express cerulean fluorescent protein (CFP) separated from Gluctag by an internal ribosomal entry site (IRES) element as a marker for transduction efficiency (Fig. 1a and Methods section). We employed human U87 glioblastoma cells stably expressing Fluc and the mCherry fluorescent protein6 (U87-FM) for validation of the Gluctag multiplex system. These cells were transduced with each of the lentiviral vectors to produce ten different cell lines, each stably expressing a different Gluctag and CFP (U87-FM-Gluctag-CFP). Fluorescent microscopy analysis for mCherry and CFP showed that these cells were nearly 100% transduced with the reporter vectors (Fig. 1b). We first demonstrated that all of the individual U87-FM-Gluctag-CFP cells expressed the corresponding tags by immunostaining using specific antibodies, except for GlucAu1 that was not detectable by immunostaining attributed to anti-Au1 antibody properties (Fig. 1c). As a control we stained cells expressing Glucctrl with the different tag antibodies, which did not show any staining (data not shown). We confirmed Gluctag expression by Western blot using anti-tag antibodies (Sup. Fig. 1a) and RT-PCR of the tag region (Sup. Fig. 1b and c). Next, we determined the effect of the tags on Gluc by measuring luciferase activity at different time points in an aliquot of conditioned medium from U87-FM cells expressing each Gluctag reporter construct using the Gluc substrate coelenterazine. We demonstrated that fusion of epitope tags to Gluc does not dramatically affect the expression, secretion or activity of Gluc (Fig. 1d and supplemental Fig. 1a–c). We then used the conditioned media containing the different Gluctag constructs to optimize antibody-tag immunobinding in a 96-well plate format (Methods). Plates were first coated with monoclonal antibodies against the different tags and incubated with different amounts of conditioned medium allowing Gluctag immunobinding. Subsequently, the wells were washed to remove unbound Gluctag and subjected to the Gluc assay by direct addition of coelenterazine to the wells and measuring photon counts using a plate luminometer. As a control, an aliquot of conditioned medium was assayed directly for total Gluc activity. We determined that six of the ten individual Gluctag constructs, i.e. GlucFlag, GlucHis, GlucHA, GlucAcV5, GlucV5, GlucGlu, reported with high-efficiency after immunobinding in the antibody-coated wells, as compared to total Gluctag activity (Fig. 1e). The GlucMyc, GlucKt3, GlucAu1, and GlucE2 reporters showed lower activity upon immunobinding, attributed to suboptimal antibody-tag interactions. In order to demonstrate the use of the Gluctag system for multiplex applications, we mixed equal numbers of the ten different U87-FM-Gluctag-CFP cells and plated them in a single well. One, three and five days later, we measured the total Gluc level in aliquots of the conditioned medium. The levels of the individual Gluctag reporters in the mixed culture medium were analyzed by immunobinding with the different tag specific antibodies. We were able to monitor in real-time the growth of the individual U87-FM-Gluctag-CFP cell populations in the mixed culture (Fig. 1f). Immunostaining confirmed the expression of each tag in the corresponding cells in the mixed culture, again except for GlucAu1 (Fig. 1g). In order to determine whether we can measure the activity of ten different Gluctag constructs in the same cell, we transduced U87-FM cells with all ten different lentiviral gluctag reporters. Immunostaining of the U87-FM-Gluc10× tagCFP glioblastoma cells with tag specific antibodies showed that almost all cells expressed all ten different Gluctag constructs (Fig. 1h). The tag expression as demonstrated by immunostaining is less intens in Fig. 1h as compared to Fig. 1c, which we attribute to the transduction pool of ten different lentivectors used in Fig. 1h. We used these cells to monitor cell viability in the presence and absence of the alkylating agent temozolomide (TMZ), the standard chemotherapy for glioblastoma patients. Gluctag immunobinding assays were used to determine the expression of all ten reporters in the presence or absence of TMZ. We treated the U87-FM-Gluc10× tagCFP cells with TMZ and measured Gluctag activity after six days (Fig. 1i). Precipitation of all ten different Gluctag reporters showed a ~75% decrease in Gluc bioluminescence signal after TMZ treatment, similar to the total Gluc signal and confirmed by CFP immunofluorescence analysis (Fig. 1j). Altogether, these results demonstrate that we can measure biologically relevant differences using our multiplex Gluctag system.
Figure 1. Gluctag multiplex assay development and validation in vitro.
(a) Schematic of the lentiviral vector constructs encoding the luciferase reporters. (b) Fluorescence microscopy analysis of a representative U87-FM-Gluctag-CFP cells (using GlucFlag reporter) showing mCherry levels (in red; marker for tumor cells) and CFP expression (in blue; marker for transduction efficiency). (c) Immunostaining against the various tags in U87-FM cells expressing different Gluctag. (d) Gluc activity in U87 cells expressing different Gluctag with respect to cell proliferation over time and cell number. The dashed line represents GlucCtrl activity. (e) Gluctag immunobinding assay versus total Gluc activity using serial dilutions of the conditioned medium from a 1-day culture of U87-FM cell lines expressing the individual Gluctag reporters. (f-g) Equal mixture of ten cell lines each expressing a different Gluctag reporter. (f) Immunobinding assay of Gluctag from a mixed population. (g) Immunostaining for the ten different tags expression in the mixed population of U87-FM cell culture. (h-i) U87-FM cells expressing all ten different Gluctag reporters. (h) Immunostaining for the ten different tags in one cell line. The insert panels show staining of control parental cells. Percentages of positively stained cells are indicated. (i) Immunobinding assay of all ten Gluctag reporters in one cell line after TMZ treatment (black bars) compared to untreated control (white bars). (j) CFP fluorescence microscopy of cells in (h–i) with or without 600 μM TMZ. Size bar (in b,c, g and h) = 200 μm. Size bar (i) = 400 μm. N.d. = not detectable.
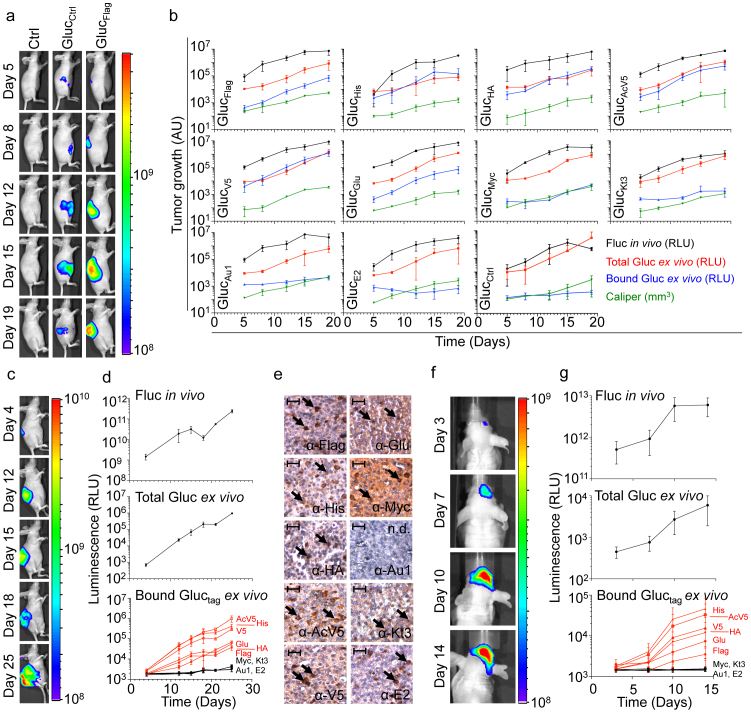
In order to determine the in vivo applicability of the Gluctag reporter assay, we injected the ten individual U87-FM cell cultures expressing either one of the Gluctag or GlucCtrl reporters subcutaneously in nude mice for singleplex application. In another set, we injected mice with PBS as a negative control (12 groups; n = 3/group). Tumor growth was monitored over time by calliper measurement, and Fluc bioluminescence imaging (Fig. 2a,b). Blood was collected from mice at different time points and 5 μl of whole blood (optimum amount for Gluc blood assay16) was assayed for total Gluc activity. At the same time, 25 μl of blood was analyzed for Gluc activity after immunobinding on wells coated with the corresponding tag antibody (Fig. 2b). All of the Gluctag reporters, except GlucE2, allowed blood monitoring of tumor growth over time in a singleplex assay. We compared the tumor growth as measured by calliper, Fluc imaging and Gluc blood assay to signals obtained from the ten different Gluctag reporters after immunobinding (Fig. 2b). The six Gluctag reporters GlucFlag, GlucHis, GlucHA, GlucAcV5, GlucV5, GlucGlu were more sensitive in detecting tumor growth than the others, possibly due to a higher affinity of the antibodies to these tags. To demonstrate the use of the Gluctag system for multiplex applications, we mixed equal numbers of U87-FM cells expressing the ten different Gluctag and implanted the heterogeneous cell pool subcutaneously in nude mice (n = 5). We monitored tumor growth by Fluc bioluminescence imaging and total Gluc blood assay (Fig. 2c,d). Importantly, we were able to efficiently monitor the growth of the individual subpopulation of the U87-FM cells expressing the different high-efficiency Gluctag reporters (GlucFlag, GlucHis, GlucHA, GlucAcV5, GlucV5, GlucGlu) in the same tumor over time (Fig. 2d). We confirmed the expression of all tags (except for GlucAu1) in the same tumor by immunostaining (Fig. 2e). To determine the applicability of the Gluctag multiplex system in deep tissues, the mixture of the ten different U87-FM-Gluctag-CFP cells were implanted orthotopically in the brain of nude mice (n = 5). We monitored tumor growth by Fluc bioluminescence imaging and total Gluc blood assay (Fig. 2f,g). Again, we were able to monitor the intracranial growth of the individual subpopulations of U87-FM cells expressing the high-efficiency Gluctag reporters within the same brain tumor in real-time, by blood sampling and the application of the Gluctag multiplex reporter system (Fig. 2g).
Figure 2. Gluctag multiplex assay in vivo.
(a) U87-FM cells expressing different Gluctag were implanted subcutaneously in nude mice (n = 3) and tumor growth was monitored overtime. Shown are representative Fluc bioluminescence images of the U87-FM-Gluctag-CFP tumors expressing the GlucFlag reporter. (b), Quantitation of the different U87-FM tumors expressing different Gluctag reporters; Fluc activity (black), calliper measurement (green), total Gluctag activity in the blood (red), and bound Gluctag activity after immunobinding (blue). (c–d) U87-FM cells expressing the different Gluctag reporters were mixed in equal amounts and injected subcutaneously in the same mouse (n = 5). Tumor growth was monitored using Fluc imaging and total Gluc blood assay as in (a–b). Individual cells subpopulation within the same tumor were monitored using the multiplex Gluctag immunobinding assay. (e) Immunostaining for the ten different tags expression in the mixed U87-FM-Gluctag-CFP tumor. Black arrows indicate positive staining for cells expressing the corresponding Gluctag. Size bar = 200 μm. N.d. = not detectable. (f–g) Mixed population of U87-FM cells expressing all ten different tags were implanted intracranially in the brain of nude mice (n = 5). Tumor growth was monitored with Fluc imaging and Gluc total blood assay, and different tumor cell subpopulations were monitored using the multiplex Gluctag assay as in (c–d). Fluc bioluminescence imaging of a representative mouse is shown in (f). Data shown in (b,d, and g) as average relative light units (RLU) ± standard deviation. Heatscales in (a, c and f) indicate relative photon levels (photons/sec) as measured by the CCD camera.
Discussion
Here we demonstrated the functionality of the Gluctag multiplex reporter system by the parallel monitoring of mixed cell cultures in vitro, and of subpopulations in mixed subcutaneous and intracranial tumors ex vivo. The individual epitope tags could also be employed to localize expression by immunostaining. We used ten different Gluctag reporters, but this tag library can be readily extended allowing for increased multiplexing. In addition, antibody binding of the low-efficiency tags in the Gluctag immunobinding assay might be further optimized to achieve higher sensitivity since these tags, except for GlucAU1, were efficiently detected by immunostaining. Real-time monitoring of individual cells in a heterogeneous mixture may allow, for instance, multiplexed RNAi screening or measurement of drug responses of multiple cell populations in parallel. The Gluctag multiplex system might be extended to monitor different variables in a single cell type. For instance, engineering each individual Gluctag under the control of different transcription response elements for multiplex transcription factor activity measurements7, constructing microRNA (miRNA)-binding sequences in the 3′-UTR of the Gluctag constructs for multiplex monitoring of miRNA activity10 or by inserting different protease cleavage sites into the Gluctag gene for multiplex protease activity measurement17. This reporter assay provides a versatile tool to study complex processes with different variables in systems biology.
Methods
Cells
For in vitro and in vivo validation, we used the glioblastoma cell line U87, stably co-expressing Firefly luciferase and mCherry fluorescent protein. Cell lines were maintained in DMEM high glucose complemented with sodium pyruvate, stable glutamine, 10% FBS and pen/strep (all PAA), incubated under standard cell culture conditions of 37°C and 5% CO2.
Lentivirus vector construction and transduction
First, we amplified Gluc by PCR using Accuprime Pfx DNA polymerase (Life Technologies) from CSCW-Gluc-IRES-CFP6 with specific primers that incorporate an XbaI site downstream of the Gluc cDNA (Forward primer: 5′- GCGTGTACGGTGGGAGGTCT-3′, reverse primer: 5′-TCTCGAGTAGAGATCTGTCACCACCGGCCCCCTT-3′). The reverse primer also covers the original stop-codon that is thereby removed. The Gluc cDNA, now containing a unique XbaI site, was ligated back into the CSCW-Gluc-IRES-CFP vector with T4 DNA ligase (Life Technologies), after removal of the wild-type Gluc using NheI and XhoI (Life Technologies). Plasmid DNA was transformed in XL-10 Gold ultracompetent cells (Agilent Technologies), cultured overnight in LB agar containing 50 μg/ml Ampicillin. We isolated DNA using a DNA plasmid mini kit (Qiagen) and verified successful transformation by XbaI restriction analysis. The Gluc construct was then digested with XbaI and XhoI to insert the different epitope tags. The epitope tags were designed with an XbaI site upstream, a stop codon and XhoI site downstream. A total of 20 μM of both single strand oligonucleotides of the acquired tag DNA (Life Technologies, see Table S1 for sequences) was annealed in annealing buffer (100 mM Tris-HCl pH 7.5, 1 M NaCl and 10 mM EDTA) by heating to 65°C for 10 minutes and slow cooling to room temperature. The epitope tag was then inserted into the vector using T4 DNA ligase and transformed in XL-10 Gold ultracompetent cells. Bacteria were cultured and DNA was isolated. We verified the Gluctag constructs by sequencing using BigDye Terminator v3.1 Cycle Sequencing kit (Life Technologies, primer: 5′-GCCAACGTGCAGTGTTC-3′). The Gluctag construct was co-transfected with a third generation lentiviral packaging mix (pMDLg/pRRE, pRSV-Rev and pMD2.G, Addgene) in HEK293T cells using Lipofectamine 2000 (Life Technologies). Virus was harvested two and three days after transfection and cell debris was spun down for five minutes at 1,000 × g. U87 cells were transduced overnight with lentivirus using a multiplicity of infection of 100 transducing units per cell in the presence of 8 μg/ml polybrene in standard culture conditions.
Fluorescence microscopy
Successful transduction was verified by visualizing CFP (co-expressed with the Gluctag construct) and mCherry fluorescent protein (co-expressed with Fluc) using fluorescent microscopy (Leica).
Immunostaining and antibodies
For in vitro immunostaining, cells were fixed in 3.7% formaldehyde for 20 minutes, washed with PBS and permeabilized with PBS containing 0.1% Triton X-100. Cells were then washed three times in PBS containing 5% FBS and blocked with PBS containing 5% FBS. Primary mouse-anti-tag monoclonal antibody (10 μg/ml in 100 μl of PBS, Table S1) was added and incubated for one hour at room temperature. Cells were washed again and incubated with 1:100 goat-anti-mouse horseradish peroxidase (HRP) conjugate (Dako) for one hour. After washing, cells were stained with DAB+ (Life Technologies). For mouse tissue staining, tumors were removed and embedded in paraffin. Microtome sections of 5 micron on glass slides were deparaffinized in xylene and rehydrated in ethanol series of 100%, 96%, and 70% ethanol. Endogenous peroxide was blocked with 0.3% H2O2 in methanol for 30 minutes. After rinsing with water, antigens were retrieved with citrate buffer (pH 6) with 0.05% Tween 20 using a microwave (Bosch, five minutes 900 watt, 10 minutes 360 watt). After slowly cooling, tissues were washed three times with PBS and incubated with primary antibody (10 μg/ml), for one hour at room temperature. After washing three times again, tissues were incubated with REAL Envision Rabbit anti-mouse (Dako and REAL DAB+ stained (Dako). Tissue sections were dehydrated with ethanol series as before and fixed in xylene. Cells and sections were imaged and photographed by light microscopy (Leica).
Western blot
U87-FM-Gluctag-CFP cells were lysed in RIPA buffer and mixed with Laemli buffer containing β-mecaptoethanol (80 μl/ml). The samples were separated using a 10% SDS-PAGE gel and the XCell-II blot module (Invitrogen). Primary antibodies (Table S1) were used in a 1:3,000 dilutions and goat-anti-mouse HRP immunoglobuline (Dako) secondary antibody was used in a 1:2,000 dilution. Incubation times were 1 hour at room temperature.
RT-PCR
U87-FM-Gluctag-CFP RNA was isolated using Trizol (Invitrogen) according to manufacturer's guidelines. cDNA was generated using Omniscript RT kit (Qiagen) and PCR was performed in triplo using FastStart SYBR Green Master (Rox) mix (Roche). As a forward primer we used 5′- GCCAACGTGCAGTGTTC-‘3, which is located in the Gluc gene. We used the tag antisense oligonucleotides as tag specific reverse primers (Table S1).
In vitro gluc activity assay
For Gluctag activity measurement over time, 50,000 cells were plated in a 24-well plate and incubated overnight. A total of 10 μl conditioned medium was harvested from cells and Gluc activity was measured by adding 50 μl (5 μg/ml) coelenterazine (Nanolight Technologies) in PBS and 0.1% Triton X-100). Before addition to the sample, the substrate was incubated at room temperature for 30 minutes for stabilization. Photon counts were determined over 10 seconds in a luminometer (Berthold Technologies). For the measurement of Gluctag activity with respect to cell number, indicated numbers of cells were plated in a 24-well plate and incubated overnight. Gluctag activity in medium was determined as described above.
Gluctag immunobinding assay
White goat-anti-mouse-coated 96-well plates (Thermo Scientific) were washed three times with PBS containing 0.05% Tween 20 and incubated with 50 μl (10 μg/ml) of each of the mouse monoclonal antibody directed against the specific epitope tags (Table S1). Incubation time was two hours while centrifuging at 500 × g at 4°C. Wells were washed and blocked three times with PBS containing 5% FBS and 0.05% Tween 20 on a plate shaker at 65–75 rpm. A total of 30 μl of Gluctag conditioned medium or Gluctag mouse blood was added to the well and incubated for two hours at room temperature on a microplate shaker at 65–75 rpm. Wells were washed five times for five minutes on a plate shaker at 65–75 rpm. A total of 50 μl Gluc substrate (5 μg/ml coelenterazine in PBS and 0.1% Triton X-100) was added to the well and photon counts were determined in a microplate luminometer (Tecan) at 0.1 second per well. For the comparison of the bound Gluc to the total Gluc activity, aliquots of conditioned medium or 5 μl mouse blood were transferred to a white 96-wells microplate and assayed using 50 μl (5 μg/ml) coelenterazine in PBS and 0.1% Triton X-100.
In vivo experiments
All in vivo experiments were subject to ethical committee approval and are in accordance with VU University Medical Center and national regulations. For subcutaneous tumor xenograft model, six weeks old athymic Nude-Foxn1nu mice (Harlan) were implanted with 5 × 105 U87-FM-Gluctag-CFP cells in 50 μl DMEM and 50 μl Matrigel (BD Biosciences). For intracranial tumor implantation, a stereotactic frame (Harvard Biosciences) was used to inject cells vertically into the right hemisphere. General anesthesia was induced by subcutaneous injection of buprenorphine hydrochloride (0.1 mg/kg). The mice were further anesthetized with oxygen containing 2.5% isofluorane. After skin incision, lidocaine drops (5 mg/ml in PBS) were administered topically. A small drill was used to drill a hole into the skull. Coordinates used for injection were X = 0.5 mm, Y = 2 mm, Z = −2 mm from the bregma. A total of 2 × 105 cells in 5 μl of DMEM was injected vertically. On indicated days after injection, tumor size was monitored using a calliper (for subcutaneous) and bioluminescence in vivo Fluc imaging, by injecting D-luciferin (100 mg/kg) intraperitoneally. Imaging was performed with an IVIS CCD camera and analyzed with Living Image software (Caliper Life Sciences). We collected 300 μl mouse blood aliquots from the tail vein in Microvette CB300 EDTA capillary tubes (Sarstedt) for ex vivo whole Gluc measurements and Gluc immunobinding assays.
Author Contributions
JN and TW conceived this study and SVR, BAT, JN and TW designed the experimental setup, with help of DN and WPV. SVR performed most of the experiments. SVR, JN, and TW wrote the manuscript, which was edited and reviewed by all the authors.
Supplementary Material
Supplementary information
Acknowledgments
This work was supported partly by grants from NWO-VIDI (TW), and NIH/NINDS P30NS045776 and R01NS064983 (BAT). SVR and JN are supported by CCA/VICI Fellowships. We are thankful to Priscilla Jainandunsing, Laura Jonkman, Stephanie van Hoppe, Tonny Lagerweij and Laurine Wedekind for technical support.
References
- Gross S. & Piwnica-Worms D. Spying on cancer: molecular imaging in vivo with genetically encoded reporters. Cancer cell 7, 5–15 (2005). [DOI] [PubMed] [Google Scholar]
- Pittet M. J. & Weissleder R. Intravital Imaging. Cell 147, 983–991 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contag C. H. & Ross B. D. It's not just about anatomy: in vivo bioluminescence imaging as an eyepiece into biology. Journal of magnetic resonance imaging/: JMRI 16, 378–87 (2002). [DOI] [PubMed] [Google Scholar]
- Bhaumik S. & Gambhir S. S. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proceedings of the National Academy of Sciences of the United States of America 99, 377–82 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous B. A., Kim D.-E., Fernandez J. L., Weissleder R. & Breakefield X. O. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Molecular therapy/: the journal of the American Society of Gene Therapy 11, 435–43 (2005). [DOI] [PubMed] [Google Scholar]
- Wurdinger T. et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nature methods 5, 171–3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhang H.-eun C., Gabrielson K. L., Laterra J., Fisher P. B. & Pomper M. G. Tumor-specific imaging through progression elevated gene-3 promoter-driven gene expression. Nature medicine 17, 123–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr C. E., Hewett J. W., Breakefield X. O. & Tannous B. A. A highly sensitive assay for monitoring the secretory pathway and ER stress. PloS one 2, e571 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y. et al. Development of a dual-luciferase reporter system for in vivo visualization of MicroRNA biogenesis and posttranscriptional regulation. Journal of nuclear medicine/: official publication, Society of Nuclear Medicine 49, 285–94 (2008). [DOI] [PubMed] [Google Scholar]
- Tian W. et al. High-Throughput Functional MicroRNAs Profiling by Recombinant AAV-Based MicroRNA Sensor Arrays. PloS one 7, e29551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillin D. W. et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nature medicine 16, 483–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr C. E. & Tannous B. A. Bioluminescence imaging: progress and applications. Trends in Biotechnology 29, 624–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Steinbach P. A. & Tsien R. Y. A guide to choosing fluorescent proteins. Nature methods 2, 905–9 (2005). [DOI] [PubMed] [Google Scholar]
- Filonov G. S. et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nature biotechnology 29, 757–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deliolanis N. C. et al. In vivo tomographic imaging of red-shifted fluorescent proteins. Biomedical optics express 2, 887–900 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous B. a Gaussia luciferase reporter assay for monitoring biological processes in culture and in vivo. Nature protocols 4, 582–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niers J. M., Kerami M., Pike L., Lewandrowski G. & Tannous B. A. Multimodal in vivo imaging and blood monitoring of intrinsic and extrinsic apoptosis. Molecular therapy/: the journal of the American Society of Gene Therapy 19, 1090–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information