Abstract
The influenza virus mRNAs are structurally similar to cellular mRNAs nevertheless; the virus promotes selective translation of viral mRNAs despite the inhibition of host cell protein synthesis. The infection proceeds normally upon functional impairment of eIF4E cap-binding protein, but requires functional eIF4A helicase and eIF4G factor. Here, we have studied whether the presence of cis elements in viral mRNAs or the action of viral proteins are responsible for this eIF4E-independence. The eIF4E protein is required for viral mRNAs translation in vitro, indicating that cis-acting RNA sequences are not involved in this process. We also show that PB2 viral polymerase subunit interacts with the eIF4G protein. In addition, a chimeric mRNA containing viral UTRs sequences transcribed by the viral polymerase out of the infection is successfully translated independently of an impaired eIF4E factor. These data support that the viral polymerase is responsible for the eIF4E independence of influenza virus mRNAs translation.
INTRODUCTION
Influenza virus uses an unusual transcription mechanism in which capped and polyadenylated viral mRNAs are synthesized by the viral polymerase, a heterotrimer composed of three subunits named PA, PB1 and PB2 (Elton et al., 2005). Viral mRNA synthesis is primed by short-capped oligonucleotides of around 10 to 12 nucleotides that are generated from host cell nuclear mRNAs by a viral endonuclease activity (Plotch et al., 1981). The cap recognition and binding is achieved by the PB2 subunit (Blaas, Patzelt, and Keuchler, 1982; Ulmanen, Broni, and Krug, 1981), while the PA subunit seems to be required for the cleavage of the cap-oligonucleotides (Dias et al., 2009; Yuan et al., 2009). In addition, the 3′- end of viral mRNAs is polyadenylated by the reiterative copy of a U5-7 track present near the 5′ end of the genomic negative sense viral RNA (Luo et al., 1991). Consequently, although synthesized by different pathways, cellular and viral mRNAs are both structurally similar.
Influenza virus efficiently shuts off host cell protein synthesis (Garfinkel and Katze, 1993). Moreover, upon infection, viral mRNAs are selectively translated (Garfinkel and Katze, 1993; Park and Katze, 1995), while the initiation and elongation of cellular mRNAs translation are inhibited (Katze, DeCorato, and Krug, 1986). The translation initiation eIF4F complex plays a pivotal role in the translation of capped-mRNAs. It is a heterotrimer formed by eIF4E, the cap-binding factor that is required for cap-dependent translation, the eIF4A helicase and the scaffolding eIF4G factor. The eIF4G protein binds to eIF3, which in turn, mediates the recruitment of the 40S ribosomal subunit triggering the translation initiation of the mRNAs bound to the eIF4F complex (see for a review (Gingras, Raught, and Sonenberg, 1999) ). Since viral and cellular mRNAs are formally equivalent, influenza virus must have developed sophisticated strategies to discriminate and favor translation of its own mRNAs. Among the key factors that have been related with viral translation regulation, NS1 protein plays an important role contributing to the selective translation of viral mRNAs in the infected cell, especially for those produced later in the virus life cycle. This activation is mediated by its functional interaction with the 5′ UTR of the viral mRNAs that are conserved in every viral mRNA (de la Luna et al., 1995; Enami et al., 1994; Park and Katze, 1995). In addition, the interaction of NS1 with the eIF4GI factor (Aragón et al., 2000) and with the polyA binding protein I (Burgui et al., 2003) appear to be essential for this process. However, viral mRNAs are selectively translated in a mutant virus lacking NS1 protein, suggesting that other viral factors might be involved in the preferential translation of viral mRNAs that takes place within the infected cells (Salvatore et al., 2002).
Regarding the eIF4F complex, influenza virus infection alters the phosphorylation state of eIF4E and eIF4G, and these changes have been related with the inhibition of host-cell protein synthesis and the selective translation of viral mRNAs (Feigenblum and Schneider, 1993). In agreement with these data, we have previously shown that the translation of influenza virus mRNAs and the viral infection proceed efficiently when the eIF4E cap-binding protein is functionally impaired, even when a recombinant influenza virus lacking NS1 protein is used (Burgui et al., 2007). In addition, we have recently characterized that the other two components of the eIF4F complex, eIF4A and eIF 4G, are essential for viral translation both in in vivo and in vitro analysis and, hence, should not be related with selective translation in the infected cell (Yángüez et al., 2011).
Among the possible transacting proteins that could be involved in viral protein synthesis, we also described that the influenza virus polymerase binds to translation preinitiation complexes and that the infection increases the binding of the eIF4GI factor to cap-structures under conditions of eIF4E-eIF4GI disassociation triggered by overexpression of a non-phosphorylatable 4E-BP1 protein (Burgui et al., 2007). These data suggest a role for the viral polymerase in overriding the dependence of viral mRNA translation on the eIF4E factor, as it could behave as the cap-binding factor that mediates eIF4G recruitment to the viral mRNA. Here, we have assessed whether the viral polymerase or the presence of cis structural elements in viral mRNAs are the responsible for the eIF4E independence. This process could be part of the mechanism underlying the selective translation of viral mRNAs that takes place in the infected cell.
RESULTS
Study of the presence of structural cis elements in influenza virus mRNAs
As mentioned, influenza virus infection proceeds normally in the absence of functional eIF4E factor. Thus, rapamycin treatment, eIF4E gene silencing and overexpression of constitutively hypophosphorylated 4E-BP1, which provokes eIF4E-eIF4G dissociation, do not impair viral mRNAs translation in the infected cells (Burgui et al., 2007). Common structural determinants within influenza virus mRNAs could mediate their independence for the cap-binding factor as, for instance, internal ribosome entry sites (IRES) that are capable of directly recruiting the translation machinery (Kieft, 2008; Martinez-Salas et al., 2008). Therefore, we carried out in vitro experiments to examine whether influenza virus mRNAs contain sequences that would confer eIF4E independence.
Requirements for eIF4E in vitro
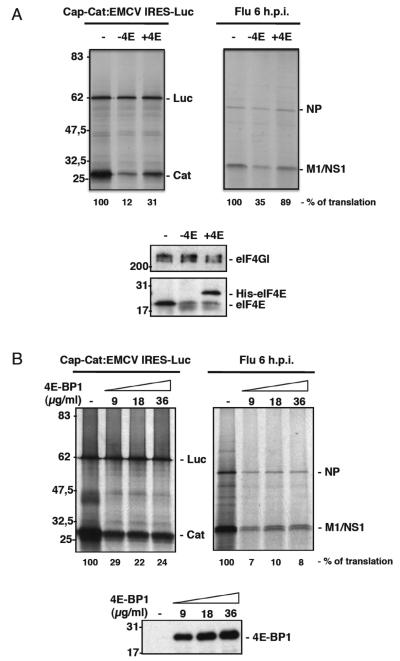
We compared the translation efficiency of a dicistronic cap-CAT-EMCV:IRES-luciferase RNA containing the encephalomyocarditis disease virus IRES, with isolated RNAs from influenza virus infected cells in conditions of limited eIF4E availability. Accordingly, in vitro translation preparations were depleted of eIF4E by adding purified 4E-BP1 protein followed by incubation with a cap-Sepharose resin. After removal of the bound complexes by centrifugation, the eIF4E-depleted lysates were used to translate in vitro transcribed cap-CAT:EMCV-IRES-Luc RNA or purified cytoplasmic RNA from infected c ells (Fig. 1A). The depletion of eIF4E produced a clear decrease in cap-dependent CAT protein synthesis, while IRES-driven luciferase synthesis remained unaffected. Similarly, the synthesis of viral proteins diminished under these conditions. Further addition of purified recombinant His-eIF4E protein partially recovered the translation of CAT and viral proteins in the eIF4E-depleted preparations.
Figure 1. Functional impairment of eIF4E inhibits the in vitro synthesis of influenza virus proteins from isolated viral RNAs.
(A); Rabbit reticulocyte extracts were depleted of eIF4E by the addition of purified 4E-BP1 protein and incubation with a 7mGTP resin. After removal of the bound complexes, the eIF4E-depleted lysates were used to assess the translation of in vitro transcribed cap-CAT:EMCV-IRES-Luc RNA or purified cytoplasmic RNA from infected cells (− 4E lanes). Subsequently, purified His-eIF4E recombinant protein was added (+ 4E lanes). The bottom panel shows the amounts of the indicated proteins in the eIF4E-depleted preparations and after the addition of recombinant His-eIF4E protein. The synthesized proteins were metabolically labeled and analyzed by SDS-PAGE. (B); Cytoplasmic RNA from HEK293T infected cells isolated after 6 hpi and dicistronic cap-CAT:EMCV IRES-Luc RNA obtained by in vitro transcription were used for in vitro translation in reticulocyte lysates, with or without increasing concentrations of purified 4EBP1. The bottom panel shows the amounts of 4EBP1 added to the reactions. The synthesized proteins were processed as described in part (A).
We also used a different approach to analyze the possible presence of specific structures in the viral mRNAs that might be involved in the low dependence on eIF4E. Accordingly, both in vitro transcribed dicistronic cap-CAT:EMCV-IRES-Luc RNA and cytoplasmic RNA from infected cells were then assayed in the presence or absence of increasing concentrations of purified 4E-BP1 to inhibit the interaction of eIF4E with eIF4G. The proteins synthesized were metabolically labeled and analyzed by SDS-polyacrylamide gels (SDS-PAGE) (Fig. 1B). Translation of the IRES-driven luciferase was not affected by the addition of 4E-BP1 even at the highest doses. By contrast, the translation of the CAT gene, which occurs in a cap-dependent manner, clearly diminished in the presence of 4E-BP1. Similarly, the addition of purified 4E-BP1 significantly reduced the translation of viral mRNAs. These results indicate that isolated influenza virus mRNAs behave like standard capped-mRNAs in terms of eIF4E dependence in vitro. Therefore, the low requirement for functional eIF4E observed in vivo is not due to an inherent property of the viral mRNA (e. g. the presence of cis elements), but it is more likely the consequence of specific factors acting in trans.
The PB2 influenza virus polymerase subunit interacts with the eIF4GI factor
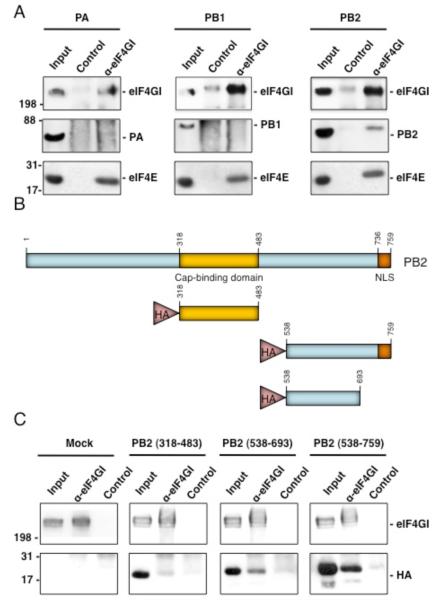
We previously reported that the influenza virus polymerase complex, both from influenza virus infected cells or expressed from cloned cDNAs, co-immunoprecipitates with the translation initiation factor eIF4GI (Burgui et al., 2007). To further characterize this interaction, viral polymerase subunits were individually expressed and their ability to associate with eIF4GI was evaluated. Thus, HEK293T cells were transfected with pCMV plasmids expressing each of the subunits and 24 h later, cell extracts were analyzed by co-immunoprecipitation using specific antibodies against eIF4GI or with a control serum. The PB2 subunit was present in the immunocomplexes with eIF4GI, whereas the PA or PB1 subunits were undetected. In addition, the presence of eIF4E in the eIF4GI immunoprecipitates was e valuated as positive control of interacting proteins (Fig. 2A). Next, we mapped the region in the PB2 subunit required for this association. This was achieved by using clones of a pCDNA-HA plasmid containing PB2 N-truncated fragments. The different fragments used are soluble when expressed in Escherichia coli as they were obtained by a high-throughput screening method known as expression of soluble proteins by random incremental truncation (ESPRIT) (Tarendeau et al., 2007; Yumerefendi et al., 2010). HEK293T cells were transfected with these constructs and 24 h later, the proteins were immunoprecipitated with specific anti-eIF4GI or an unrelated antibody and analyzed in Western blots probed with an anti-HA antibody. The results indicate that the PB2 sequence that mainly mediates the association with eIF4GI resides in its C-terminal region between amino acids 538-693 (Fig. 2 B-C), a domain that maps out of the minimal region involved in cap association (318-483) (Guilligay et al., 2008).
Figure 2. PB2 influenza virus polymerase subunit associates with eIF4GI.
(A); HEK293T cells were transfected with plasmids expressing PA, PB1, or PB2 and 24 hours later, cytosolic extracts were prepared and used for immunoprecipitation assays using anti-eIF4GI or a control antibody. The immunoprecipitates were separated by SDS-PAGE, and Western blots were probed with antibodies against the indicated proteins. (B); Scheme of the HA-tagged PB2 proteins used in part C. (C); HEK293T cells were transfected with the corresponding plasmids and immunoprecipitations assays were performed as indicated in (A). The Western blot was probed with anti eIF4GI and anti HA antibodies.
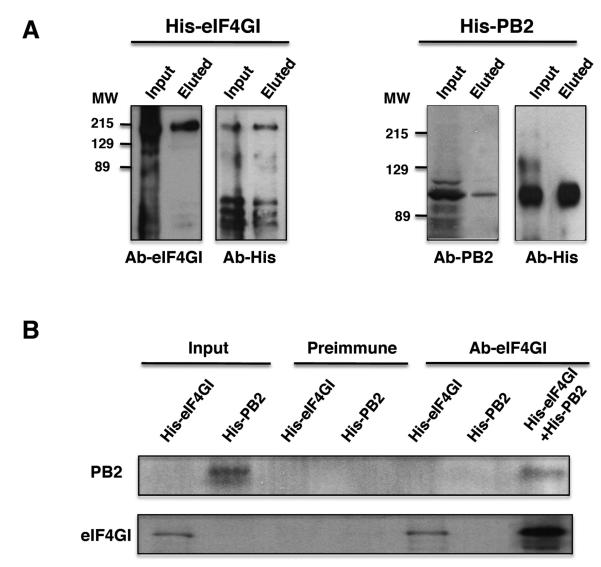
Many cellular proteins associate with the eIF4GI translation initiation factor and therefore they could mediate its interaction with PB2 and the viral polymerase. To discriminate between a direct or indirect association of eIF4GI and PB2, both proteins were expressed as His tagged-proteins in bacteria and after purification they were eluted from the affinity resins and their association was examined. Purification of His-eIF4GI and His-PB2 proteins was evaluated by Western blots using both specific antibodies against the corresponding proteins and anti-His antibodies. The results are presented in Fig. 3A; both proteins were expressed as entire recombinant proteins, although degradation pro ducts or fragments generated by premature termination were also present. For coimmunoprecipitation analysis His-eIF4GI and His-PB2 were incubated to allow their interaction and then immunoprecipitation assays were performed with antibodies against eIF4GI or the preimmune serum. The specific anti-eIF4GI antibody immunoprecipitated His-eIF4GI as well as His-PB2 when it was incubated with eIF4GI (Fig. 3B). By contrast, the preimmune serum did not immunoprecipitated either His-eIF4GI or His-PB2 from the incubation mixture and the eIF4GI antibody did not immunoprecipitate the His-PB2 alone. These results indicate that eIF4GI and PB2 establish a direct association not mediated by other cellular proteins.
Figure 3. PB2 and eIF4GI interact directly.
A); Recombinant His-eIF4GI (left) and His-PB2 (right) proteins expressed and purified from bacteria were analyzed by Western blotting with specific (Ab-eIF4GI and Ab-PB2) and anti-His antibodies. (B), Interaction of eIF4GI and PB2 proteins. Purified His-eIF4GI and His-PB2 proteins were tested for immunoprecipitation with preimmune or anti-eIF4GI antibodies when they were incubated alone (His-eIF4GI or His-PB2) or when both proteins were incubated together (His-eIF4GI+His-PB2) and the immunocomplexes analyzed by Western blotting with the indicated antibodies.
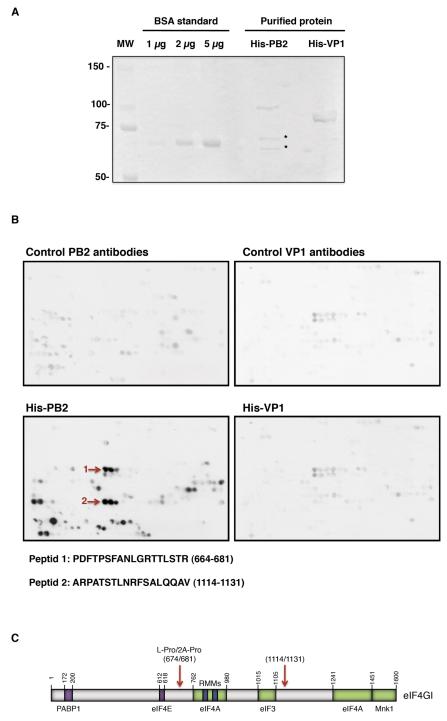
To further characterize the region of eIF4GI that interacts with PB2, a pep-spot analysis was carried out. This technique was previously described as a method to detect linear epitopes recognized by antibodies (Kaikkonen et al., 1999; Valle et al., 1999) and it can be used to map protein-protein interactions when a particular structural conformation is not required for the interaction (Huarte et al., 2001). Thus, a nitrocellulose membrane was prepared containing sequential overlapping peptides of 13 amino acids covering the entire sequence of eIF4GI, with a 3-amino-acids shift with the next peptide. Recombinant His-PB2 protein expressed and purified from bacteria was incubated with the membrane and its possible binding was examined by Western blot. As a control, recombinant His-VP1 protein from infectious bursal disease virus expressed and purified from bacteria (Garriga et al., 2007) was used (Fig. 4A). Prior to probing with His-PB2 and His-VP1, the membrane was probed with the corresponding primary and secondary antibodies to detect the unspecific spots (Fig. 4B, top panels) and subsequently, the membrane was incubated with the His-tagged proteins. The sequence of incubations is described in Experimental Procedures and the results are presented in Figure 4B. To avoid possible false-positives resulting from the specific characteristics of individual peptide spots, we have used restrictive criteria, assuming only as positive interacting regions those having positive signals in at least 19 amino acids (3 consecutive spots). With these criteria, purified His-PB2 protein binds to at least two regions of eIF4GI, between amino acids 664-681 and 1114-1131 (marked by arrows) (Fig. 4B, bottom panels). It should be noted that in this assay the peptides are distributed in the membrane according to their linear sequence location, but separated positive peptides could be close in the native folded protein. The first positive sequence includes the recognition and processing site of enterovirus 2A and picornaviruses L proteases (Foeger, Glaser, and Skern, 2002; Gradi et al., 2004; Lamphear et al., 1995; Sousa, Schmid, and Skern, 2006) indicating that this sequence should be exposed and accessible. The second putative site resides within a region where no other eIF4GI-binding proteins have been described. No specific His-VP1-eIF4GI interaction was detected. These data indicate that the interaction eIF4GI-PB2 is direct and not mediated by viral RNA or any o f the previously described eIF4G-interacting factors.
Figure 4. Mapping of PB2 interaction within the eIF4GI sequence.
(A); Coomassie staining of His-PB2 and His-VP1 proteins expressed and purified from bacteria. Asterisk represent PB2 degradation products or PB2 fragments generated from premature termination since they are detected with the anti-PB2 antibody (data not shown). (B); A collection of 122 overlapping peptides representing the complete eIF4GI factor was synthesized on a cellulose membrane, which was incubated sequentially in the presence or absence of purified His-protein s and the corresponding primary and secondary antibodies, and assayed in Western blots. After each incubation the membrane was stripped and used for the next detection. The sequence of incubation was as follows: primary and secondary antibodies used to detect PB2 (top, left); incubation with His-PB2 protein followed by its corresponding primary and secondary antibodies (bottom, left); primary and secondary antibodies used to detect VP1 (top, right); and finally His-VP1 followed by its corresponding primary and secondary antibodies (bottom, right). The arrows represent eIF4GI-PB2 interacting peptides. (C); Representation of the PB2 interaction sites in the eIF4GI sequence.
The role of the viral polymerase in viral mRNAs translation
Several observations such as: I) the cap-binding properties of the viral polymerase; II) the fact that the influenza virus polymerase complex binds to the highly conserved 5′UTR sequence common to all viral genes in vitro (Shih and Krug, 1996); III) the association of the viral polymerase with eIF4GI and the direct interaction of the PB2 cap-bin ding subunit with this factor; IV) the recruitment of eIF4GI and the polymerase to cap analogues upon infection under conditions of eIF4E-eIF4G disassociation (Burgui et al., 2007); and V) the hypophosphorylation of eIF4E factor that takes place during the infection and should decrease the eIF4E-eIF4G association (Feigenblum and Schneider, 1993), have been considered. Taken together, these data suggest a role for the viral polymerase in viral translation modulation, probably replacing the eIF4E function during the translation of viral mRNAs.
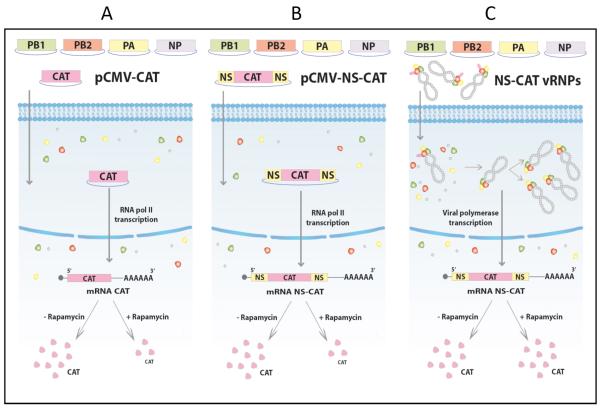
To address this possibility, we compared the requirements for eIF4E on the translation of a reporter mRNA expressing CAT, containing viral 5′ and 3′ UTRs sequences when it is expressed either by the viral polymerase or by the cellular RNA polymerase II. The experimental design is shown in Fig. 5. Briefly, HEK293T cells were transfected with plasmids expressing the viral RNP components PA, PB1, PB2 and NP, and incubated for 18 h to allow the accumulation of the se proteins. Then, the cells were maintained in the presence or absence of rapamycin. Some cells were subsequently transfected with a plasmid expressing the CAT gene driven by an RNA polymerase II promoter, with or without the 5′ and 3′ UTR sequences of the NS segment of viral mRNA (pCMV-NS-CAT and pCMV-CAT respectively: Fig. 5 A-B). Other cells were next transfected with purified, active viral RNPs that were reconstituted in vivo with a vRNA-like CAT gene that contains the same 3′ and 5′ UTRs of the vRNA o f NS segment (prepared as described in Supp. Inf. 1) (Jorba, Coloma, and Ortin, 2009) (Fig. 5C). At 7 h post transfection, samples were collected and used for CAT analysis and RNA detection. In summary, the cells in the three experimental conditions express the viral RNP components, but two of them express a CAT mRNA transcribed from the RNA polymerase II promoter (with or without UTR viral sequences), whilst the other expresses the CAT mRNA by the action of the viral polymerase.
Figure 5. Scheme used for the evaluation of eIF4E dependence on CAT expression driven by RNA polymerase II or by the viral polymerase.
In Part A and B we evaluated whether the presence in trans of the viral polymerase confers rapamycin resistance to a cellular-like CAT mRNA (pCMV-CAT) or a viral-like CAT mRNA (pCMV-NS-CAT), both transcribed by the cellular RNA polymerase II. In Part C we examined whether the translation of a viral-like CAT mRNA transcribed by the viral polymerase is rapamycin insensitive. In this case, the antisense CAT vRNA was previously used to reconstitute and purify viral RNPs (NS-CAT-vRNPs). These NS-CAT-vRNPs were then transfected in cells expressing the polymerase and the NP proteins and, therefore, the corresponding CAT-mRNA was synthesized by the influenza virus polymerase.
First, we evaluated whether similar polymerase complexes are formed under the different experimental conditions depicted in Fig. 5B and 5C. To that aim, some HEK293T cells were transfected with plasmids expressing PA, PB1 and a His-tagged PB2 protein, incubated for 18 h and after that, transfected with the pCMV-NS-CAT plasmid. Other cells were transfected with plasmids that express PA, PB1 and PB2-His with or without NP, incubated for 18 h and transfected with the pHH-NS-CAT plasmid used to reconstitute active viral RNPs. After transfection, the viral polymerase complexes were purified by 2+Ni-NTA-agarose affinity chromatography and the presence of polymerase components and NP evaluated by SDS-gels followed by Western blots. Under the different experimental conditions the amount of polymerase subunits that associate and form polymerase complexes is similar, independent of the nature of the flu-like viral RNA (sense or antisense) expressed and the formation of viral RNPs that should occur when PA, PB1, PB2-His, NP and pHH-NS-CAT are used for transfection (Supp. Inf. 2).
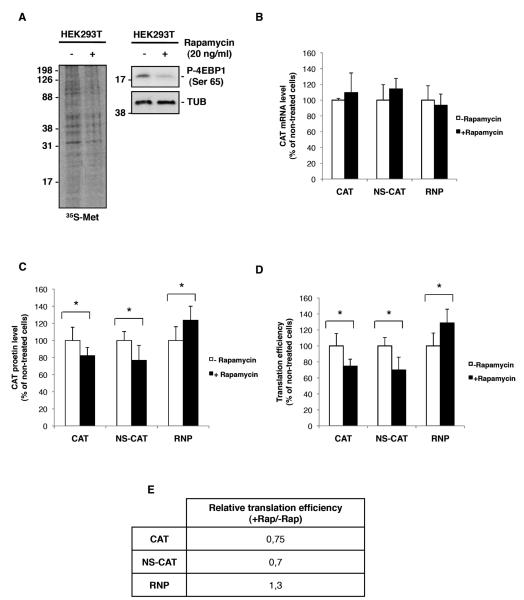
To evaluate the efficacy of the rapamycin treatment treated and untreated cells were labeled in vivo and the proteins were resolved by SDS-PAGE. As expected, rapamycin treatment reduced the phosphorylation of 4E-BP1 protein at Ser65 (Fig. 6A), which has been correlated with an increase in its affinity for eIF4E (Gingras, Raught, and Sonenberg, 1999) and the disruption of eIF4E-eIF4G association (Gingras, Raught, and Sonenberg, 2004). Consequently, rapamycin produced a reduction of around 30% in the 35S-Met incorporation, which is similar to that observed in NIH3T3 cells (Maeshima et al., 2002). Together, the reduced incorporation of 35S-Met and the pattern of 4E-BP1 phosphorylation, indicate that rapamycin produced a decrease in eIF4E-dependent translation. Under these conditions, no major differences in the accumulation of the RNP components were found in the rapamycin treated cells compared with the untreated cells, as a significant amount of these proteins was accumulated during 18 h previously to rapamycin addition (data not shown).
Figure 6. Viral polymerase confers eIF4E independence.
HEK293T cells were processed as described in Fig. 5. (A); Metabolic labeling of treated and untreated cells with rapamicyn. The right panel shows 4E-BP1 phosphorylation on Ser 65. (B); CAT mRNA accumulation measured by RT-PCR. (C); CAT protein accumulation in the different conditions described in Fig. 5. (D); Rates of CAT protein/CAT mRNA accumulation in each condition described in Fig. 5. Standard deviations are indicated by bars, asterisks (*=p<0.05) indicate statistical significance determine d by Student’s T-test. (E); Representation of the relative translation efficiency presented in part (D)
In order to discard possible differences in CAT protein after rapamycin treatment as a consequence of variation in the mRNA amount, the levels of CAT encoding mRNA were determined by qRT-PCR in the different experimental conditions. No significant variations on CAT mRNAs were found between treated or untreated cells in an y of the experimental conditions (Fig 6B). Next, we tested the CAT protein produced under the different conditions and the results are presented in Figure 6 C. Exposure to rapamycin resulted in a reduction of around 20% in the amount of CAT protein generated from the expression of pCMV-CAT and pCMV-NS-CAT plasmids. This reduction is in agreement with the observed inhibition in the general protein synthesis by 35S-Met incorporation (Fig. 6A) and with previous publications on the effect of rapamycin on translation inhibition, since, depending on the system the degree of inhibition ranges from nearly undetectable (Feldman et al., 2009; Thoreen et al., 2009) to 50% in the most severe cases (Beretta et al., 1996; Kumar et al., 2000; Maeshima et al., 2002). By contrast, CAT protein accumulation was increased by around 20% when produced from an mRNA transcribed by the viral polymerase (results were obtained consistently and repeatedly in more than six independent experiments, each performed in triplicate).
Finally, to obtain the actual translational efficiency in the different situations, the corresponding CAT protein/CAT mRNA ratios were obtained (Fig. 6D). As can be seen, the translation of a CAT mRNA transcribed by the viral polymerase is insensitive to the functional impairment of eIF4E factor induced by the rapamycin treatment. Moreover, CAT mRNA translation is augmented in this situation and this increase could reflect a reduction in the competition for eIF4GI binding, as capped mRNAs are not being efficiently translated and therefore, for the translation initiation complexes.
It is important to emphasize that the presence in trans of the protein components of the RNPs together with the vRNA-like model (NS-CAT condition) transcribed by the RNA polymerase II, do not confer rapamycin independence (Fig. 6D). Therefore, transcription of the chimeric viral mRNA by the viral polymerase is required to confer eIF4E independence.
To evaluate the involvement of viral polymerase on viral mRNA translation by a different approach, we performed similar experiments blocking cap-dependent translation by overexpression of the 4E-BP1 protein by plasmid transfection, instead of treating with rapamycin. Under these conditions the translational rates were similar to those obtained upon rapamycin treatment, but variations in the accumulated mRNAs were observed making more difficult the acquisition of accurate results (data not shown).
DISCUSSION
Most of the mechanisms involved in translational modulation are focused on the translation initiation step. Among them, regulation of the eIF4F complex through its phosphorylation, which modulates the association of its various components, is frequently observed. Within the eIF4F complex, the eIF4G factor plays a pivotal role acting as a scaffolding protein that triggers the ribosome recruitment to the mRNAs to which it is bound. Numerous viruses encode proteins that play an important role in the translational control of their own mRNAs, interfering with cellular mRNA translation and indeed some of them supplant the function of some eIF4F components. A good example is the NSP3 protein of rotavirus, which binds to eIF4G and the 3′ end of viral mRNAs, disrupting eIF4G–PABP1 binding and inhibiting the translation of cellular mRNA (Piron et al., 1998). Adenovirus also offers an example of a protein that obstructs cellular translation. It encodes a 100k protein that binds to eIF4G and disrupts the eIF4G-M nk1 interaction, thereby inhibiting the phosphorylation of eIF4E, which cooperate in the inhibition of host protein synthesis (Cuesta, Xi, and Schneider, 2000). Other viruses encode proteins that mimic different components of translation related factors. The N protein of hantaviruses, which use an orthomyxovirus-like cap-snatching mechanism to yield mRNAs with 5′ caps derived from cellular mRNAs (Dunn et al., 1995; Hutchinson, Peters, and Nichol, 1996), replaces, not only eIF4E function, but also eIF4A and eIF4G (Mir and Panganiban, 2008). During viral translation, the N protein binds to the cap structures of viral mRNAs and to the 43S pre-initiation complex, facilitating loading of ribosomes onto viral capped mRNA. This list of such proteins is now expanded with the contribution of the influenza virus polymerase, which seems to replace the cellular cap-binding factor eIF4E for viral mRNAs translation.
Influenza virus mRNA translation
Translation of cellular mRNAs is strongly inhibited in influenza virus-infected cells (Skehel, 1972). The dephosphorylation of eIF4E triggered by the infection (Feigenblum and Schneider, 1993; Katze, Chen, and Krug, 1984), which strongly correlates wit h decreased rate of translation in many systems (Scheper and Proud, 2002), could be at least in part, involved in this phenomenon. Despite the capped nature of influenza virus mRNAs, t heir translation seems to be independent of functional eIF4E protein within the infected cells (Burgui et al., 2007). Furthermore, influenza virus infection efficiently progresses in adenovirus infected cells, despite the strong dephosphorylation of eIF4E induced by adenovirus infection (Zhang, Feigenblum, and Schneider, 1994). Therefore, translation of influenza virus mRNAs may escape from the viral-induced inhibition of cap-dependent initiation. However translation of influenza virus mRNAs requires the participation of the two other components of the eIF4F complex, the eIF4A helicase and the eIF4G scaffold protein (Yángüez et al., 2011). These results discard a potential direct recruitment of the 40S ribosomal subunit to the viral mRNAs, which would confer eIF4F-independence and indicate that coupling of influenza virus mRNAs to eIF4G is absolutely required for efficient viral translation.
Role of the viral polymerase
We have provided evidence that the viral polymerase interacts with translation initiation complexes (Burgui et al., 2007). The viral polymerase co-immunoprecipitates with eIF4GI both in influenza virus infected cells and when recombinant polymerase subunits were expressed by transfection (Burgui et al., 2007). Furthermore, using experimental conditions in which the association of eIF4GI to the cellular cap-binding factor eIF4E protein is reduced, the influenza virus infection specifically increases the association of eIF4GI with the cap structures (Burgui et al., 2007). Here, we show that the PB2 cap-binding subunit interacts with eIF4GI (Figs. 3 and 4) and therefore, it could mediate the association of the polymerase complex with the translation initiation eIF4F complex.
The characterization of the role of the influenza virus polymerase in the modulation of viral protein synthesis has shown that the translation of a viral flu-like mRNA, whose expression is driven by the viral polymerase, behaves similarly to that of the viral mRNAs in the infected cells in terms of eIF4E independence. In fact, an increase in the translation of this viral flu-like mRNA occurs under conditions of eIF4E-eIF4G dissociation (Fig. 6). The availability of eIF4E appears to play a critical role in the switch from cap-dependent to IRES-mediated translation in picornavirus-infected cells. It has been reported that in picornavirus-infected cells where both capped and IRES-containing mRNAs are present, a decrease in the amount of eIF4E associated with the eIF4F complex elicits a striking increase in IRES-mediated viral mRNA translation (Svitkin et al., 2005). This effect is not observed in translation extracts depleted of capped mRNAs, indicating that capped mRNAs compete with IRES-containing mRNAs for translation (Svitkin et al., 2005). These data parallel the observations during influenza virus infection since the reported eIF4E dephosphorylation should decrease the eIF4E-eIF4G association and viral mRNAs might associate with the eIF4G/4A subcomplex via the PB2-eIF4G interaction, resembling the behavior of the IRES.
Model for viral mRNA translation
As suggested by different authors, preferential translation of viral mRNAs could rely on the specific interaction of the viral 5′-UTR with a selective factor present in the infected cell. Viral polymerase interacts with the 5′UTR sequences common to all viral segments in vitro (Shih, Nemeroff, and Krug, 1995). This interaction has been proposed as a way to protect the viral mRNAs during the cap-snatching process. Moreover, the polymerase complex from extracts of infected cells binds to cap structures with greater affinity than eIF4E does and accordingly, m7GTP is 200-fold less potent cap binding inhibitor for the influenza virus polymerase than for the eIF4E factor (Hooker et al., 2003). Therefore, the viral polymerase could associate to the capped 5′UTR sequences of the viral transcripts, even in the presence of the cap-binding complex (CBC) or eIF4E. This is feasible and in fact, the polymerase bind s to the cap-structures of cellular pre-mRNAs in the presence of the CBC complex during the initiation of viral transcription. On the other hand, the presence of influenza virus polymerase complexes free of nucleoprotein, both in the nucleus and cytoplasm of the infected cells, has been reported (Detjen et al., 1987).
As described above although viral mRNAs can overcome the viral-induced eIF4E dephosphorylation they need a functional eIF4G factor to be successfully translated (Burgui et al., 2007; Yángü ez et al., 2011). Therefore, collectively, the data support a model in which influenza virus polymerase, bound to the cap and the 5′-UTR common viral sequence, would replace eIF4E function and specifically recruit translation machinery to the viral mRNAs. The association of the viral polymerase and eIF4GI may be involved in the preferential translation of viral mRNAs during influenza infection. In addition, the interaction of NS1 with the translation initiation factors eIF4GI (Aragón et al., 2000) and PABP1 (Burgui et al., 2003) could promote the formation of a “closed loop” between the 5′ and 3′ ends of the viral mRNA.
Finally, as for the aforementioned examples of other viruses, influenza virus seems to enlarge the list of viruses that possess proteins that can modulate the translation of viral proteins by specifically associating with translation initiation complexes. In this particular case two viral proteins, PB2 and NS1, and two cellular translation related factors, eIF4GI and PABP1, appear to facilitate the specific translation of viral mRNAs, contributing to the optimal synthesis of proteins during the viral cycle.
MATERIALS AND METHODS
Biological materials
The HEK293T cell line and the influenza virus A/Victoria/3/75 (VIC) strain were used throughout these studies, as were the plasmids pCMV-PA, pCMV-PB1, pCMV-PB2 and pCMV-NP (Falcón et al., 2004). J.J. Sanz-Ezquerro supplied plasmid pRSET-PB2 that expresses a recombinant His-PB2 protein. Plasmids pCMV-PB2-His and pRSET-eIF4GI157-1553 expressing recombinant PB2-His and an N-terminal truncated His-eIF4GI proteins have been previously described (Jorba, Coloma, and Ortin, 2009, Aragón, 2000 #1070). Plasmids pCDNA3-HA-PB2 (318-483), pCDNA3-HA-PB2 (538-693) and pCDNA3-HA-PB2 (538-759) were provided by D. Hart. The plasmid expressing, by an RNA poymerase I promoter, the CAT gene flanked with the UTRs sequences of influenza virus segment 8 in antisense orientation (pHH-NS-CAT) was kindly provided by A.Rodriguez. The plasmid expressing the CAT gene with the UTRs sequences of segment NS expressed by the RNA polymerase II promoter (pC MV-NS-CAT) was constructed by insertion of the NS-CAT fragment from pHH-NS-CAT into the pCDNA5 plasmid. E. Martínez-Salas supplied the plasmid expressing the CAT protein without the viral UTRs (pCMV-CAT). Complete protease inhibitors and RNase (human placenta RNAse inhibitor) inhibitor were obteined from Roche and rapamycin was bought from Calbiochem.
Transfection and virus infection
All infections were carried out at a multiplicity of infection of 3 PFU/cell and when necessary, HEK293T were previously transfected by the calcium-phosphate method (Wigler et al., 1979). At different times post-infection, cell were used for immunoprecipitation, metabolic labeling, RT-PCR or CAT Elisa (Roche) studies.
Western blotting
Western blotting was performed as described previously (Aragón et al., 2000). The following primary antibodies were used: for translation initiation factor eIF4GI, a mixture of four rabbit polyclonal antibodies (1:8.000 each) (Aragón et al., 2000); for eIF4E, a monoclonal antibody from Transduction Laboratories (1:2.000); for PA, monoclonal antibodies 2 and 9 (1:20 each) (Bárcena et al., 1994); for PB1, a rat polyclonal antibody (1:1.000) (Coloma et al., 2009); for PB2, monoclonal antibodies 8 and 28 (1:100 each) (Bárcena et al., 1994); for NP, a rabbit polyclonal antibody (1:5.000) (Coloma et al., 2009); for the His tag, a rabbit polyclonal antibody (1:10.000) from Sigma; for VP1, a rabbit polyclonal antibody (1:1000) (a gift from J. F. Rodríguez); for ß-tubulin, a mouse monoclonal antibody (1:50.000) from Sigma; for HA, a mouse monoclonal antibody (1:1.000) from Abcam and for 4E-BP1 a goat polyclonal antibody (1:200); and 4E-BP1 P-Ser65, a rabbit polyclonal antibody (1:200) both from Santa Cruz Biotechnology.
Expression and Purification of Recombinant Proteins
Recombinant plasmids expressing the His-4E-BP1 or His-eI F4E proteins were generously supplied by S. Morley (University of Sussex) and S. Curry (Imperial College London), respectively. The recombinant proteins were purified by affinity chromatography on a Hitrap chelating column (GE Healthcare).
To perform eIF4GI-PB2 interaction and pep-spot analysis, the His-PB2 protein and a His-eIF4GI protein that contains the eIF4GI sequence lacking the first 157 amino acids (Aragón et al., 2000), were expressed in E. coli BL21DE3 pLysS cells harboring the corresponding pRSET recombinant plasmids. After induction for 27 h at 16°C with IPTG (10 μM) for His-PB2 expression or 2 h at 37°C for His-eIF4G I expression with IPTG (1M), the cells were resuspended in a buffer containing 50 mM Tris-HCl (pH-8.0), 500 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% NP40 and 100 mM imidazol (supplemented before use with “Complete” and 10 mM 2ME) and they were sonicated. After removal of the cell debris by centrifugation, the supernatant was incubated overnight at 4°C with 2+Ni-NTA-agarose resin (Invitrogen) equilibrated in the same buffer with gentle rocking. After extensive washes with 20 mM Tris-HCl (pH-8.0), 0.1 M KCl, 5 mM MgCl2, 10 % glycerol, 10 mM 2ME and 50 mM imidazole (washing buffer), the proteins were eluted with 1 M imidazole in washing buffer. The control protein His-VP1 was purified as described previously (Garriga et al., 2007).
Analysis of viral polymerase subunits associated with eIF4GI factor
HEK293T cells w ere transfected with the plasmids pCMVPB1, pCMVPB2 or pCMVPA and 24 hours later, cytosolic extracts were prepared in buffer A (150 mM NaCl, 1.5 mM MgCl2 10 mM Tris-HCl (pH 8.5), 0.2%, Igepal) with Complete protease inhibitor and phosphatase inhibitors (5 mM Na3VO4, 5 mM ß-glycerophosphate, 5 mM sodium molibdate) plus human placenta RNAse inhibitor (1:1.000). After centrifugation at 10.000 x g, the supernatants were collected and used for immunoprecipitation studies. The cell extracts were incubated with a specific anti-eIF4GI antibody or pre-immune serum as reported previously (Aragón et al., 2000) and after incubation, they were washed 10 times with buffer A and analyzed by SDS-PAGE and Western blotting.
To map the interaction domain of the PB2 subunit with eIF4GI, HEK293T cells were mock transfected or transfected with pCDNA3-HA-PB2 (318-483), pCDNA3-HA-PB2 (538-693) or pCDNA3-HA-PB2 (538-759) and, 24 hours later, cytosolic extracts were prepared in buffer A containing proteases, phosphatases, and RNase inhibitors. After centrifugation at 10.000 × g, the supernatants were collected and used for co-immunoprecipitation studies as described above.
To examine the eIF4GI-PB2 interaction, purified His-PB2 and His-eIF4GI proteins were incubated in buffer A with 0.5% Igepal over night at 4°C. After that the proteins were processed for immunoprecipitation with antibodies against eIF4GI o r the pre-immune serum, washed 10 times with buffer A with 0.5% Igepal and analyzed by SDS-PAGE and Western blotting.
For pep-spot analysis, a collection of 122 overlapping peptides corresponding to the eIF4GI protein was synthesized on a cellulose membrane as described (Valle et al., 1999). Each peptide contained 13 amino acids and had three amino acids shift with the next. The membrane was blocked with 3% low fat milk in PBS overnight at room temperature (RT) and it was then incubated with the primary and secondary antibodies used to detect PB2. Subsequently, the membrane was incubated with His-PB2 protein (5 μg/ml) in PBS for 2 h at RT followed by its corresponding primary and secondary antibodies. The membrane was then incubated with the primary and secondary antibodies used to detect VP1 and finally with His-VP1 (5 μg/ml) in PBS for 2 h at RT followed by its corresponding primary and secondary antibodies. After each round of incubation Western blots were carried out and after each Western blot assay the membrane was stripped three times in solution A (8M urea, 1% SDS, 0,5% β-mercaptoethanol in PBS pH-7) for 5 min at 40°C in a sonication bath, followed by treatment with stripping solution B (acetic acid, EtOH, H2O 10:50:40) with agitation. Before the next incubation round, the stripped membrane was examined by ECL and developed.
In vitro translation
For in vitro translation reactions, transcription of control capped dicistronic mRNA was performed from XhoI linearized pGEM-Cap-CAT:EMCV-IRES-Luc (which expresses the IRES element of the encephalomyocarditis virus) (Pisarev et al., 2004) using the Megascript transcription system (Ambion). Addition of the 7-mGTP cap 0 structure was performed using ScriptCaptm m7G Capping System (Epicentre Biotechnologies) and mRNA was poly-adenylated using poly-A polymerase (PAP) following the suppliers’ recommendations. To obtain influenza virus RNAs for in vitro translation reactions, cytosolic extracts of infected cells were obtained 6 hpi in buffer A as described above. Total RNA was isolated from the extracts using Ultraespec reagent (Biotecx Laboratories).
In vitro translation reactions were performed using the Flexi rabbit reticulocyte lysate (RRL) system (Promega), using 200 μg/ml of cytosolic RNA from influenza infected cells or 10 μg/ml of control dicistronic RNAs. In reactions that required the addition of either recombinant His-4E-BP1 or His-eIF4E the reactions were preincubated with the recombinant proteins at 30 °C for 15 min prior to the addition of RNA. The reactions were terminated after 90 min. by the addition of SDS-PAGE sample buffer and the products were subsequently resolved on 12.5% SDS-gels. In some experiments eIF4E protein was depleted from RRL as previously described (McKendrick et al., 2001).
Amplification and purification of recombinant polymerase complexes and RNPs
Recombinant RNPs containing the NS-CAT genomic RNA were generated and amplified in vivo by transfection of plasmids, pCMV-PB1, pCMV-PB2-His, pCMV-PA, pCMV-NP and pHH-NS-CAT in HEK293T cells as described previously (Jorba, Coloma, and Ortin, 2009). For RNP purification, clarified cell extracts were incubated overnight at 4°C with 2+Ni-NTA-agarose resin in a buffer containing 50 mM Tris-HCl (pH-8.0), 100 mM KCl, 5 mM MgCl2, 0.5% Igepal, 20 mM imidazol and 1 U/μl RNAsin-EDTA free protease inhibitors cocktail. The resin was washed with 80 volumes of this buffer and RNPs were eluted in the same buffer containing 150 mM imidazol. In vivo reconstituted RNPs using pCMV-PB2 (untagged) were used as control.
Recombinant polymerase complexes were obtained by transfection of plasmids pCMV-PB1, pCMV-PB2-His and pCMV-PA in HEK293T cells that were processed and purified as described above.
Effect of rapamicyn in CAT expression driven by the RNA polymerase II or by the viral polymerase
To examine the effect of rapamicyn treatment on the translation of a CAT gene transcribed by the RNA polymerase II or the viral polymerase, the scheme outline d in Fig. 5 was followed. HEK293T cells were first transfected with pCMV-PB1, pCMV-PB2, pCMV-PA and pCMV-NP plasmids and 12 h later, the cells were maintained in the presence or absence of rapamicyn for 4 h before the transfection with pCMV-CAT, pCMV-NS-CA T or recombinant His-tagged NS-CAT RNPs purified as described above. At 7 hours post transfection, total cell extracts were prepared and used for CAT determination by ELISA assays. In each condition, cytolplasmic RNA was isolated from the cells extracts using TRI reagent (Sigma) to evaluate the CAT and β-actin mRNA content by quantitative RT-PCR using the Applied Biosystems kit.
qRT-PCR mRNA detection
Cytosolic extracts were prepared as described and the RNA was isolated using TRIzol (Invitrogen). RNA (2-5 μg) was then treated with Turbo DNA-free kit (Ambion) and reverse transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer’s instructions. Real-time PCR was performed with SYBR green (Applied Biosystems) using an ABI Prism 7700 thermocycler with fluorescence detection (Applied Biosystems). The following pair of oligos was used for CAT mRNA detection: FW (5′-CTGGCGATTCAGGTTCATC-3′) and RV (5′-TTTTTTTTTTTTTTTTCAGATCTATTACG-3′). Beta-actin mRNA was detected with: FW (5′-CCCAGCACAATGAAGATCAA-3′ and RV (5′ CGATCCACACGGAGTACTTG-3′) and used for normalization. Appropriate ) controls were included in each reaction, and dissociation analysis was performed at the end of each run to confirm the specificity of the reaction.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to J. Ortin and A. Rodriguez for critical review of the manuscript. The technical assistance of F. Roncal, Y. Fernández and N. Zamarreño is gratefully acknowledged. I. Goodfellow is a Wellcome Senior Fellow. This work was supported by Ministerio de Educacion y Ciencia, Plan Nacional de Investigacion Científica, Desarrollo e Innovacion Tecnologica (BFU2008-00448) and Ciber de Enfermedades Respiratorias.
REFERENCES
- Aragón T, de la Luna S, Novoa I, Carrasco L, Ortín J, Nieto A. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 2000;20(17):6259–6268. doi: 10.1128/mcb.20.17.6259-6268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárcena J, S. d. l. L., Ochoa M, Melero JA, Nieto A, Ortín J, Portela A. Monoclonal antibodies against the influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J. Virol. 1994;68:6900–6909. doi: 10.1128/jvi.68.11.6900-6909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. Embo J. 1996;15(3):658–664. [PMC free article] [PubMed] [Google Scholar]
- Blaas D, Patzelt E, Keuchler E. Identification of t he cap binding protein of influenza virus. Nucl. Acids Res. 1982;10:4803–4812. doi: 10.1093/nar/10.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgui I, Aragón T, Ortín J, Nieto A. PABP1 and eIF4GI associate to influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 2003;84:3263–3274. doi: 10.1099/vir.0.19487-0. [DOI] [PubMed] [Google Scholar]
- Burgui I, Yángüez E, Sonenber N, Nieto A. Influenza mRNA translation revisited: is the eIF4E cap-binding factor required for viral mRNA translation? J. Virol. 2007;81(22):12427–12438. doi: 10.1128/JVI.01105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coloma R, Valpuesta JM, Arranz R, Carrascosa JL, Ortin J, Martin-Benito J. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 2009;5(6):e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Xi Q, Schneider RJ. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. Embo J. 2000;19(13):3465–3474. doi: 10.1093/emboj/19.13.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Luna S, Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 1995;69(4):2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detjen BM, St Angelo C, Katze MG, Krug RM. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J Virol. 1987;61(1):16–22. doi: 10.1128/jvi.61.1.16-22.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458(7240):914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- Dunn EF, Pritlove DC, Jin H, Elliott RM. Transcription of a recombinant bunyavirus RNA template by transiently expressed bunyavirus proteins. Virology. 1995;211(1):133–143. doi: 10.1006/viro.1995.1386. [DOI] [PubMed] [Google Scholar]
- Elton D, Digard P, Tiley L, Ortín J. Structure and function of the influenza virus RNP. In: Kawaoka Y, editor. Contemporary topics in influenza virology. Horizon Scientific Press; Norfolk: 2005. [Google Scholar]
- Enami K, Sato TA, Nakada S, Enami M. Influenza virus NS1 protein stimulates translation of the M1 protein. J. Virol. 1994;68(3):1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón A, Marión R, Zürcher T, Gomez P, Portela A, Nieto A, Ortín J. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 2004;78:3880–3888. doi: 10.1128/JVI.78.8.3880-3888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenblum D, Schneider RJ. Modification of eukaryotic initiation factor 4F during infection by influenza virus. J. Virol. 1993;67(6):3027–3035. doi: 10.1128/jvi.67.6.3027-3035.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7(2):e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foeger N, Glaser W, Skern T. Recognition of eukaryotic initiation factor 4G isoforms by picornaviral proteinases. J Biol Chem. 2002;277(46):44300–44309. doi: 10.1074/jbc.M208006200. [DOI] [PubMed] [Google Scholar]
- Garfinkel MS, Katze MG. Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5′-untranslated region. J. Biol. Chem. 1993;268(30):22223–22226. [PubMed] [Google Scholar]
- Garriga D, Navarro A, Querol-Audi J, Abaitua F, Rodriguez JF, Verdaguer N. Activation mechanism of a noncanonical RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 2007;104(51):20540–20545. doi: 10.1073/pnas.0704447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. mTOR signaling to translation. Curr Top Microbiol Immunol. 2004;279:169–197. doi: 10.1007/978-3-642-18930-2_11. [DOI] [PubMed] [Google Scholar]
- Gradi A, Foeger N, Strong R, Svitkin YV, Sonenberg N, Skern T, Belsham GJ. Cleavage of eukaryotic translation initiation factor 4GII within foot- and -mouth disease virus-infected cells: identification of the L-protease cleavage site in vitro. J Virol. 2004;78(7):3271–3278. doi: 10.1128/JVI.78.7.3271-3278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol. 2008;15(5):500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- Hooker L, Sully R, Handa B, Ono N, Koyano H, Klumpp K. Quantitative analysis of influenza virus RNP interaction with RNA cap structures and comparison to human cap binding protein eIF4E. Biochemistry. 2003;42(20):6234–6240. doi: 10.1021/bi027081r. [DOI] [PubMed] [Google Scholar]
- Huarte M, Sanz-Ezquerro JJ, Roncal F, Ortín J, Nieto A. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J. Virol. 2001;75(18):8597–8604. doi: 10.1128/JVI.75.18.8597-8604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson KL, Peters CJ, Nichol ST. Sin Nombre virus mRNA synthesis. Virology. 1996;224(1):139–149. doi: 10.1006/viro.1996.0515. [DOI] [PubMed] [Google Scholar]
- Jorba N, Coloma R, Ortin J. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 2009;5(5):e1000462. doi: 10.1371/journal.ppat.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen L, Lankinen H, Harjunpaa I, Hokynar K, Soderlund-Venermo M, Oker-Blom C, Hedman L, Hedman K. Acute-phase-specific heptapeptide epitope for diagnosis of parvovirus B19 infection. J Clin Microbiol. 1999;37(12):3952–3956. doi: 10.1128/jcm.37.12.3952-3956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze MG, Chen YT, Krug RM. Nuclear-cytoplasmic transport and VAI RNA-independent translation of influenza viral messenger RNAs in late adenovirus-infected cells. Cell. 1984;37(2):483–490. doi: 10.1016/0092-8674(84)90378-7. [DOI] [PubMed] [Google Scholar]
- Katze MG, DeCorato D, Krug RM. Cellular mRNA translation is blocked at both initiation and elongation after infection by influenza virus or adenovirus. J. Virol. 1986;60(3):1027–1039. doi: 10.1128/jvi.60.3.1027-1039.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS. Vira l IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33(6):274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Sabatini D, Pandey P, Gingras AC, Majumder PK, Kumar M, Yuan ZM, Carmichael G, Weichselbaum R, Sonenberg N, Kufe D, Kharbanda S. Regulation of the rapamycin and FKBP-target 1/mammalian target of rapamycin and cap-dependent initiation of translation by the c-Abl protein-tyrosine kinase. J Biol Chem. 2000;275(15):10779–10787. doi: 10.1074/jbc.275.15.10779. [DOI] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornavirus proteases-Implication for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Luo GX, Luytjes W, Enami M, Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J. Virol. 1991;65(6):2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima Y, Sudhakar A, Lively JC, Ueki K, Kharbanda S, Kahn CR, Sonenberg N, Hynes RO, Kalluri R. Tumstatin, an endothelial cell-specific inhibitor of protein synthesis. Science. 2002;295(5552):140–143. doi: 10.1126/science.1065298. [DOI] [PubMed] [Google Scholar]
- Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J Gen Virol. 2008;89(Pt 3):611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol Cell Biol. 2001;21(11):3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT. A protein that replaces the entire cellular eIF 4F complex. EMBO J. 2008;27(23):3129–3139. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YW, Katze MG. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 1995;270(47):28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J Virol. 2004;78(9):4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- Salvatore M, Basler CF, Parisien J-P, Horvath CM, Bourmakina S, Zheng H, Muster T, Palese P, García-Sastre A. Effects of influenza A virus NS1 protein on protein expression:the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 2002;76(3):1206–1212. doi: 10.1128/JVI.76.3.1206-1212.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, Proud CG. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur J Biochem. 2002;269(22):5350–5359. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SR, Krug RM. Surprising function of the three influenza vira l polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology. 1996;226(2):430–435. doi: 10.1006/viro.1996.0673. [DOI] [PubMed] [Google Scholar]
- Shih SR, Nemeroff ME, Krug RM. The choice of alternative 5′ splice sites in influenza virus M1 mRNA is regulated by the viral polymerase complex. Proc Natl Acad Sci U S A. 1995;92(14):6324–6328. doi: 10.1073/pnas.92.14.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ. Polypeptide synthesis in influenza-virus infected cells. Virology. 1972;49:23–36. doi: 10.1016/s0042-6822(72)80004-7. [DOI] [PubMed] [Google Scholar]
- Sousa C, Schmid EM, Skern T. Defining residues involved in human rhinovirus 2A proteinase substrate recognition. FEBS Lett. 2006;580(24):5713–5717. doi: 10.1016/j.febslet.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol. 2005;25(23):10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol. 2007;14(3):229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I, Broni BA, Krug RM. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc. Natl. Acad. Sci. USA. 1981;78:7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Muñoz M, Kremer L, Valpuesta JM, Martinez C, Carrascosa JL, Albar JP. Selection of antibody probes to correlate protein sequence domains with their structural distribution. Protein Science. 1999;8:883–889. doi: 10.1110/ps.8.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. DNA-mediated transfer of the adenine phosphoribosyltranferase locus into mammalian cells. Proc. Natl. Acad. Sci. USA. 1979;76(3):1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yángüez E, Castello A, Welnowska E, Carrasco L, Goodfellow I, Nieto A. Functional impairment of eIF4A and eIF4G factors correlate with inhibition of influenza virus mRNA translation. Virology. 2011;25:93–102. doi: 10.1016/j.virol.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458(7240):909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- Yumerefendi H, Tarendeau F, Mas PJ, Hart DJ. ESPRIT: An automated, library-based method for mapping and soluble expression of protein domains from challenging targets. J Struct Biol. 2010;172:66–74. doi: 10.1016/j.jsb.2010.02.021. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feigenblum D, Schneider RJ. A late adenovirus factor induces eIF-4E dephosphorylation and inhibition of cell protein synthesis. J Virol. 1994;68(11):7040–7050. doi: 10.1128/jvi.68.11.7040-7050.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.