Abstract
Several vertebrate positive-sense RNA viruses, namely the Picornaviridae and Caliciviridae have evolved to use a protein-primed mechanism of genome replication. This results in the covalent linkage of a virus encoded protein, VPg (viral protein genome-linked), to the 5′ end of viral RNA. Recent studies have highlighted the pivotal role VPg plays in the life cycle of these viruses, which in the case of the Caliciviridae, includes a role in viral protein synthesis. This article provides an overview of the current knowledge of the functions of vertebrate RNA virus VPg proteins, illustrating their diverse function and the parallels they share with plant virus VPg proteins.
Introduction
Early studies with a number of vertebrate positive-sense (+) RNA genomes highlighted that RNA isolated from virus particles contained a peptide or protein covalently linked to the viral RNA [1-3]. This protein, subsequently shown to be encoded by the viral genome and named VPg for virus protein, genome linked, is now known to play essential functions in the viral life cycle. The VPg protein has been extensively characterized from only two families of vertebrate (+) RNA viruses, namely members of the Picornaviridae and Caliciviridae. Computational studies have indicated a possible VPg protein present in the Astroviridae [4], although experimental evidence on the presence of VPg linked to astrovirus RNA has yet to be provided and warrants further study.
This review will focus on the published literature on the role of vertebrate virus VPg proteins, namely members of the Picornaviridae and Caliciviridae and their key roles in viral protein synthesis and genome replication. Whilst a comprehensive review of the Caliciviridae and Picornaviridae family translation and replication mechanisms is beyond the scope of the current review, recent articles that cover many aspects of the calicivirus and picornavirus replication cycle are readily available (e.g. [5,6] and references therein). This article should be read in combination with the accompanying chapter on plant virus VPg proteins to allow a full appreciation of the diverse function of viral VPg proteins but where possible, common themes will be highlighted in the text below.
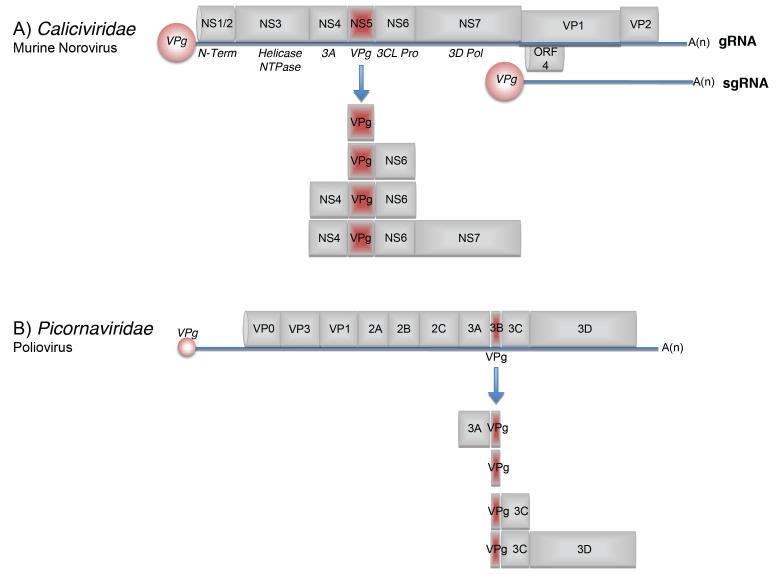
Whilst the Caliciviridae represent one of the most poorly characterized family of (+) RNA viruses, the Picornaviridae represent some of the best characterized of all viruses. The genome layout of each family is somewhat different (Figure 1), although they share some similarities in the presence of a large polyprotein that is posttranslationally cleaved by virus-encoded proteases. In contrast to the Picornaviridae that express only a single open reading frame, members of the Caliciviridae typically have three ORFS (Figure 1A). Some divergence from this exists within the Caliciviridae however because as illustrated in Figure 1, murine norovirus possesses four ORFs (Figure 1A). The synthesis of the calicivirus capsid proteins VP1 and VP2, and ORF4 in the case of MNV, occurs via the production of the multicistronic subgenomic RNA (sgRNA). As in vitro cell culture systems exist for only a few members of the Caliciviridae, a number of the animal viruses tend to be used as model systems with which to characterize calicivirus biology [5].
Figure 1. Schematic of the genomes of representative members of the Caliciviridae and Picornaviridae.
Viral RNA is illustrated as the blue line with the individual mature proteins boxed in grey. The VPg proteins linked to the 5′ end of the viral RNA are shown as red filled circles. (A) The murine norovirus genome is shown highlighting the four open reading frames note however that the majority of caliciviruses possess only three open reading frames. In addition to the viral genomic RNA (gRNA), calicivirus replication results in the production of a ~2.5Kb subgenomic RNA (sgRNA), which in the case of murine norovirus, expresses the major and minor capsid proteins, VP1 and VP2 respectively. The NS1-7 nomenclature is used to highlight the mature nonstructural proteins made from the processing of the ORF1 polyprotein. Other synonyms widely used in the literature are displayed below the viral genome in italics. Note that in addition to the mature nonstructural proteins, a variety of precursors are produced during virus replication. For simplicity however, only those containing VPg are displayed. (B) The poliovirus genome is shown as a representative member of the Picornaviridae family. The large viral polyprotein is displayed along with the various mature proteins produced as a result of polyprotein processing. Note that for simplicity only the VPg containing precursor proteins are shown. The positions of the cis-acting RNA replication elements is not shown but are illustrated in figure 4. The figure is not drawn to scale.
VPg as a proteinaceous cap substitute
Whilst the calicivirus VPg proteins share little or no sequence homology with the VPg peptides/proteins from picornaviruses and plant viruses, their larger size (13-15kDa) make them a closer relative of plant virus VPg proteins than those from picornaviruses, typically 22 amino acids. Early studies illustrated a disparity in the relative requirement for the VPg protein for viral infectivity for vertebrate (+) RNA viruses. Whereas treatment of the purified virion RNA from the calicivirus vesicular exanthema virus (VESV) with proteinase K, which removed covalently linked VPg, ablated infectivity [7], the VPg protein linked to the poliovirus genome was not required for infectivity [8,9] and in fact in vitro transcribed uncapped poliovirus RNA is infectious [10]. These data indicated that the VPg protein of caliciviruses plays an essential role in the initial stages of virus infection, whereas the linkage of the picornavirus VPg to RNA is not required for infectivity of purified viral RNA. Evidence that this was due to a role for the calicivirus VPg protein in viral translation was first confirmed for feline calicivirus (FCV) [11], indicating that, as described in the accompanying chapter for plant virus VPg proteins, that the calicivirus VPg may function as a proteinaceous cap substitute, a unique feature for any vertebrate RNA virus. The VPg protein of members of the Picornaviridae is not required for viral translation and instead they use an internal ribosome entry site (IRES) within the 5′ UTR of the viral RNA [12]. Studies with FCV confirmed that the VPg protein interacts directly with the cap-binding protein eIF4E and that this interaction is essential for viral translation [13]. Unlike the plant virus VPg-eIF4E interaction however, the calicivirus VPg-eIF4E interaction does not affect the binding of eIF4E to the cap structure present on cellular mRNAs. In fact cap-sepharose chromatography can be readily used to isolate the calicivirus VPg-eIF4E complex from infected cells [13,14]. This approach also highlighted that in addition to the mature form of VPg, typically 13-15kDa, a variety of VPg containing precursors (illustrated in figure 1) can also potentially bind eIF4E. The role of the VPg precursor-eIF4E interaction is yet to be determined but one might expect that as the concentration of viral VPg-containing precursors accumulates in the cell, their interaction with eIF4E may contribute to host protein shut off or allow the sequestration of initiation factors to the sites of viral RNA translation and synthesis.
The role of the VPg-eIF4E interaction is clearly defined for FCV, a member of the Vesivirus genus of the Caliciviridae, as the addition of recombinant 4E-BP1 to prevent the eIF4E-4G interaction [13] or the depletion of eIF4E [14] inhibits FCV translation. In the latter case this can be clearly restored via the addition of recombinant eIF4E. In contrast, the role of the VPg-eIF4E interaction in protein synthesis for the only member of the Norovirus genus for which this has been studied, namely murine norovirus (MNV), is less clear. A reproducible interaction between purified recombinant MNV VPg and eIF4E can be observed in vitro by capture ELISA [14] and the proteomic characterization of VPg containing complexes from cells indicates that eIF4E is present (our unpublished data), yet despite this, depletion of eIF4E or the addition of 4E-BP1 has no effect on MNV translation in vitro [14]. This may be due to the increased levels of other translation initiation factors in the rabbit reticulocyte lysates used for these studies i.e. functional complementation is occurring during in vitro translation. However, our recent unpublished studies would indicate that whilst the MNV VPg protein interacts with eIF4E, it also interacts directly with eIF4G and that mutations in VPg that affect the VPg-eIF4G interaction, without affecting the VPg-eIF4E interaction, are lethal to the virus (our unpublished data). Whilst further studies are required to validate our preliminary observations, these data are also paralleled in the plant virus Rice yellow mottle virus (discussed in more detail in the accompanying chapter) where a direct interaction between VPg and the central domain of the rice eIF(iso)4G1 factor has also been documented and this interaction is essential for viral virulence [15]. Cleavage of eIF4GI and II is observed during FCV replication in cell culture, although the cleavage products retain the ability to bind to eIF4E [16]. The function of this eIF4G cleavage is unknown but the kinetics of processing correlates with an observed decrease in host cell protein synthesis.
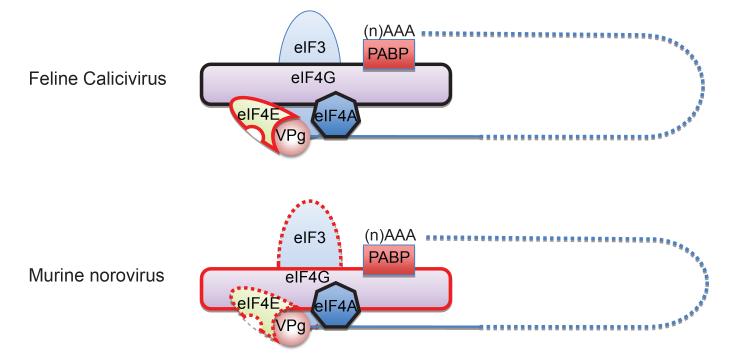
A role for the RNA helicase component of the eIF4F complex, namely eIF4A has also been demonstrated for FCV and MNV [14], which again displays similarities with the plant virus VPg (see the accompany chapter for further details, [17]). Dominant negative forms of eIF4A inhibit both FCV and MNV translation in vitro and the small molecule eIF4A inhibitor hippuristanol, potently inhibits MNV translation and replication in cell culture, with a lesser (but still significant) effect on FCV [14]. A direct interaction between the human norovirus VPg protein and the eIF3d component of the eIF3 complex has also been detected using the yeast two hybrid system, however a functional role for this interaction has yet to be confirmed [18]. It is likely that the calicivirus VPg has many interacting partners, forming a hub for the assembly of the translation and replication complex, as illustrated in the accompanying chapter for plant virus VPg proteins. Based on the limited published data available for the role of the calicivirus VPg protein in translation initiation, it is possible to propose a model of the components of the calicivirus translation initiation complex (Figure 2). Future studies will undoubtedly lead to the identification of additional host factors involved in the novel mechanism of calicivirus VPg-dependent protein synthesis.
Figure 2. Diagrammatic model of the calicivirus VPg-containing translation initiation complex.
Models based on published and unpublished data for two members of the Caliciviridae, namely feline calicivirus and murine norovirus are illustrated. Initiation factors that interact directly with VPg and play a functional role in viral protein synthesis are highlighted with a solid red surrounding line, whereas those that bind VPg directly but the function remains to be determined are highlighted with a dashed red surrounding line. Factors that do not interact directly with VPg or for which no data exists, yet are functionally important for calicivirus translation initiation are displayed with a solid black surrounding line. The interaction of poly A binding protein (PABP) with the 3′ poly A tail is inferred from published literature on other (+) RNA viruses and the presence of a poly A tail on the calicivirus genome.
VPg as a protein primer
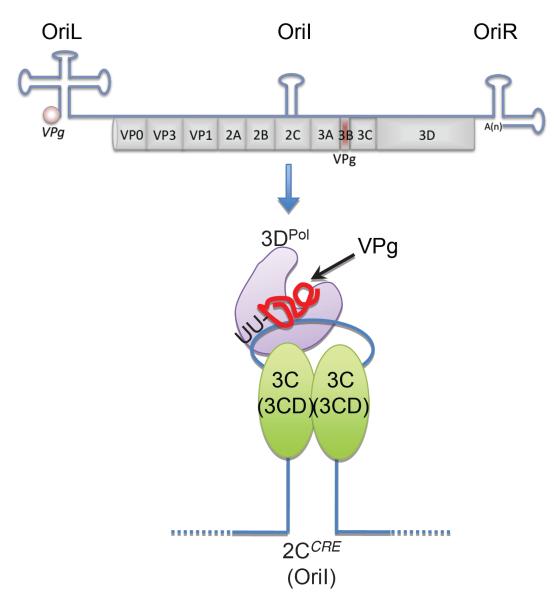
VPg is linked to picornavirus and calicivirus viral RNA, including the sgRNA produced during calicivirus infection [11]. This linking occurs during viral replication via the function of VPg as a protein primer for the viral RNA polymerase. The viral RNA-dependent RNA polymerase is encoded by the 3D region of the picornavirus genome and NS7 region of the calicivirus genome (Figure 1) and whilst they have the ability to initiate RNA synthesis de novo, viral RNA synthesis occurs in a VPg-dependent manner. The role of VPg as a protein primer for picornaviruses has been the focus of much study over many years (reviewed in [19]), in contrast the process was relatively poorly understood for the Caliciviridae until quite recently. The 5′ terminal nucleotides of the genomes of picornaviruses are UU whereas all calicivirus genomes begin with the sequence GU, hence the linkage of VPg to picornavirus genomes occurs via uridylylation of VPg whereas linkage to the calicivirus (+) RNA will occur guanylation. The generation of the picornavirus VPg containing protein primer VPg-pUpU-OH occurs in a template dependent manner in a complex which contains a small RNA stem loop structure first identified in the 2C coding region of the poliovirus genome [20,21] but located at various positions in other picornavirus genomes [19], the viral RNA-dependent RNA polymerase 3Dpol, two copies of the viral 3C protease (possibly in the precursor form of 3CD) and VPg, either in it’s mature or precursor forms (Figure 3) [19]. The mechanism by which this protein primer is translocated from its site of synthesis i.e. the middle of the viral RNA genome to the sites of RNA initiation i.e. the 3′ ends of the sense and anti-sense RNAs, remains to be determined. It has been proposed that this may occur via the use of precursor forms of the VPg protein [6] as although the mature form of VPg is readily detected during both calicivirus and picornavirus infection, a number of stable precursors are also observed (illustrated in Figure 1). Studies with picornaviruses has now clearly established that the precursor forms of VPg play essential roles in the viral life cycle (reviewed in [6]) and that a precursor form of VPg can be used for the initiation of viral genome replication [22]. Whilst not yet studied in detail for members of the Caliciviridae, the unique or additional functions of the precursors effectively allows (+) RNA viruses to overcome the limited coding capacity of their small RNA genomes, again a feature shared with plant (+) RNA viruses.
Figure 3. Model of the picornavirus VPg-uridylylation complex.
The positions of the three well characterized cis-acting RNA replication signals are illustrated as OriL, Oril and OriR, for Origin Left, Internal and Right respectively. For simplicity the minimal components of the uridylylation complex are shown. The VPg protein primer is synthesized by the Oril (also referred to as the 2CCRE) templated addition of two U residues to tyrosine 3 in VPg (shown in red). In addition, the reaction requires the viral RNA polymerase (3DPo1) and two copies of the 3C protease in its mature or precursor form (3CD). Note that experimental evidence suggest that precursor forms of the various components play an essential role in the formation of the VPg-pUpU-OH primer. A More in-depth description and analysis of this complex can be found in published literature ([6,22,50] and references therein).
The precise site of nucleotide linkage in the VPg proteins has been defined as tyrosine 3 for Picornaviruses (Figure 4A). In contrast, the site of RNA linkage to the calicivirus VPg has yet to be defined using similar rigorous experimental approaches but a number of studies have made preliminary steps in the identification of the site of RNA linkage. Biochemical studies with purified recombinant calicivirus RNA polymerase and VPg has determined that in vitro nucleotidylation can occur in a template independent manner. Tyrosine 21 of the rabbit hemorrhagic disease virus VPg [23] and the homologous position in human norovirus VPg, namely tyrosine 27 [24] can be nucleotidylated in vitro in similar reactions. Based on sequence alignments the equivalent position in the FCV VPg protein is tyrosine 24, mutation of which by reverse genetics is lethal [25]. All calicivirus VPg proteins share an invariant E/DEYDEΩ motif around the proposed site of RNA linkage where Ω represents any aromatic residue (Figure 4B). Whilst amino acid conservation would indicate that position 26 of the MNV VPg is the likely site of RNA linkage, in vitro studies have demonstrated that mutation of position 117 reduces VPg nucleotidylation at least in vitro [26]. Whether or not this position represents the authentic site of viral RNA linkage clearly requires additional studies. Whilst template independent nucleotidylation can occur in vitro, evidence for presence of RNA sequence or sequences that stimulate this reaction has been published. Studies with human norovirus have indicated that sequences near the 3′ end of the viral genome stimulate the nucleotidylation of VPg [24]. In MNV, the positive or minus sense RNA from the viral sgRNA also stimulates VPg nucleotidylation [26]. Another alternative interpretation of this data, is that rather than functioning as a template for VPg nucleotidylation, these regions of the viral genome contains RNA sequences that stabilize the viral polymerase or stimulate its activity. Either way, there is a clear need for additional experimental data to determine if the calicivirus genome contains a sequence which functions as a template for VPg nucleotidylation in a similar manner to that found in the Picornaviridae.
Figure 4. Features of the picornavirus and calicivirus VPg proteins.
Sequence alignment of the VPg proteins from representative members of the Picornaviridae (A) and Caliciviridae (B). The site of RNA linkage in the picornavirus VPg is highlighted in green. Abbreviations: PV1 poliovirus type 1, CVB3 Coxsackievirus type 3, FMDV foot and mouth disease virus. B) The site of RNA linkage in the calicivirus VPg highlighted in turquoise with position 117 of the murine norovirus (MNV) VPg protein, known to be important for nucleotidylation in vitro highlighted in yellow. The absolutely conserved signature motif of the calicivirus VPg proteins is shown (E/DEYDEΩ). Basic residues in the N-terminus of calicivirus VPg proteins are shown in red. Abbreviations: MNV murine norovirus, Norwalk human norovirus GI, FCV feline calicivirus, RHDV rabbit hemorrhagic disease virus.
Sometimes one is just not enough
Whereas all members of the Caliciviridae and the majority of the Picornaviridae express only a single VPg protein, foot and mouth disease virus (FMDV) possesses three [27] (Figure 4A), all of which are found liked to viral to viral RNA [28]. The function of the multiple copies of FMDV VPg has yet to be fully elucidated, but studies have illustrated a possible role of VPg in host range as a recombinant virus expressing only 1 VPg was attenuated in swine [29]. Recent data would however question these studies as FMDV with a single VPg protein displayed identical virulence in cattle [30]. A role for the FMDV VPg in virus release and/or maturation has also been demonstrated as viruses expressing only VPg1, replicated viral RNA efficiently but did not result in cytopathic effect or the production of extracellular virus [31]. Surprisingly, compensatory mutations in VPg itself or the 2C or 3A proteins restored the ability of the single VPg-expressing virus to form plaques, indicating either a direct or indirect interaction with VPg.
VPg, many other possible functions
Recent studies have illustrated that viruses have evolved many way of avoiding or inhibiting the many anti-viral sensing mechanisms present in cells [32,33]. Whilst many (+) RNA viruses produce viral RNA with a 5′ triphopshate group, which could be detected by the cytoplasmic sensors RIG-I and PKR known to detect 5′ triphosphorylated RNA [34-36], by virtue of the presence of VPg covalently linked to the 5′ end of viral RNA, it is possible that caliciviruses and picornaviruses do not. Whilst definitive experimental evidence does not exist as yet, it is possible that one of the benefits of using a protein-primed (i.e. VPg-dependent) mechanism of viral RNA synthesis is the ability to prevent the formation of 5′ triphosphorylated RNA during infection. Picornaviruses have however developed a variety of methods for antagonizing the innate immune response (e.g. cleavage of RIG-I [37] and other components of the innate immune response [38,39], as well as reference therein). It is also worth noting that an enzymatic activity found in cells often referred to as ‘VPg-unlinkase’, which cleaves the picornavirus VPg from the viral RNA has been described [40]. The function of this ‘unlinkase’ in the viral life cycle has yet to be determined but it has been suggested that it may play a role in distinguishing viral RNA used for translation, replication and encapsidation [41]. Whether the ‘unlinkase’ activity functions on calicivirus RNA is not known, however some specificity of the activity has been reported as the enzyme purified from mouse ascites cells was unable to cleave the plant virus VPg from comoviral RNA [42]
The calicivirus VPg proteins typically contain a positive charged N-terminus, rich in arginine and lysine (Figure 4B). Given this, it is possible that the N-terminus contributes to RNA-binding activity. Recombinant calicivirus VPg proteins can be readily purified using phosphocellulose (P11) or heparin-affinity chromatography [43], a feature of many RNA binding proteins. Our unpublished observations would also indicate that both the FCV and MNV VPg proteins possess the ability to interact with RNA in vitro but whether this is sequence specific and plays any role in the calicivirus life cycle requires further study. Whilst no experimental evidence exists to suggest that the picornavirus VPg protein interacts directly with RNA, a stable precursor form of VPg found in the infected cell, namely 3AB (3A-VPg, Figure 1), has RNA chaperone activity and mutations in the 3B (VPg) domain reduce this activity [44]. The precise function of the chaperone activity has yet to be determined, but 3AB stimulates RNA polymerase activity under certain conditions [45]. The 3AB precursor may well function to stabilize the replication complex-cell membrane interactions (reviewed in [6]). The ability to interact with RNA is also shared with plant viruses as the potyvirus VPg protein interacts with RNA in a sequence independent manner [46,47], although again the precise role for this interaction has yet to be determined.
A possible role for VPg in the encapsidation process of calicivirus RNA into virus particles has also been proposed as yeast-two hybrid analysis suggested a direct interaction occurs between the FCV VPg protein and the major capsid protein VP1 as well as the viral RNA polymerase NS6/7 (Figure 1, [48]). This would provide a mechanism to allow the specific incorporation of viral VPg-linked RNA in the presence of high concentrations of cellular mRNA. As discussed above, the VPg protein from the picornavirus FMDV, may also play a role in the release of viral RNA from cells via a mechanism involving the 2C NTPase/helicase as well as 3A [31]. Recent data shed new insights into the process of picornavirus genome encapsidation involving 2C [49]. Plant virus VPg proteins are also known to play a role in cell to cell spread (see the accompanying chapter for further details).
Conclusions
The VPg protein plays a critical role in the life cycle of members of the Picornaviridae and Caliciviridae families of small (+) RNA viruses. As described in the accompanying chapter for plant (+) RNA viruses, the many interactions and functions of the VPg protein from vertebrate (+) RNA viruses suggest that it functions as a hub around which virus replication, and in the case of caliciviruses, virus translation occurs. As a result of these critical roles, VPg forms a focal point through which many aspects of the virus life cycle can be tightly regulated either through the interaction of viral or cellular proteins with VPg, or through the availability of VPg and it’s precursors. There are clear parallels between the function of plant and vertebrate virus VPg proteins; whilst the main function of the picornavirus VPg appears to be in priming viral genome replication, calicivirus VPg proteins appear to be closer orthologues of the plant virus VPg proteins playing additional roles in viral translation initiation. Further studies will undoubtedly provide new insights into the additional functions of VPg from vertebrate RNA viruses, identifying additional parallels with plant viral VPgs.
Highlights.
The viral VPg protein is linked to viral RNA via a protein-primed mechanism of genome replication
The picornavirus VPg protein is primarily involved in viral genome replication
Calicivirus VPg plays an essential role in viral protein synthesis and viral genome replication
The calicivirus VPg shares some functional similarities to the VPg proteins from plant viruses
Acknowledgments
We apologize to all colleagues whose contributions we could not adequately discuss due to space constraints. Work in the Goodfellow laboratory is supported by the Wellcome Trust, Medical Research Council, European Union Marie Curie Networks and the Biotechnology and Biological Sciences Research Council. IG is a Wellcome Senior Fellow.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Schaffer FL, Ehresmann DW, Fretz MK, Soergel MI. A protein, vpg, covalently linked to 36s calicivirus rna. J Gen Virol. 1980;47(1):215–220. doi: 10.1099/0022-1317-47-1-215. [DOI] [PubMed] [Google Scholar]
- 2.Flanegan JB, Petterson RF, Ambros V, Hewlett MJ, Baltimore D. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate rnas of poliovirus. Proc Natl Acad Sci U SA. 1977;74:961–965. doi: 10.1073/pnas.74.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YF, Nomoto A, Detjen BM, Wimmer E. The genome-linked protein of picornaviruses i. A protein covalently linked to poliovirus genome rna. Proc Natl Acad Sci U S A. 1977;74:59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Mutairy B, Walter JE, Pothen A, Mitchell DK. Genome prediction of putative genome-linked viral protein (vpg) of astroviruses. Virus Genes. 2005;31(1):21–30. doi: 10.1007/s11262-004-2196-1. [DOI] [PubMed] [Google Scholar]
- 5.Vashist S, Bailey D, Putics A, Goodfellow I. Model systems for the study of human norovirus biology. Future Virology. 2009;4(4):353–367. doi: 10.2217/fvl.09.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron CE, Suk Oh H, Moustafa IM. Expanding knowledge of p3 proteins in the poliovirus lifecycle. Future Microbiol. 2010;5(6):867–881. doi: 10.2217/fmb.10.40. ** A general overview of the role of precursor proteins in the picornavorus life cycle.
- 7.Burroughs JN, Brown F. Presence of a covalently linked protein on calicivirus rna. J Gen Virol. 1978;41(2):443–446. doi: 10.1099/0022-1317-41-2-443. [DOI] [PubMed] [Google Scholar]
- 8.Nomoto A, Kitamura N, Golini F, Wimmer E. The 5′-terminal structures of poliovirion rna and poliovirus mrna differ only in the genome-linked protein vpg. Proc Natl Acad Sci D S A. 1977;74(12):5345–5349. doi: 10.1073/pnas.74.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanegan J, Petersen R, Ambros V, Hewlett M, Baltimore D. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion & ri rna’s of poliovirus. Proc Natl Acad Sci D S A. 1977;74:961–965. doi: 10.1073/pnas.74.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van der Werf S, Bradley J, Wimmer E, Studier FW, Dunn JJ. Synthesis of infectious poliovirus rna by purified t7 rna polymerase. Proc Natl Acad Sci. 1986;83:2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbert TP, Brierley I, Brown TD. Identification of a protein linked to the genomic and subgenomic mrnas of feline calicivirus and its role in translation. J Gen Virol. 1997;78(Pt 5):1033–1040. doi: 10.1099/0022-1317-78-5-1033. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgerald KD, Semler BL. Bridging ires elements in mrnas to the eukaryotic translation apparatus. Biochim Biophys Acta. 2009;1789(9-10):518–528. doi: 10.1016/j.bbagrm.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberte JF, Roberts L. Calicivirus translation initiation requires an interaction between vpg and eif 4 e. EMBO Rep. 2005;6(10):968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhry Y, Nayak A, Bordeleau ME, Tanaka J, Pelletier J, Belsham GJ, Roberts LO, Goodfellow IG. Caliciviruses differ in their functional requirements for eif4f components. J Biol Chem. 2006;281(35):25315–25325. doi: 10.1074/jbc.M602230200. [DOI] [PubMed] [Google Scholar]
- 15.Hebrard E, Poulicard N, Gerard C, Traore O, Wu HC, Albar L, Fargette D, Bessin Y, Vignols F. Direct interaction between the rice yellow mottle virus (rymv) vpg and the central domain of the rice eif(iso)4g1 factor correlates with rice susceptibility and rymv virulence. Mol Plant Microbe Interact. 2010;23(11):1506–1513. doi: 10.1094/MPMI-03-10-0073.* Evidence for a direct interaction between VPg and eIF4G.
- 16.Willcocks MM, Carter MJ, Roberts LO. Cleavage of eukaryotic initiation factor eif4g and inhibition of host-cell protein synthesis during feline calicivirus infection. J Gen Virol. 2004;85(Pt 5):1125–1130. doi: 10.1099/vir.0.19564-0. [DOI] [PubMed] [Google Scholar]
- 17.Huang TS, Wei T, Laliberte JF, Wang A. A host rna helicase-like protein, atrh8, interacts with the potyviral genome-linked protein, vpg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol. 2010;152(1):255–266. doi: 10.1104/pp.109.147983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daughenbaugh KF, Fraser CS, Hershey JW, Hardy ME. The genome-linked protein vpg of the norwalk virus binds eif3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22(11):2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steil BP, Barton DJ. Cis-active rna elements (cres) and picornavirus rna replication. Virus Res. 2009;139(2):240–252. doi: 10.1016/j.virusres.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodfellow IG, Polacek C, Andino R, Evans DJ. The poliovirus 2c cis-acting replication element-mediated uridylylation of vpg is not required for synthesis of negative-sense genomes. J Gen Virol. 2003;84(Pt 9):2359–2363. doi: 10.1099/vir.0.19132-0. [DOI] [PubMed] [Google Scholar]
- 21.Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. Identification of an rna hairpin in poliovirus rna that serves as the primary template in the in vitro uridylylation of vpg. J Virol. 2000;74(22):10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathak HB, Oh HS, Goodfellow IG, Arnold JJ, Cameron CE. Picornavirus genome replication: Roles of precursor proteins and rate-limiting steps in orii-dependent vpg uridylylation. J Biol Chem. 2008;283(45):30677–30688. doi: 10.1074/jbc.M806101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machin A, Alonso JM Martin, Parra F. Identification of the amino acid residue involved in rabbit hemorrhagic disease virus vpg uridylylation. J Biol Chem. 2001;276(30):27787–27792. doi: 10.1074/jbc.M100707200. [DOI] [PubMed] [Google Scholar]
- 24.Belliot G, Sosnovtsev SV, Chang K, McPhie P, Green KY. Nucleotidylylation of the vpg protein of a human norovirus by its proteinase-polymerase precursor protein. Virology. 2008;374(1):33–49. doi: 10.1016/j.virol.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitra T, Sosnovtsev SV, Green KY. Mutagenesis of tyrosine 24 in the vpg protein is lethal for feline calicivirus. J Virol. 2004;78(9):4931–4935. doi: 10.1128/JVI.78.9.4931-4935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han KR, Choi Y, Min BS, Jeong H, Cheon D, Kim J, Jee Y, Shin S, Yang JM. Murine norovirus-1 3dpol exhibits rna-dependent rna polymerase activity and nucleotidylylates on tyr of the vpg. J Gen Virol. 2010;91(Pt 7):1713–1722. doi: 10.1099/vir.0.020461-0. [DOI] [PubMed] [Google Scholar]
- 27.Forss S, Schaller H. A tandem repeat gene in a picornavirus. Nucleic Acids Res. 1982;10:6441–6450. doi: 10.1093/nar/10.20.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King A, Sangar D, Harris T, Brown F. Heterogeneity of the genome-linked protein of fmdv. J Virol. 1980;34:627–634. doi: 10.1128/jvi.34.3.627-634.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pacheco JM, Henry TM, O’Donnell VK, Gregory JB, Mason PW. Role of nonstructural proteins 3a and 3b in host range and pathogenicity of foot-and-mouth disease virus. J Virol. 2003;77(24):13017–13027. doi: 10.1128/JVI.77.24.13017-13027.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pacheco JM, Piccone ME, Rieder E, Pauszek SJ, Borca MV, Rodriguez LL. Domain disruptions of individual 3b proteins of foot-and-mouth disease virus do not alter growth in cell culture or virulence in cattle. Virology. 2010;405(1):149–156. doi: 10.1016/j.virol.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 31.Arias A, Perales C, Escarmis C, Domingo E. Deletion mutants of vpg reveal new cytopathology determinants in a picornavirus. PLoS One. 2010;5(5):e10735. doi: 10.1371/journal.pone.0010735. * Evidence for a role of VPg in virus release and cytopathic effect.
- 32.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22(1):41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randall RE, Goodbourn S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 34.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of pkr by rnas with short stem-loops. Science. 2007;318(5855):1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 35.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. Rig-i-mediated antiviral responses to single-stranded rna bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 36.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, et al. 5′-triphosphate rna is the ligand for rig-i. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 37.Barral PM, Sarkar D, Fisher PB, Racaniello VR. Rig-i is cleaved during picornavirus infection. Virology. 2009;391(2):171–176. doi: 10.1016/j.virol.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K, Lemon SM. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci D S A. 2007;104(17):7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang TY, Coyne CB. The coxsackievirus b 3c(pro) protease cleaves mavs and trif to attenuate host type i interferon and apoptotic signaling. Plos Pathogens. 2011;7(3) doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ambros V, Baltimore D. Purification and properties of a hela cell enzyme able to remove the 5′ terminal protein from poliovirus rna. J Biol Chem. 1980;255 [PubMed] [Google Scholar]
- 41.Rozovics JM, Virgen-Slane R, Semler BL. Engineered picornavirus vpg-rna substrates: Analysis of a tyrosyl-rna phosphodiesterase activity. PLoS One. 2011;6(3):e16559. doi: 10.1371/journal.pone.0016559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drygin YF, Shabanov AA, Bogdanov AA. An enzyme which specifically splits a covalent bond between picornaviral rna and vpg. FEBS Lett. 1988;239:343–346. doi: 10.1016/0014-5793(88)80948-7. [DOI] [PubMed] [Google Scholar]
- 43.Chaudhry Y, Skinner MA, Goodfellow IG. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing t7 rna polymerase. J Gen Virol. 2007;88(Pt 8):2091–2100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gangaramani DR, Eden EL, Shah M, Destefano JJ. The twenty-nine amino acid c-terminal cytoplasmic domain of poliovirus 3ab is critical for nucleic acid chaperone activity. RNA Biol. 2010;7(6):820–829. doi: 10.4161/rna.7.6.13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeStefano JJ. Effect of reaction conditions and 3ab on the mutation rate of poliovirus rna-dependent rna polymerase in a alpha-complementation assay. Virus Res. 2010;147(1):53–59. doi: 10.1016/j.virusres.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rantalainen KI, Eskelin K, Tompa P, Makinen K. Structural flexibility allows the functional diversity of potyvirus genome-linked protein vpg. J Virol. 2011;85(5):2449–2457. doi: 10.1128/JVI.02051-10. *Structural analysis of plant virus VPg
- 47.Merits A, Guo D, Saarma M. Vpg, coat protein and five non-structural proteins of potato a potyvirus bind rna in a sequence-unspecific manner. J Gen Virol. 1998;79(12):3123–3127. doi: 10.1099/0022-1317-79-12-3123. [DOI] [PubMed] [Google Scholar]
- 48.Kaiser WJ, Chaudhry Y, Sosnovtsev SV, Goodfellow IG. Analysis of protein-protein interactions in the feline calicivirus replication complex. J Gen Virol. 2006;87(Pt 2):363–368. doi: 10.1099/vir.0.81456-0. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Wang C, Mueller S, Paul AV, Wimmer E, Jiang P. Direct interaction between two viral proteins, the nonstructural protein 2c and the capsid protein vp3, is required for enterovirus morphogenesis. PLoS Pathog. 2010;6(8) doi: 10.1371/journal.ppat.1001066. ** Direct evidence for a role of the 2C protein in picornavirus genome encapsidation.
- 50.Oh HS, Pathak HB, Goodfellow IG, Arnold JJ, Cameron CE. Insight into poliovirus genome replication and encapsidation obtained from studies of 3b-3c cleavage site mutants. J Virol. 2009;83(18):9370–9387. doi: 10.1128/JVI.02076-08. * Evidence that precuros proteins play an essnetial role in virus replication and encapsidation.