Abstract
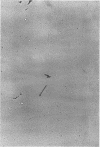
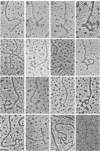
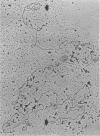
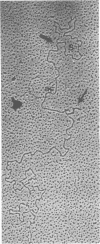
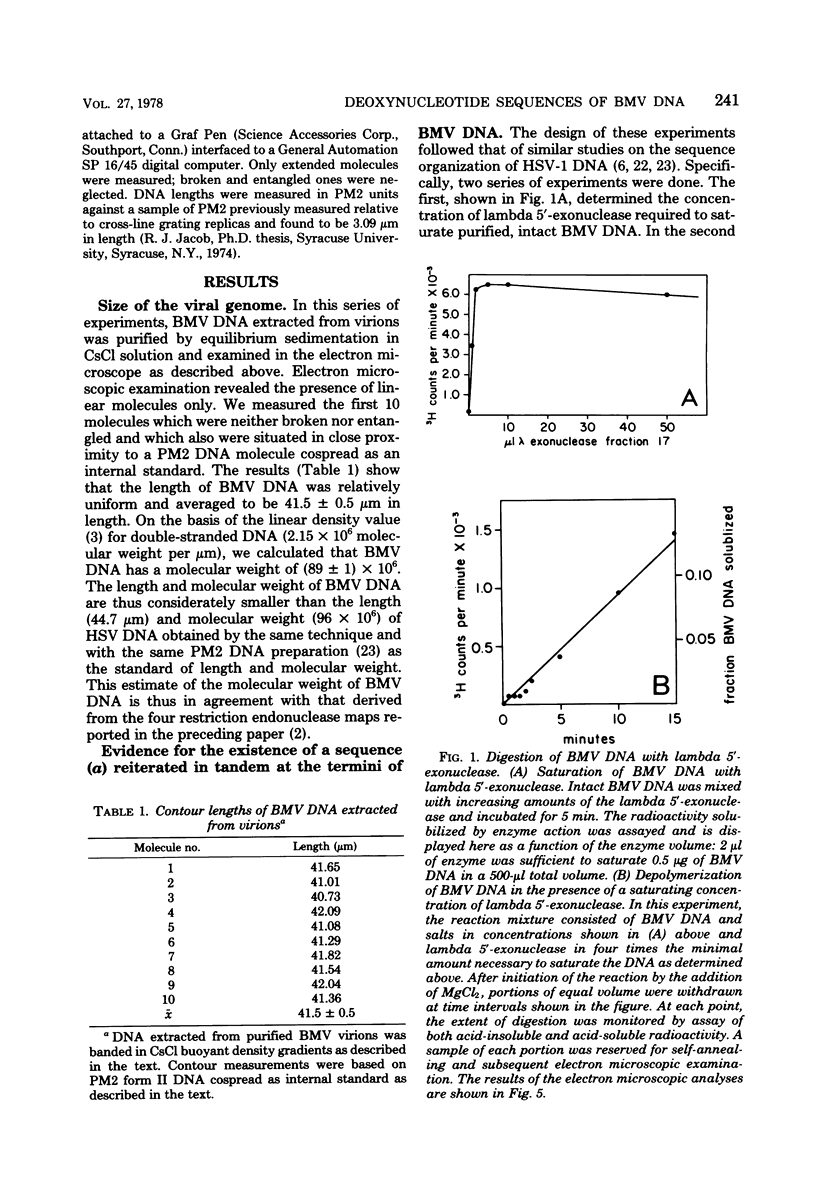
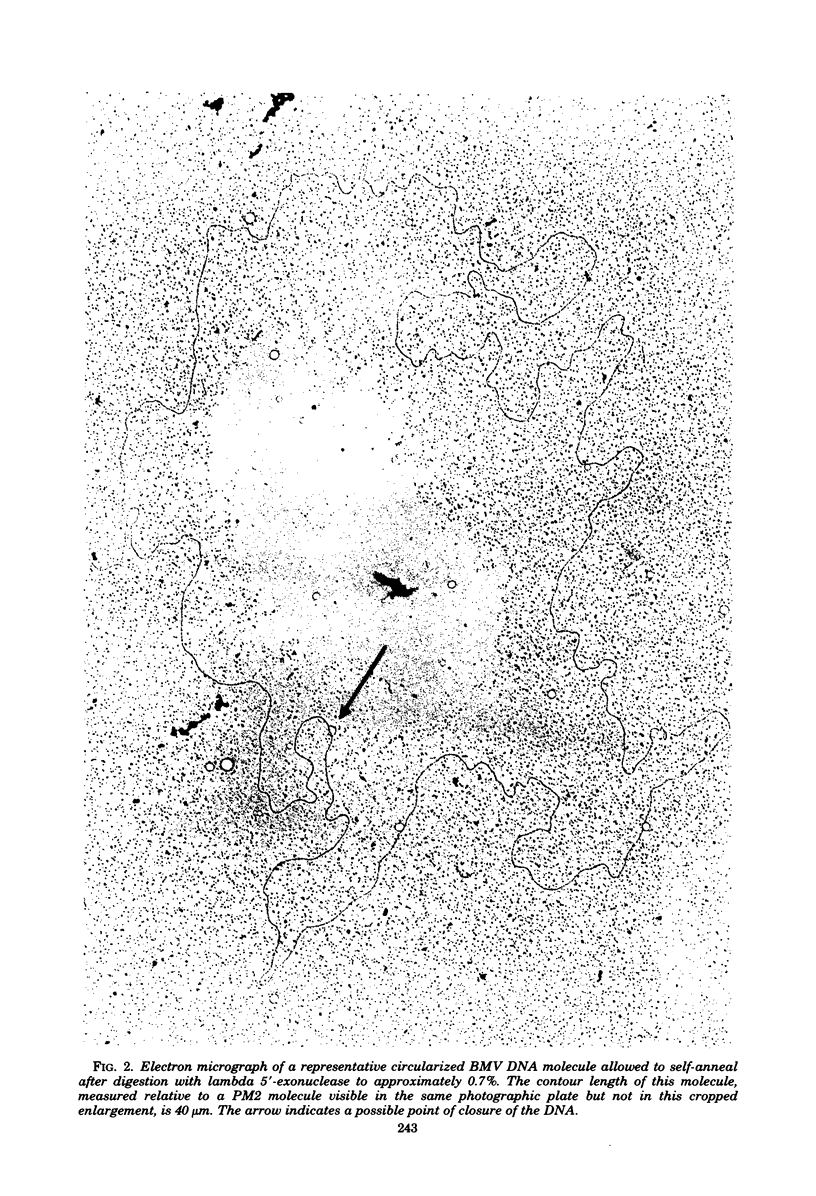
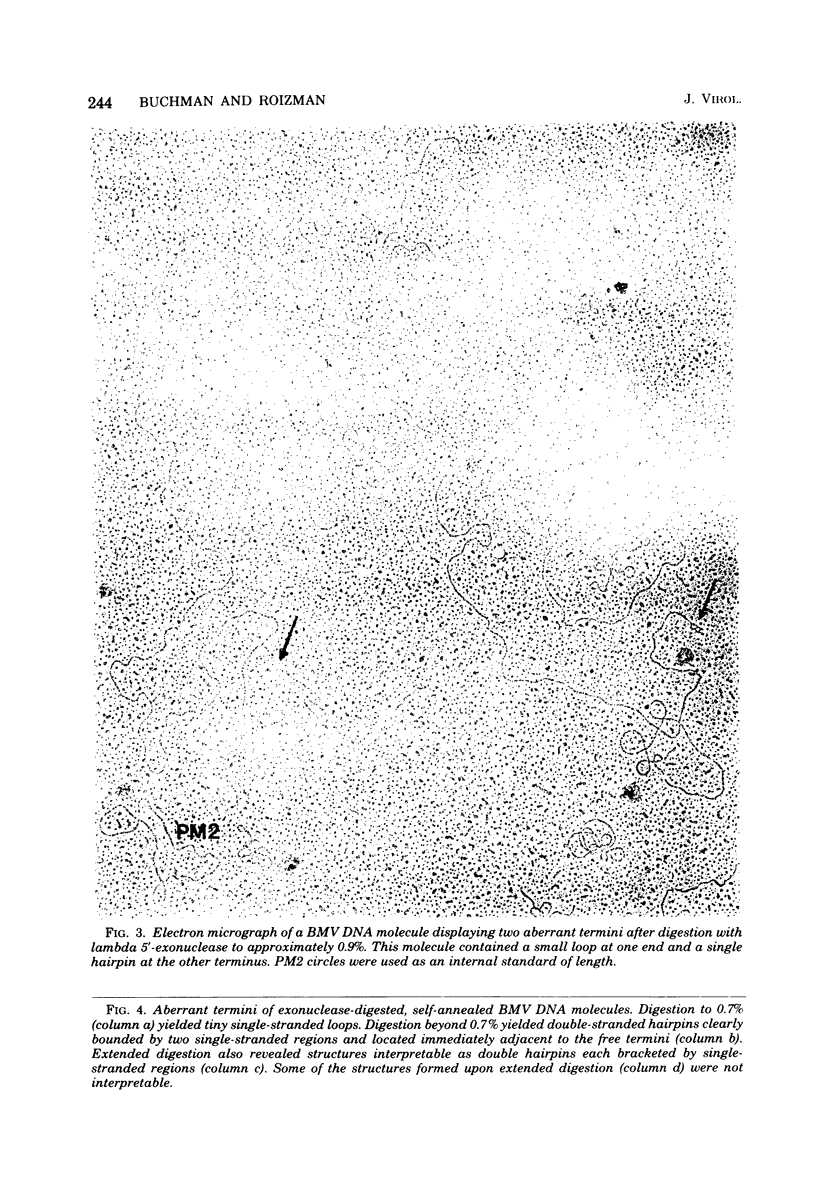
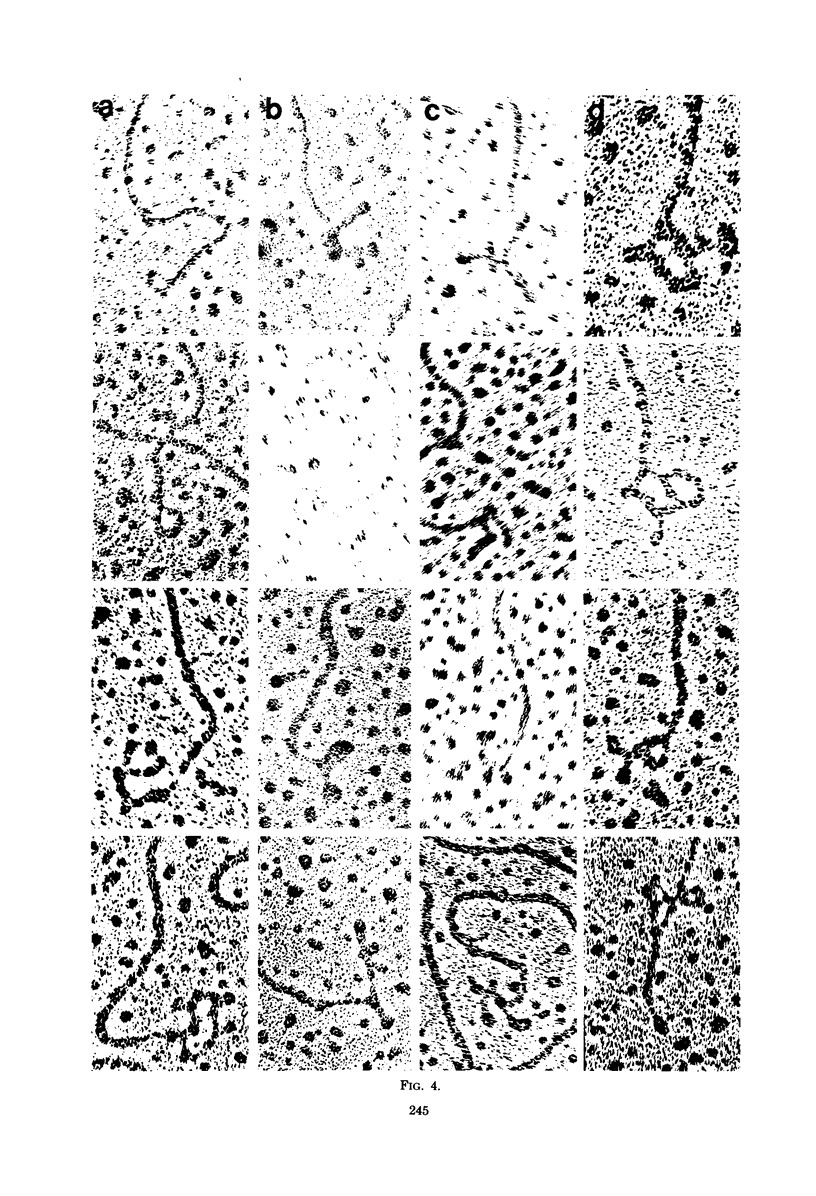
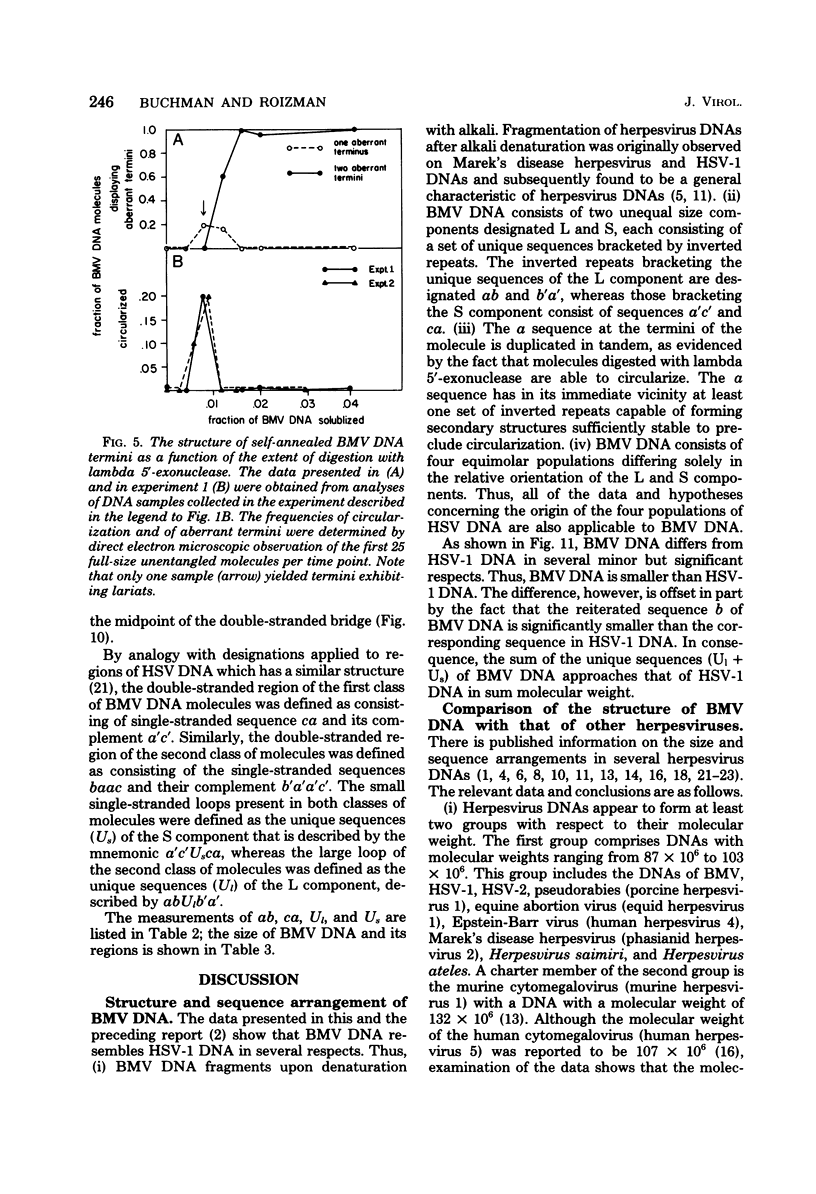
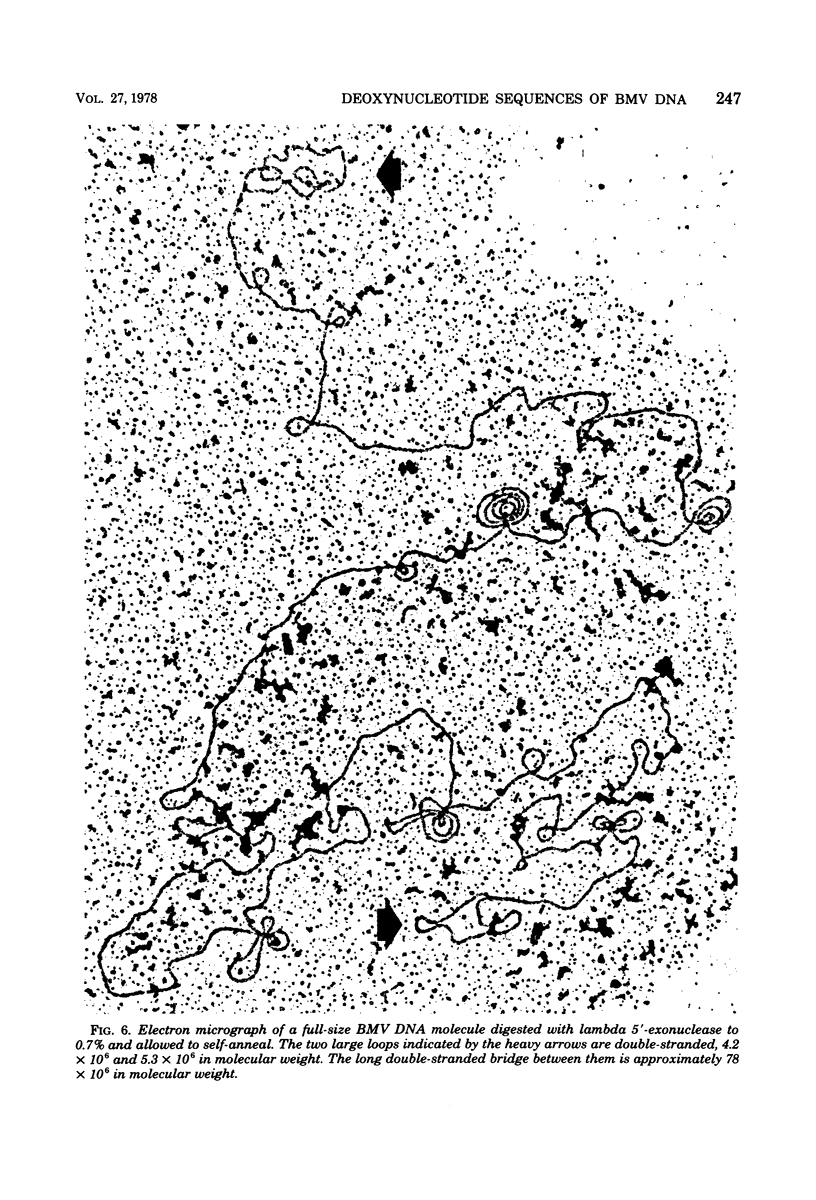
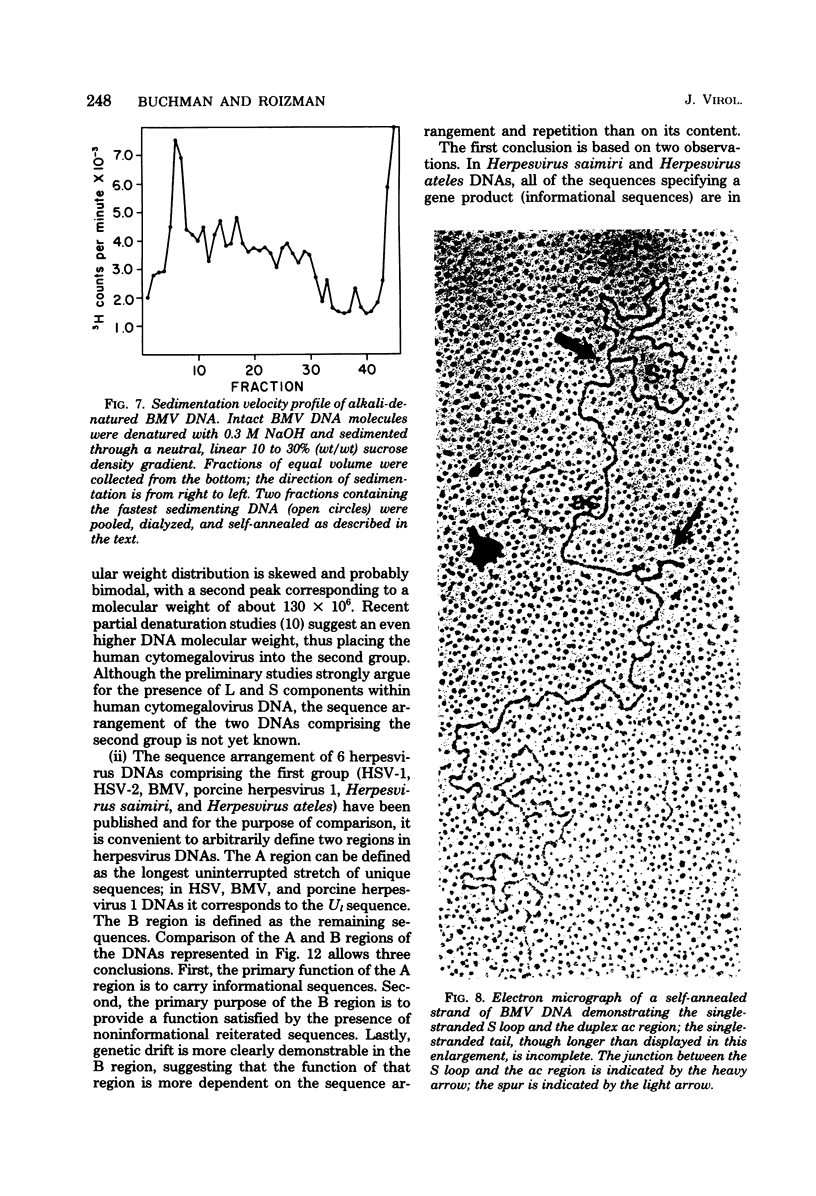
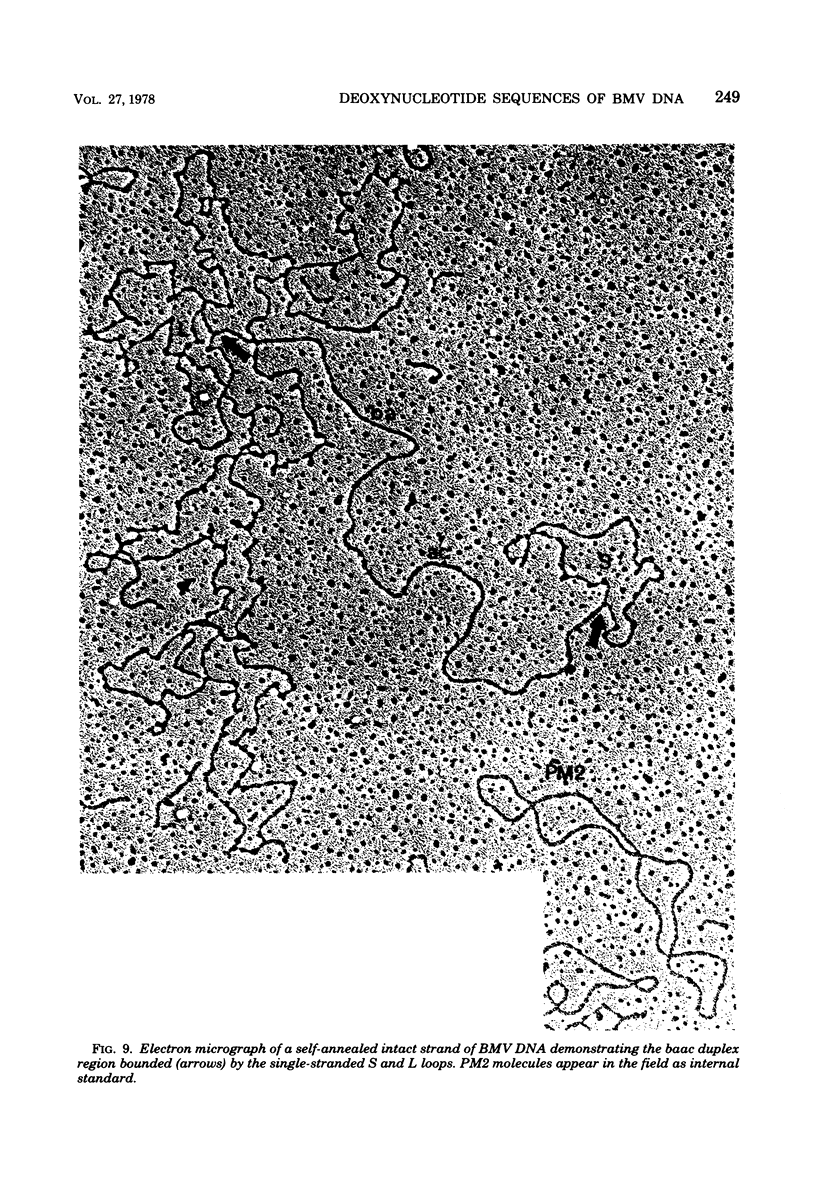
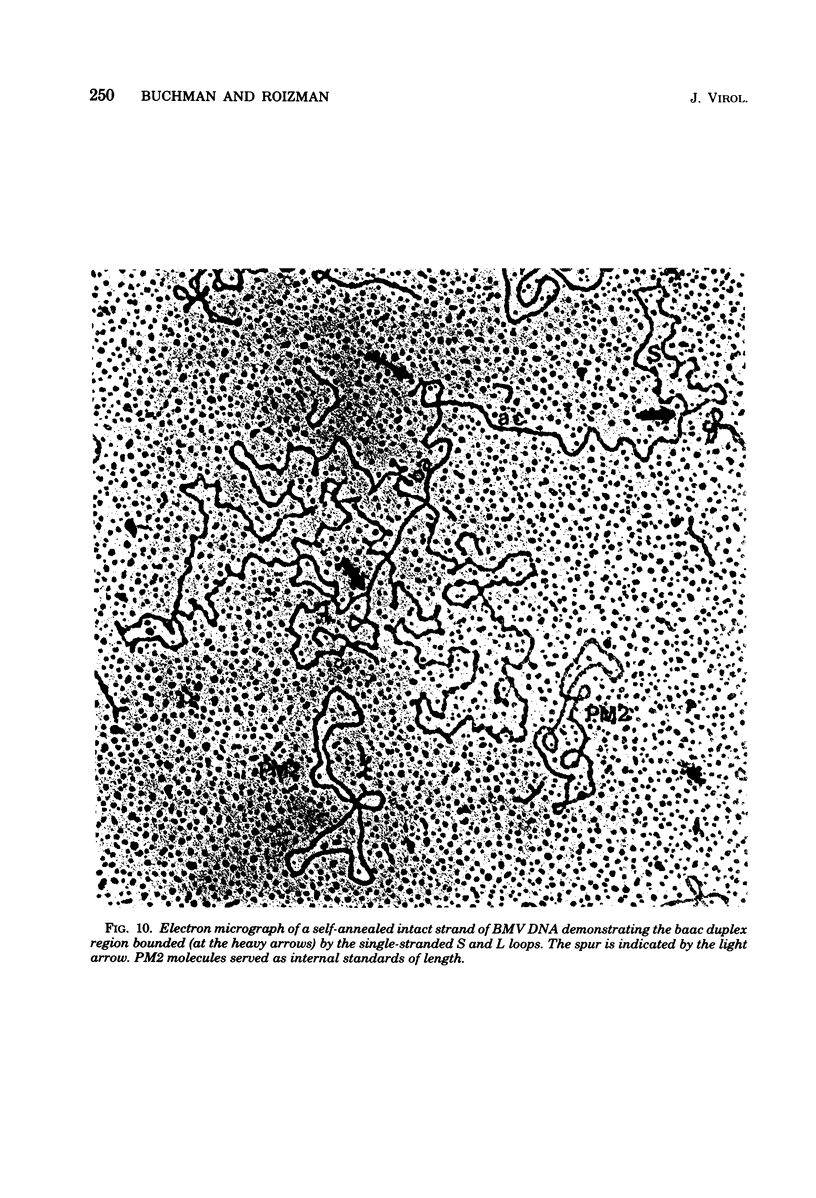
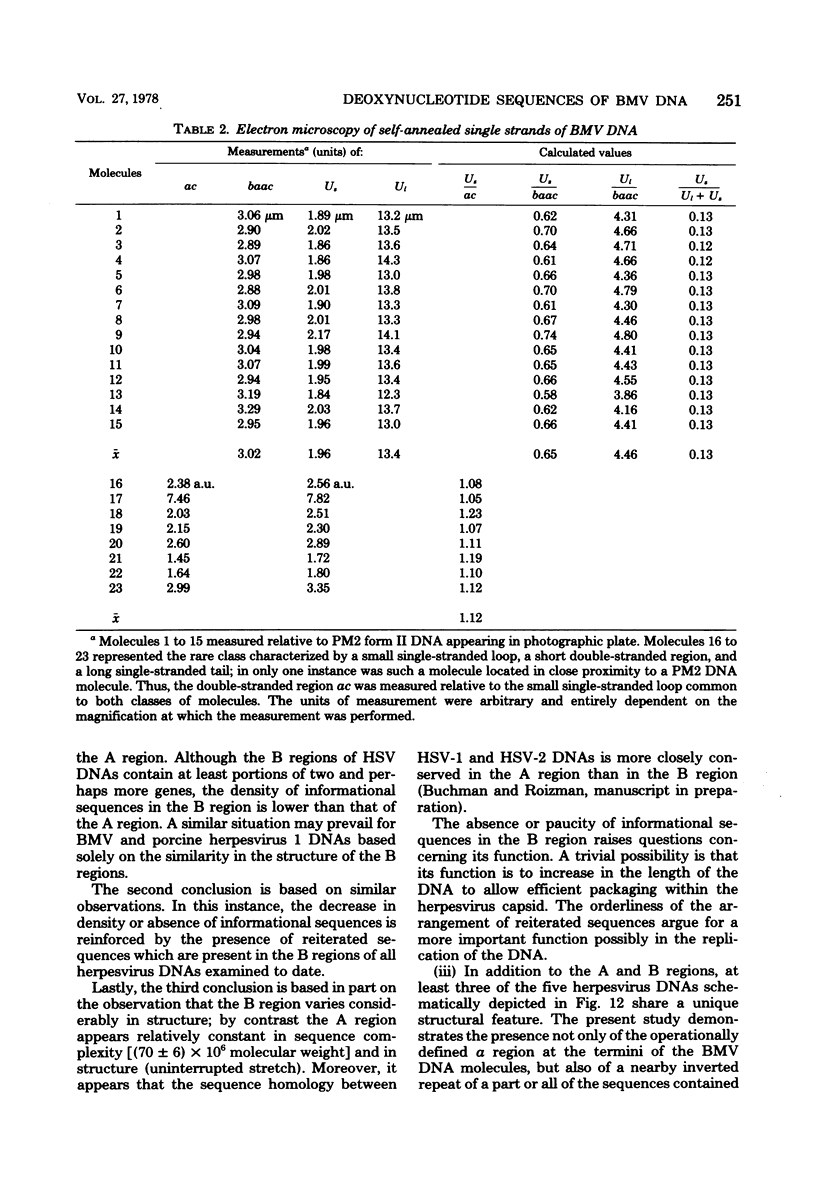
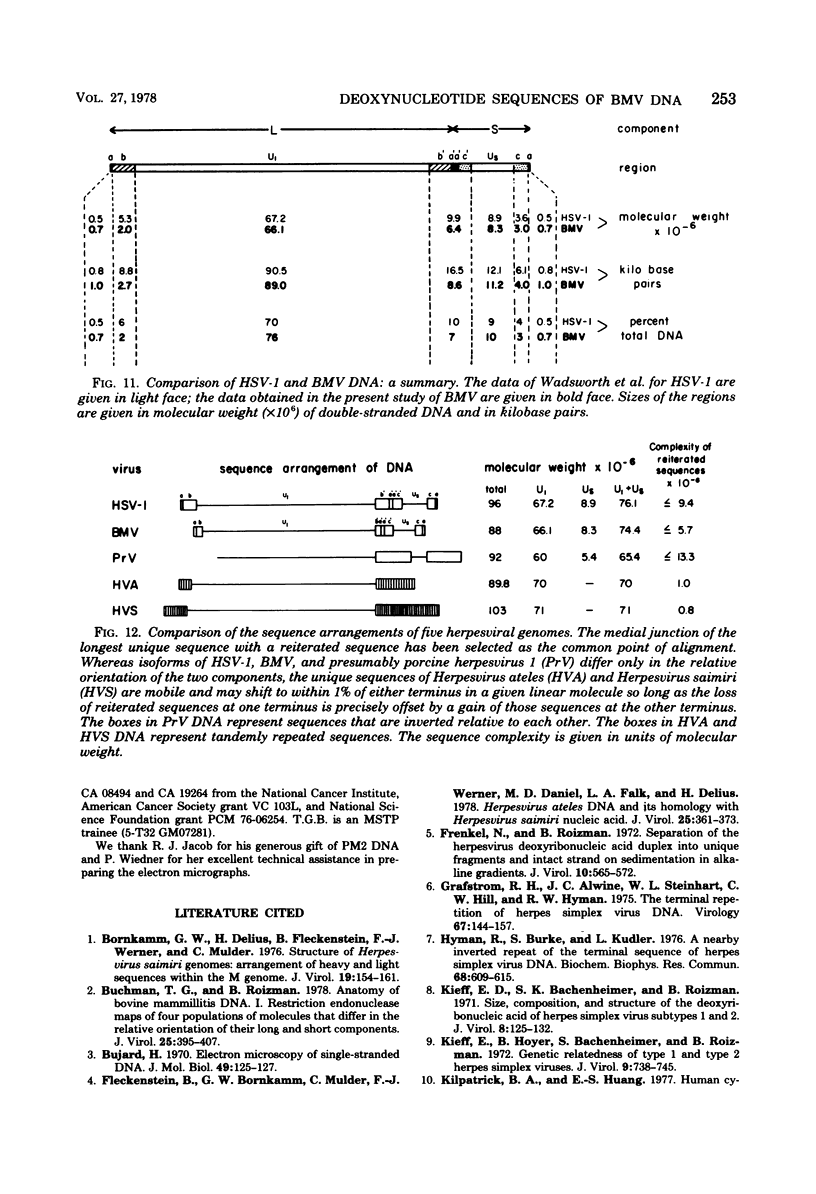
We previously reported that bovine mammillitis virus (BMV) DNA consists of two covalently linked components designated L and S and estimated to be 71.5 x 10(6) and 15.7 x 10(6) in molecular weight, respectively; the components invert relative to each other, giving rise to four equimolar populations differing soley in the relative orientation of the two components. We now report that (i) BMV DNA has a contour length corresponding to a molecular weight of 89 x 10(6). (ii) Component L consists of a unique sequence (Ul) bracketed by sequences ab and its inverted repeat b'a', estimated to be of molecular weights 66.1 x 10(6), 2.7 x 10(6), and 2.7 x 10(6), respectively. (iii) Component S consists of a unique sequence (Us) bracketed be sequence ca and its inverted repeat a'c', estimated to be of molecular weights 8.3 x 10(6), 3.7 x 10(6), and 3.7 x 10(6), respectively. (iv) The a sequences present at the termini of a complete linear molecule (abUlb'a'a'c'Usca) are arranged in tandem so that the DNA can circularize after limited digestion with arranged in tandem so that the DNA can circularize after limited digestion with lambda 5'-exonuclease. The size of the a sequences was estimated to be 0.7 x 10(6) in molecular weight. (v) At least portions of the a sequences are repeated in an inverted orientation immediately adjacent to or near the a sequence. Thus, BMV DNA mimics herpes simplex virus type 1 DNA with respect to the arrangement but not size of deoxynucleotide sequences. The evolutionary relationship of BMV DNA relative to other herpesvirus DNAs is discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bornkamm G. W., Delius H., Fleckenstein B., Werner F. J., Mulder C. Structure of Herpesvirus saimiri genomes: arrangement of heavy and light sequences in the M genome. J Virol. 1976 Jul;19(1):154–161. doi: 10.1128/jvi.19.1.154-161.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman T. G., Roizman B. Anatomy of bovine mammillitis DNA. I Restriction endonuclease maps of four populations of molecules that differ in the relative orientation of their long and short components. J Virol. 1978 Jan;25(1):395–407. doi: 10.1128/jvi.25.1.395-407.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujard H. Electron microscopy of single-stranded DNA. J Mol Biol. 1970 Apr 14;49(1):125–137. doi: 10.1016/0022-2836(70)90381-5. [DOI] [PubMed] [Google Scholar]
- Fleckenstein B., Bornkamm G. W., Mulder C., Werner F. J., Daniel M. D., Falk L. A., Delius H. Herpesvirus ateles DNA and its homology with Herpesvirus saimiri nucleic acid. J Virol. 1978 Jan;25(1):361–373. doi: 10.1128/jvi.25.1.361-373.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N., Roizman B. Separation of the herpesvirus deoxyribonucleic acid duplex into unique fragments and intact strand on sedimentation in alkaline gradients. J Virol. 1972 Oct;10(4):565–572. doi: 10.1128/jvi.10.4.565-572.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W., Hyman R. W. The terminal repetition of herpes simplex virus DNA. Virology. 1975 Sep;67(1):144–157. doi: 10.1016/0042-6822(75)90412-2. [DOI] [PubMed] [Google Scholar]
- Hyman R. W., Burke S., Kudler L. A nearby inverted repeat of the terminal sequence of herpes simplex virus DNA. Biochem Biophys Res Commun. 1976 Jan 26;68(2):609–615. doi: 10.1016/0006-291x(76)91189-x. [DOI] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E., Hoyer B., Bachenheimer S., Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972 May;9(5):738–745. doi: 10.1128/jvi.9.5.738-745.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. F., Kieff E. D., Bachenheimer S. L., Roizman B., Spear P. G., Burmester B. R., Nazerian K. Size and composition of Marek's disease virus deoxyribonucleic acid. J Virol. 1971 Mar;7(3):289–294. doi: 10.1128/jvi.7.3.289-294.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. R., Hudson J. B. Some properties of the genome of murine cytomegalovirus (MCV). Virology. 1973 Jul;54(1):135–149. doi: 10.1016/0042-6822(73)90123-2. [DOI] [PubMed] [Google Scholar]
- Pritchett R. F., Hayward S. D., Kieff E. D. DNA of Epstein-Barr virus. I. Comparative studies of the DNA of Epstein-Barr virus from HR-1 and B95-8 cells: size, structure, and relatedness. J Virol. 1975 Mar;15(3):556–559. doi: 10.1128/jvi.15.3.556-559.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Regulation of lambda exonuclease. I. Properties of lambda exonuclease purified from lysogens of lambda T11 and wild type. J Mol Biol. 1966 Jul;18(2):235–250. doi: 10.1016/s0022-2836(66)80243-7. [DOI] [PubMed] [Google Scholar]
- SOEHNER R. L., GENTRY G. A., RANDALL C. C. SOME PHYSICOCHEMICAL CHARACTERISTICS OF EQUINE ABORTION VIRUS NUCLEIC ACID. Virology. 1965 Jul;26:394–405. doi: 10.1016/0042-6822(65)90003-6. [DOI] [PubMed] [Google Scholar]
- Sarov I., Friedman A. Electron microscopy of human cytomegalovirus DNA. Arch Virol. 1976;50(4):343–347. doi: 10.1007/BF01317960. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Spear P. G., Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972 Jan;9(1):143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterz H., Ludwig H., Rott R. Immunologic and genetic relationship between herpes simplex virus and bovine herpes mammillitis virus. Intervirology. 1974;2(1):1–13. doi: 10.1159/000149398. [DOI] [PubMed] [Google Scholar]
- Stevely W. S. Inverted repetition in the chromosome of pseudorabies virus. J Virol. 1977 Apr;22(1):232–234. doi: 10.1128/jvi.22.1.232-234.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. V. Terminally repetitive sequences. J Virol. 1976 Feb;17(2):503–512. doi: 10.1128/jvi.17.2.503-512.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]