Abstract
Induced pluripotent stem (iPS) cells generated using Yamanaka factors have great potential for use in autologous cell therapy. However, genomic abnormalities exist in human iPS cells, and most mouse iPS cells are not fully pluripotent, as evaluated by the tetraploid complementation assay (TCA); this is most likely associated with the DNA damage response (DDR) occurred in early reprogramming induced by Yamanaka factors. In contrast, nuclear transfer can faithfully reprogram somatic cells into embryonic stem (ES) cells that satisfy the TCA. We thus hypothesized that factors involved in oocyte-induced reprogramming may stabilize the somatic genome during reprogramming, and improve the quality of the resultant iPS cells. To test this hypothesis, we screened for factors that could decrease DDR signals during iPS cell induction. We determined that Zscan4, in combination with the Yamanaka factors, not only remarkably reduced the DDR but also markedly promoted the efficiency of iPS cell generation. The inclusion of Zscan4 stabilized the genomic DNA, resulting in p53 downregulation. Furthermore, Zscan4 also enhanced telomere lengthening as early as 3 days post-infection through a telomere recombination-based mechanism. As a result, iPS cells generated with addition of Zscan4 exhibited longer telomeres than classical iPS cells. Strikingly, more than 50% of iPS cell lines (11/19) produced via this “Zscan4 protocol” gave rise to live-borne all-iPS cell mice as determined by TCA, compared to 1/12 for lines produced using the classical Yamanaka factors. Our findings provide the first demonstration that maintaining genomic stability during reprogramming promotes the generation of high quality iPS cells.
Keywords: somatic reprogramming, genomic stability, telomere, Zscan4, tetraploid complementation, iPS cells
Introduction
The inefficiency of reprogramming1 and the low quality of induced pluripotent stem (iPS) cells2 limit their potential applications in cell therapy. While many studies have described techniques for improving the efficiency of iPS cell generation3, recent evidence shows that iPS cells harbor aberrant gene expression4,5,6,7,8 and genomic aberrations2,9,10,11,12,13, raising significant concerns about the safety of iPS cells for potential clinical applications. In response to introduction of Yamanaka factors into donor cells, DNA damage response (DDR) is increased and this response is followed by activation of p53 pathway14,15, which acts as a barrier to reprogramming. The absence or mutation of p53 facilitates nuclear reprogramming; however, p53-deficient or mutant iPS cells present tumor-like features16,17. Furthermore, most mouse iPS cells fail to give rise to live mice as determined by tetraploid complementation assay (TCA), which is the most stringent test for pluripotency18,19. In contrast, nuclear transfer (NT)-derived embryonic stem (ES) cells (ntES cells), derived from somatic cells by oocyte-mediated rapid reprogramming, are broadly similar to conventional ES cells20,21,22 and generate live all-ES cell mice with high efficiency, as determined by TCA20,21,22. Together, these data support the hypothesis that DNA damage and genomic instability arise during reprogramming process induced by the Yamanaka factors, whereas factors involved in oocyte-mediated reprogramming may play important roles in stabilizing the genome during reprogramming.
Here, we demonstrate that Zscan4, a unique gene highly expressed at the zygotic genome activation stage23, in combination with the Yamanaka factors, not only dramatically decreases DDR but also significantly promotes iPS cell generation. Zscan4 can stabilize the genomic DNA in the process of somatic reprogramming, probably via rapid enhancement of telomere lengthening, resulting in high quality iPS cells as demonstrated by TCA analyses. Our results indicate that maintaining genomic stability during reprogramming is critical in generation of bona fide iPS cells.
Results
Zscan4 dramatically decreases DDR during reprogramming and enhances iPS cell generation
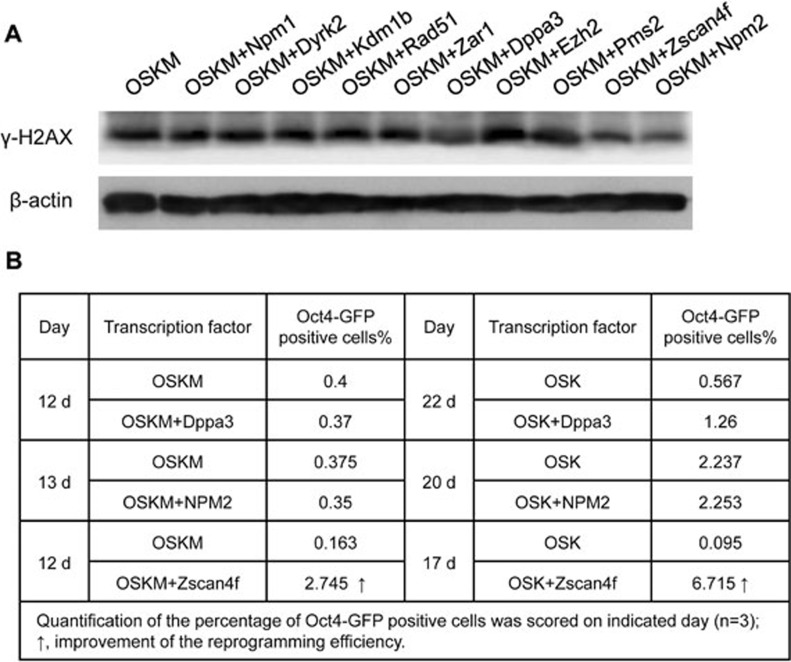
We selected 10 factors (Supplementary information, Table S1) that are highly expressed in oocytes or early cleavage-stage embryos and are essential for preimplantation embryonic development. We incorporated each of them individually with the Yamanaka factors to generate iPS cells. We analyzed DDR during iPS cell formation by testing for the presence of phosphorylated histone H2AX (γ-H2AX). γ-H2AX is formed rapidly following DNA double-strand breaks (DSBs), which are serious lesions that can cause genomic instability, ultimately leading to cancer24. The γ-H2AX assay is frequently used for basic studies in DNA damage repair pathways25 and translational studies of cancer therapies26. Therefore, in the initial screening experiments, we examined the total γ-H2AX protein in reprogramming cells collected on day 4 after viral transfection by western blot. We found that three factors, Zscan4f, Dppa3 and NPM2, in combination with Yamanaka factors, could remarkably decrease the total γ-H2AX protein level at the early stage of reprogramming compared with induction using the classical method (Figure 1A). In the second screening experiments, we tested whether these three factors could also increase efficiency of iPS cell generation by co-introducing one factor with the 'OSK' or 'OSKM' transcription factors into mouse embryonic fibroblasts (MEFs) carrying an enhanced green fluorescence protein (EGFP) reporter driven by Oct4 promoter. Reprogramming efficiency was evaluated by fluorescence-activated cell sorting (FACS) analysis of EGFP-positive cells and Oct4-EGFP-positive colonies in each population on day 12 post-infection. We found that Zscan4f strongly enhanced reprogramming efficiency in combination with OSK or OSKM (Figure 1B), while the other two factors did not enhance iPS cell formation. This is consistent with a recent independent study, which also showed the enhancement of reprogramming efficiency by Zscan427.
Figure 1.
Screening analyses indicate that Zscan4 markedly decreases the DNA damage response and increases reprogramming efficiency. (A) The total phospho-H2AX (γ-H2AX) protein levels were markedly decreased when Zscan4f, Dppa3 and NPM2 were used in combination with the Yamanaka factors. Total protein was extracted from reprogramming cells collected on day 4 after viral transfection. β-actin was used as a loading control. (B) The reprogramming efficiency was significantly increased when Zscan4f was used in combination with OSKM or OSK.
Zscan4 improves genomic stability during reprogramming
We next carefully evaluated the function of Zscan4 in iPS cell generation. Zscan4 gene contains a SCAN domain and four zinc finger domains, and three isoforms (Zscan4c, Zscan4d and Zscan4f) encode full-length open reading frames23. Zscan4 is expressed specifically in two-cell embryos23 and transiently in ES cells28, and plays important roles in preimplantation embryonic development23 and in the maintenance of genomic stability in ES cells28. γ-H2AX was detected in reprogramming cells two days after MEFs were transduced with either OSKM or OSKM plus Zscan4f (OSKMZ) (Figure 2A). Interestingly, the total γ-H2AX protein level was reduced in OSKMZ-infected MEFs at days 4 and 6 after reprogramming process was initiated compared to OSKM-infected MEFs (Figure 2A). We next performed immunofluorescence analyses and found that although the percentage of γ-H2AX-positive cells among cells undergoing reprogramming did not differ between the two groups (37.00% ± 2.96% in OSKM vs 34.19% ± 1.34% in OSKMZ, P = 0.389) (Figure 2B), γ-H2AX-positive foci were more abundant in OSKM-infected cells than in OSKMZ-infected cells (Figure 2B). Furthermore, colocalization of γ-H2AX and Trf1, indicative of telomere-induced DNA damage foci28,29,30, was significantly reduced in OSKMZ-infected cells as compared to OSKM-infected cells (Figure 2C). Because γ-H2AX is a marker for DSBs31, we tested general sister chromatid exchange (SCE)32,33 in reprogramming cells. As expected, the rates of spontaneous SCE in OSKMZ-infected MEFs were lower than those in OSKM-infected MEFs during the reprogramming process (Figure 2D and 2E). Together, these data show that overexpression of Zscan4 reduces both DNA damage at telomeres and the incidence of genomic SCE (G-SCE), thus stabilizing the genomes of reprogrammed cells.
Figure 2.
Zscan4 attenuates the DNA damage response and improves genomic stability during reprogramming. (A) Zscan4 reduced DSB signals during reprogramming. Western blot results indicated that the total γ-H2AX level was remarkably reduced during reprogramming. β-actin was used as a loading control. (B) Left, γ-H2AX-positive foci in OSKM or OSKMZ virus-infected MEFs on day 7 post-infection. The results shown are the mean of two experiments. More than 600 cells were analyzed per sample/experiment. Error bars, s.d. Right, representative images of γ-H2AX-positive foci in OSKM or OSKMZ virus-infected MEFs on day 7 post-infection. White arrows, pan-nuclear γ-H2AX. Scale bars, 50 μm. (C) Left, telomere-induced DNA damage foci in OSKM or OSKMZ virus-infected MEFs on day 7 post-infection. n = number of cells with γ-H2AX-positive foci analyzed. The results are the mean of two experiments. Error bars, s.d. Right, representative images of γ-H2AX (green) and Trf1 (red) staining in OSKM or OSKMZ virus-infected MEFs 7 days post-infection. Arrows indicate co-localization events (yellow). Scale bars, 5 μm. (D) Summary of total G-SCE events in > 35 nuclei per sample from two independent experiments. (E) Representative images of OSKM (left) or OSKMZ (right) virus-infected MEFs on day 6 post-infection following the G-SCE assay, which demonstrated the reduced rate of SCE observed using the OSKMZ reprogramming system. White arrows mark SCE. The insets are magnified images of one chromosome showing SCE. Scale bars, 10 μm.
Because DDR was reduced during the reprogramming process when Zscan4 was incorporated, we investigated the p53 pathway, which can be activated by DDR, during iPS cell formation induced by Yamanaka factors with or without Zscan4f. As determined by western blot, p53 expression was markedly reduced in OSKMZ-infected MEFs compared to OSKM-infected cells during early stages of reprogramming (Figure 3A). Similarly, expression levels of p21 and bax, two p53 target genes, also decreased (Figure 3A). Inactivation of the p53 pathway markedly enhances the efficiency of iPS cell generation using the standard Yamanaka method34,35. Consistent with these results, Zscan4f significantly facilitated iPS cell formation as assessed by increased Oct4-EGFP-positive cells (Figure 3B and Supplementary information, Figure S1A) and colonies (Supplementary information, Figure S1B and S1C) on different days post-infection compared to those cells without Zscan4f. In addition, although Oct4 was typically activated 10 or 14 days after infection with OSKM or OSK, respectively, Oct4 activation was observed only 8 days following transfection of OSKMZ or OSKZ (Supplementary information, Figure S1D), suggesting that Zscan4f accelerated the reprogramming process. We also cloned another analogous gene, Zscan4c, which is transcribed predominantly in ESCs23,36, and showed that it has a similar activity during iPS cell generation (Supplementary information, Figure S1E). Furthermore, endogenous Zscan4 was expressed at a low level in MEFs before and after introduction of OSKM or OSKMZ (Supplementary information, Figure S1F). Knockdown of endogenous Zscan4 by specific small hairpin RNAs (shRNAs) significantly decreased the reprogramming efficiency in comparison with the scrambled control shRNA (Supplementary information, Figure S1G), suggesting that the basal expression level of Zscan4 is required for generation of mouse iPS cells.
Figure 3.
Zscan4 indirectly downregulates p53 during reprogramming. (A) The expression levels of p53, p21 and bax were decreased during reprogramming when Zscan4 was incorporated. Western blot analysis was performed on protein extracts from OSKM- or OSKMZ-infected MEFs at different timepoints. (B) The kinetics of the reprogramming induced by OSKM or OSKMZ virus infection. Oct4-EGFP-positive cells were scored from day 8 to day 14 (n = 3). Error bars, s.d. (C) The overexpression of p53 during reprogramming did not affect the reprogramming efficiency promoted by Zscan4. Quantification of Oct4-EGFP-positive colonies (left) and the percentage of Oct4-GFP-positive cells (middle) are shown (n = 3). The expression of p53 was examined by western blot analysis (right). β-actin was used as a loading control. Error bars, s.d. (D) Knockdown of endogenous p53 by shRNA increased reprogramming efficiency and addition of Zscan4 further promoted the efficiency. Quantification of Oct4-EGFP-positive colonies (left) and the percentage of Oct4-EGFP-positive cells (middle) are shown (n = 3). The expression of p53 was examined by western blot analysis (right). β-actin was used as a loading control. Error bars, s.d.
Suppression of p53 increases efficiency of iPS cell generation by classical Yamanaka method; however, this also carries a risk of tumor development due to compromised genome integrity16,17. Therefore, we investigated whether Zscan4 enhances reprogramming efficiency by direct inhibition of the p53 pathway. While the overexpression of p53 reduced reprogramming efficiency of iPS cell generation using OSKM, Zscan4 remarkably increased proportion of Oct4-positive colonies and cells (Figure 3C). Moreover, knockdown of endogenous p53 by shRNA significantly increased reprogramming efficiency, consistent with previous reports34,35, and the addition of Zscan4 further increased the proportion of GFP-positive colonies and cells (Figure 3D). Since previous report showed that additional inhibition of the p53 pathway directly accelerated cell division37, we then tested the effects of Zscan4 on cell cycle during reprogramming. Our results showed that cell growth did not differ between reprogramming cells with and without Zscan4 (data not shown). Taken together, these data indicate that Zscan4-promoted iPS cell generation may not target p53 directly and that downregulation of p53 and its target genes may be concomitant with the improvement of genomic stability.
Zscan4 promotes telomere elongation during reprogramming
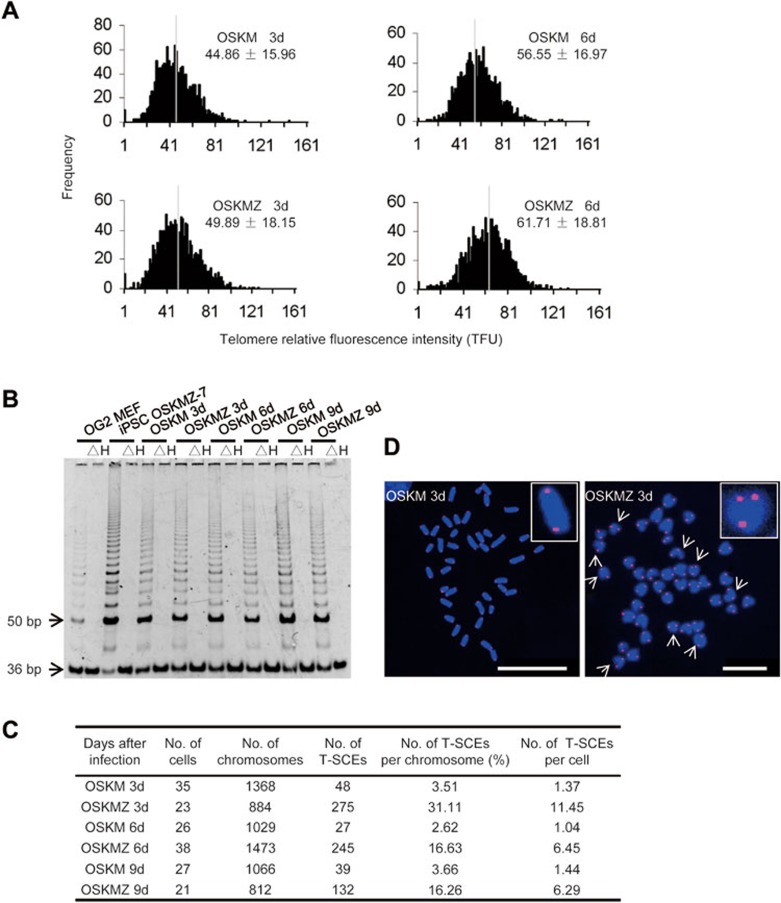
Because Zscan4 expression is associated with the maintenance of genomic stability in ES cells, likely by rapid telomere elongation28, we speculated that Zscan4 induction might contribute to telomere extension during reprogramming. We examined relative telomere length by quantitative telomere FISH (Q-FISH)38 in OSKM- and OSKMZ-infected MEFs on days 3 and 6 post-infection. Zscan4 promoted telomere elongation starting as early as day 3 following retrovirus infection (Figure 4A). The rapid telomere lengthening by Zscan4 was not associated with increased telomerase activity because telomerase activity, assessed with the telomeric repeat amplification protocol (TRAP) assay, did not differ between OSKM- and OSKMZ-infected cells (Figure 4B). These results are consistent with a previous report investigating the role of Zscan4 in normal ES cells28, and suggest that Zscan4 regulates telomere extension during reprogramming, most likely through the same mechanism as in ES cells. Because Zscan4 regulates telomere elongation in ES cells through telomere recombination28, we then investigated whether Zscan4 activated the telomere sister chromatid exchange (T-SCE) apparatus using a telomere chromosome orientation-FISH (CO-FISH) assay. As expected, the frequency of T-SCE in OSKM virus-infected MEFs was low and showed no significant difference (average 1.28 ± 0.21 per cell) among cells collected from different days post-infection (Figure 4C and 4D). In contrast, addition of Zscan4 resulted in 8.36-fold, 6.20-fold and 4.37-fold increases in T-SCE frequency in OSKMZ-infected MEFs on days 3, 6 and 9 post-infection (Figure 4C and 4D), respectively. We also analyzed expression levels of shelterin components and found that Trf2, Pot1b and Rap1, which can inhibit T-SCE after replication39,40, were repressed when Zscan4 was overexpressed (Figure 5A). Other shelterin proteins, Trf1, Pot1a, Tpp1 and Tin2, were not repressed (Figure 5B). These data suggest that Zscan4 expression promotes telomere elongation during iPS cell generation, most likely through telomere recombination involving T-SCE.
Figure 4.
Zscan4 promotes telomere elongation during reprogramming. (A) A histogram shows the distribution of the relative telomere length (in relative telomere fluorescence units (TFUs)) measured by Q-FISH in OSKM or OSKMZ virus-infected MEFs 3 and 6 days post-infection. The median telomere length (white bars) is shown as the mean ± s.d. above each panel. (B) Telomerase activity during reprogramming. ΔH: heat-inactivation served as a negative control. The 36-bp bands at the bottom of the gel are the internal standard. MEFs (passage 2) and the iPS cell line OSKMZ-7 (passage 10) served as a control. No remarkable differences in telomerase activity between the OSKM- and OSKMZ-infected MEFs were observed during reprogramming. The assays were repeated twice. (C) Summary of total T-SCE events in > 20 nuclei per sample based on two independent experiments. (D) Representative images of CO-FISH signals in OSKM (left) or OSKMZ (right) virus-infected MEFs on day 3 post-infection: chromosomes were stained with DAPI (blue), and telomeres were marked with a Cy3-conjugated telomere probe (red). White arrows mark T-SCE. The insets are enlarged images of one chromosome showing CO-FISH signals. Scale bars, 25 μm (left) and 10 μm (right).
Figure 5.
Quantitative RT-PCR for shelterin complex genes during reprogramming. (A) The expression levels of Trf2, Pot1b and Rap1, which inhibit T-SCE after replication, were repressed in OSKMZ-infected MEFs compared to OSKM-infected MEFs during reprogramming. The data are presented as the mean ± s.d. (n = 3). (B) The expression levels of Trf1, Pot1a, Tpp1 and Tin2, the other components of shelterin, were not repressed. The data are presented as the mean ± s.d. (n = 3). The relative expression levels were normalized against the average expression of OSKM virus-infected MEFs on day 3 post-infection.
Zscan4 significantly improves the quality of iPS cells
Mouse iPS cells generated by OSKM/OSK plus Zscan4 showed ES cell-like morphologies (Supplementary information, Figure S2A). Endogenous Oct4, which serves as a key pluripotency marker, was expressed at levels comparable to those observed in ES cells (Supplementary information, Figure S2B). Meanwhile, endogenous Zscan4, a specific marker for ES cells28, was activated and expressed at similar levels, while the exogenous expression of retroviral plasmids containing Zscan4 was silenced in the established iPS cell lines (Supplementary information, Figure S2C). OSKMZ- or OSKZ-induced iPS cells formed teratomas comprising all three embryonic germ layers in nude mice (Supplementary information, Figure S2D). To test the in vivo developmental potency of the iPS cells, we injected these cells into diploid blastocysts of ICR origin. Chimeric mice were generated from all tested cell lines (Table 1 and Supplementary information, Figure S2E), and iPS cells contributed to the germline (Supplementary information, Figure S2F). These data show that the iPS cells generated by OSKMZ or OSKZ are pluripotent.
Table 1. In vivo-developmental potency of iPS cells.
| Donor Cells | Factors | 2N injections |
4N injections |
|||
|---|---|---|---|---|---|---|
| Blastocysts injected | Chimeric/Total | Chimaerism (%)a | Blastocysts injected | Live pups (%) | ||
| OSKM-1 | O, S, K, M | 14 | 4/6 | 20-80 | 136 | 0 |
| OSKM-2 | O, S, K, M | ND | ND | ND | 113 | 0 |
| OSKM-3 | O, S, K, M | 22 | 4/7 | 40-70 | 125 | 0 |
| OSKM-4 | O, S, K, M | ND | ND | ND | 153 | 0 |
| OSKM-5 | O, S, K, M | 16 | 1/2 | 70 | 139 | 0 |
| Subtotal | 666 | 0 | ||||
| OSKMZ-1 | O, S, K, M, Z | 40 | 2/6 | 30, 90 | 194 | 5 (2.58) b |
| OSKMZ-2 | O, S, K, M, Z | 28 | 1/3 | 5 | 192 | 3 (1.56) b |
| OSKMZ-3 | O, S, K, M, Z | 16 | 2/3 | 10, 60 | 83 | 1 (1.20) b |
| OSKMZ-5 | O, S, K, M, Z | ND | ND | ND | 109 | 0 b |
| OSKMZ-6 | O, S, K, M, Z | ND | ND | ND | 80 | 1 (1.25) b |
| OSKMZ-7 | O, S, K, M, Z | 16 | 2/2 | 50, 80 | 120 | 2 (1.67) b |
| OSKMZ-8 | O, S, K, M, Z | ND | ND | ND | 117 | 0 |
| OSKMZ-9 | O, S, K, M, Z | ND | ND | ND | 121 | 0 |
| Subtotal | 1016 | 12 (1.18) | ||||
| OSK line1.1 | O, S, K | ND | ND | ND | 137 | 0 |
| OSK line1.4 | O, S, K | 15 | 4/7 | 70, 80 | 162 | 0 |
| OSK line2.4 | O, S, K | 17 | 2/2 | 70, 90 | 166 | 0 b |
| OSK line23 | O, S, K | 14 | 1/3 | 90 | 105 | 1 (0.95) |
| OSK-1 | O, S, K | ND | ND | ND | 115 | 0 |
| OSK-2 | O, S, K | ND | ND | ND | 155 | 0 |
| OSK-3 | O, S, K | ND | ND | ND | 141 | 0 |
| Subtotal | 981 | 1 (0.10) | ||||
| OSKZ-1 | O, S, K, Z | 15 | 2/10 | 20, 20 | 30 | 1 (3,33) |
| OSKZ-10 | O, S, K, Z | 15 | 1/6 | 30 | 148 | 2 (1.35) b |
| OSKZ-12 | O, S, K, Z | 18 | 1/6 | 70 | 104 | 0 |
| OSKZ-14 | O, S, K, Z | 26 | 1/3 | 80 | 115 | 0 b |
| OSKZ-16 | O, S, K, Z | ND | ND | ND | 143 | 0 b |
| OSKZ line8 | O, S, K, Z | 18 | 1/3 | 30 | 87 | 0 b |
| OSKZ line16 | O, S, K, Z | ND | ND | ND | 91 | 0 b |
| OSKZ line18 | O, S, K, Z | ND | ND | ND | 48 | 1 (2.08) |
| OSKZ line22 | O, S, K, Z | ND | ND | ND | 73 | 0 |
| OSKZ line24 | O, S, K, Z | 16 | 2/4 | 10, 50 | 35 | 1 (2.86) |
| OSKZc-1 | O, S, K, Z | ND | ND | ND | 40 | 2 (5) |
| Subtotal | 914 | 7 (0.77) | ||||
Z, Zscan4f; Zc, Zscan4c; O, Oct4; S, Sox2; K, Klf4; M, c-Myc;
a, the extent of chimaerism was estimated on the basis of coat colour;
b, embryonic lethal at E10-19;
ND, not determined.
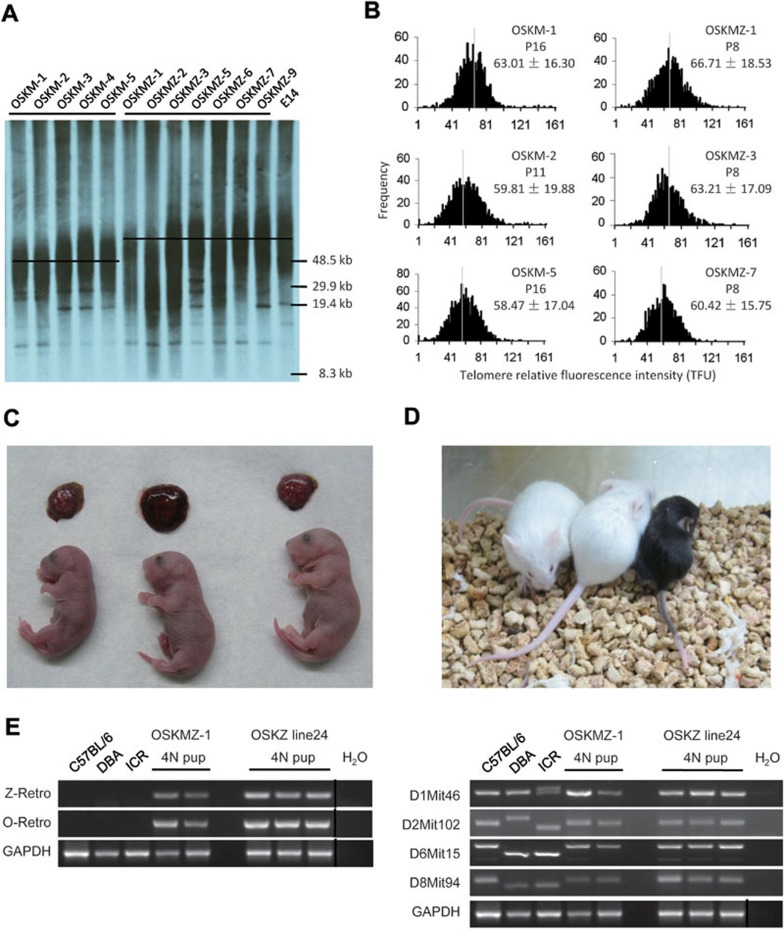
Overexpression of Zscan4 allowed rapid telomere extension and alleviated DNA damage during reprogramming. We anticipated that the quality of OSKMZ- or OSKZ-induced iPS cells might be improved significantly. We measured telomere length in iPS cells generated with OSKM and OSKMZ using two independent techniques, Southern telomere restriction analysis (TRF)41 and Q-FISH on metaphase spreads38. As expected, telomeres were longer in OSKMZ-induced iPS cells compared to the corresponding OSKM-induced iPS cells, as determined by both TRF (Figure 6A) and Q-FISH analysis (Figure 6B), indicating that Zscan4 contributes to the generation of iPS cells with longer telomeres. We next tested the in vivo developmental potential of OSKMZ iPS cells by TCA, which is the most stringent assay for pluripotency and also the most reliable functional assay to determine the quality and genomic stability of mouse iPS/ES cells42. Remarkably, 5 out of the 8 OSKMZ-induced iPS cell lines gave rise to viable all-iPS cell-derived pups at an efficiency similar to that of ES cells and ntES cells, as described in previous reports42,43 (Figure 6C and 6D and Table 1). In contrast, all 5 OSKM-induced iPS cells with the same genetic background failed to produce iPS cell-derived pups. Likewise, in 6 OSK-induced iPS cells, only 1 cell line generated one live-born pup, whereas 6 out of 11 OSKZ iPS cell lines produced live-born all-iPS cell-derived pups (Figure 6C and 6D and Table 1). Genotyping analysis showed that all-iPS cell mice carried the Zscan4 and Oct4 transgenes (Figure 6E, left). We further verified the cell origin of these mice by simple sequence-length polymorphism (SSLP) analysis and confirmed that they were from iPS cells with an inbred genetic background (C57BL/6 strain) and not the tetraploid embryo host or recipient ICR mice (Figure 6E, right). Interestingly, ES cells derived from an inbred background, such as the C57BL/6 mouse strain, were less able to generate cloned and all-ES cell pups44. These data demonstrate that the iPS cells generated by the addition of Zscan4 exhibit high quality and show improved developmental potential in vivo in contrast to iPS cells derived using classical OSK or OSKM technique.
Figure 6.
Zscan4 leads to longer telomeres in the resultant iPS cells and significantly improves the developmental potential of iPS cells. (A) TRF analysis of iPS cells and control ES cells (E14) between passages 7-10 demonstrated longer telomeres in OSKMZ-induced iPS cells compared to OSKM-induced iPS cells. (B) A histogram shows the distribution of relative telomere length (in TFUs) measured by Q-FISH in iPS cells. The median telomere length (white bars) is shown as the mean ± s.d. above each panel. The passage number was indicated. (C) Three live-born (E19.5) all-iPS cell mice generated from OSKMZ-1 cells via TCA. (D) An adult all-iPS cell mouse generated from OSKMZ-1 via TCA. (E) Genotype analyses of all-iPS cell mice. Retrovirus integration (left) and SSLP (right) analyses were performed using genomic DNA isolated from tail tips of adult mice. Genomic DNA from C57BL/6, DBA and ICR mice served as controls.
Discussion
The conversion of somatic cells to iPS cells by Yamanaka factors remains an inefficient process1,45. Although significant progress has been made in terms of improving the efficiency of iPS cell generation through the use of chemical compounds46,47 and different cell types48,49, interfering with the epigenetic status of donor cells50,51,52, or modulating different signal pathways34,53,54, the quality of iPS cells to date is not significantly improved or even getting worse when the p53 was absent to facilitate reprogramming35. On the other hand, regarding the safety concerns of iPS cells, strategies of generating integration-free iPS cells have been developed55; however, these reprogramming processes are extremely inefficient and the quality of resulting integration-free iPS cells still needs to be completely characterized56. In our study, we identified Zscan4 as a factor, essential for cleavage development of mouse embryos23, which not only remarkably promotes the efficiency of iPS cell generation, but also significantly improves the quality of iPS cells by stabilizing the genomic and telomere DNA and promoting the telomere elongation at early stage of reprogramming. Consistently, a recent independent study27 also verified that Zscan4 can improve the reprogramming efficiency.
Recently, several studies reported that genetic abnormalities are found in human iPS cells derived from fibroblasts2,9,10,11,13, implying the importance of maintaining genomic integrity during cellular reprogramming. During the first several cell cycles following fertilization, when epigenetic reprogramming occurs, the telomeres of the parental chromosomes lengthen remarkably, and 1- to 2-cell stage embryos have elongated telomeres57, while the telomeres in mature oocytes are shorter than those in somatic cells57. Also, telomeres become shorter during the pachytene spermatocyte stage until round spermaid58. This extension could be related to T-SCE57, which may be regulated by Zscan4; Zscan4 is highly expressed in late 2-cell embryos23 and can promote rapid telomere elongation in ES cells via T-SCE28. Taken together, these observations suggest that rapid telomere elongation following fertilization may protect DNA from damage induced by acute epigenetic reprogramming. These findings may also partially explain why NT can faithfully reprogram somatic cells into ES cells5,8,20,21,42. In contrast, during somatic reprogramming induced by the Yamanaka factors, telomere elongation is primarily mediated by telomerase59,60. Endogenous Zscan4 is not activated during the early stages of reprogramming induced by OKSM or OSK, as shown in our study (Supplementary information, Figure S1F) and in other report38, further supporting the hypothesis that telomerase activity is the primary mechanism of telomere re-elongation regulation during OSKM- or OSK-mediated reprogramming. However, telomere extension by telomerase is a slow process and requires a number of cell divisions to restore a normal average telomere length59,61. The delay in telomere extension by telomerase alone may not properly protect the genome, especially during the early stages of reprogramming when most epigenetic reprogramming events occur. As a result, most reported iPS cell lines are unable to generate live-born mice through TCA18,19,42. Here, we report that overexpression of Zscan4 promotes rapid telomere extension by facilitating T-SCE at early stages of iPS cell formation, resulting in not only significant improvement of reprogramming efficiency but also significant improvement of iPS cell quality. Our study provides the first evidence that factors involved in the process of ooctye-mediated reprogramming can help maintain genomic stability during reprogramming and thus promote generation of high quality iPS cells.
Materials and Methods
Cell culture
iPS cells and ES cells were cultured on irradiated feeder cells in mouse ES cell medium containing leukemia inhibitory factor (LIF), 15% FBS (Gibco), nonessential amino acids, L-glutamine, nucleosides, 2-mercaptoethanol and penicillin/streptomycin. iPS cells and ES cells were passaged every 2-3 days. MEFs, plat-E packaging cells and feeder cells were cultured in MEF medium containing 10% FBS, nonessential amino acids and penicillin/streptomycin. Feeder cells were generated by irradiating ICR MEFs. MEFs were derived from E13.5 embryos of C57BL/6 mice that were heterozygous for the Oct4-GFP transgenic allele, as previously described60,62. In this study, we used MEFs within the first three passages for iPS cell generation to avoid replicative senescence.
Generation of iPS cells
Mouse iPS cells were generated with retroviruses as previously described62. Retrovirus was produced by transfecting plat-E cells with pMXs retroviral vectors. MEFs from OG2 mice, which carry a transgenic Oct4 promoter driving GFP expression, were seeded at a density of 8 × 104 cells per well in six-well plates 12 h before infection. Virus-containing supernatants from the plat-E cultures were recovered and combined (e.g., to mix viruses for Oct4, Sox2, Klf4, c-myc and Zscan4). Virus-containing supernatants supplemented with 4 μg/ml polybrene were added to the plates of MEFs, and this day was defined as “Day 0”. The culture medium was changed 12 h later. Four days post-infection, MEFs were digested into single cells and reseeded at a density of 2 000 cells (OSKM/OSKMZ), 2 × 104 cells (OSK/OSKZ) per well in six-well plate, or 3 000 cells (OSKM/OSKMZ + p53) per well in six-well plates pre-seeded with irradiated feeder cells. The culture medium was changed into mouse ES cell medium at day 5 post-infection and was changed daily.
To quantify the reprogramming efficiency using the Oct4-GFP reporter, GFP-positive colonies were counted using an Olympus inverted fluorescent microscope. Colonies were also trypsinized and analyzed using Becton Dickinson FACS Calibur cell sorter. GFP-positive cells were gated with a control signal from the PE channel, and a minimum of 30 000 events were recorded. Numbers of GFP-positive colonies and the percentage of GFP-positive cells were scored at day 12 for OSKM/OSKMZ and day 16 for OSK/OSKZ.
Plasmid construction
Full-length cDNAs of murine Zscan4f, Npm1 and the other genes shown in Supplementary information, Table S1 were cloned into the retroviral vector pMXs. The short hairpin RNAs (shRNAs) that targeted mouse Zscan4 mRNA, p53 mRNA and the control shRNA were cloned into the pMKO.1 retroviral vector, which was obtained from Addgene (Addgene, Cambridge, MA, http://www.addgene.org). The Zscan4 shRNA target sequences were: GACCTTCAGACCAACAATTTA (Z shRNA1) and GAAATGGACTCCCTTCTTATT (Z shRNA2). The p53 shRNA target sequence was: GTACATGTGTAATAGCTCCTG (p53 shRNA3). A randomly distributed sequence was used as a control: GATTAATAGGATTGCAGAATT.
Quantitative PCR
Total RNA was isolated from the cells using TRIzol reagent (Invitrogen). One microgram of total RNA was reverse transcribed using an iScriptTM cDNA Synthesis kit (Bio-Rad). Real-time quantitative PCR reactions were set up in triplicate using iQTM SYBR Green Supermix (Bio-Rad) and run on the Bio-Rad CFX96. Primer information is provided as follows.
Endo-Oct4-F: ATTCCCAACGAGAAGAGTATGA, Endo-Oct4-R: TCCCTTGCCTTGGCTCACAG;
β-actin-F: GAAATCGTGCGTGACATCAAAG, β-actin-R: TGTAGTTTCATGGATGCCACAG;
Zscan4-F: GAGATTCATGGAGAGTCTGACTGATGAGTG, Zscan4-R: GCTGTTGTTTCAAAAGCTTGATGACTTC;
Zscan4-endo-F2: CCTATGTAGCCGTCGCTTTC, Zscan4-endo-R2: AAGTGGCAATCCACAAGCAT;
Trf2-F: GGTCTTCCAGCAGAAGATGC, Trf2-R: AAGCTGCCTTCAGAGTCCTG;
Pot1b-F: TCCTCATACGAGGGAAGGTG, Pot1b-R: GATGCTGGGATCTGGAAAAA;
Rap1-F: CCGCTACCTCAAGCACCTAC, Rap1-R: CACTCCTCCTCAGGCAAGTC;
Trf1-F: GGACAGATTCGGATTCTCCA, Trf1-R: GCTGTCACGAGTACGACGAA;
Pot1a-F: AGCTTCACTCCTCAGGACCA, Pot1a-R: GGGTTCCATCCCATACCTTT;
Tpp1-F: TCGGATCCTAGCTCTCCTCA, Tpp1-R: AAAGTCCTGGGTGGTCACTG;
Tin2-F: GGAGTTTCTGCAGTCCTTGC, Tin2-R: TCTGGACTCTGCTGGGAAGT
Teratoma formation and histological analysis
iPS cells were suspended at 1 × 107 cells/ml in MEF medium. Each nude mouse was subcutaneously injected in the dorsal flank with 100 μl of cell suspension (1 × 106 cells). Four to six weeks after the injection, teratomas were surgically dissected from the mice. Samples were fixed in PBS containing 4% formaldehyde and subsequently embedded in paraffin wax. Sections were stained with hematoxylin and eosin.
Pulsed-field gel electrophoresis and terminal restriction fragment analysis
TRF analysis was performed as described previously41. DNA was extracted from iPS cells and ES cells. A total of 15 μg DNA was digested overnight with MboI at 37 °C and electrophoresed through 1% agarose gels in 1× TAE at 14 °C using a CHEF Mapper® XA pulsed field electrophoresis system (Bio-Rad). Auto Algorithm was used to separate DNA samples with a size range from 7.5 kb to 100 kb. The gel was blotted and probed with (TTAGGG)13 using DIG high prime DNA labeling and detection starter Kit II (Roche).
Telomere Q-FISH
Telomere FISH was performed using a telomere peptide nucleic acid (PNA; 0.5 μg/ml) (Panagene, Korea) FISH Kit/FITC according to the manufacturer's instructions. Telomeres were denatured at 80 °C and hybridized with the telomere PNA probe. Chromosomes were stained with 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Fluorescence from the chromosomes and telomeres was digitally imaged on a Zeiss microscope with fluorescein isothiocyanate (FITC)/DAPI filters using AxioCam and AxioVision software 4.6. Telomere length, presented in telomere fluorescence units, was integrated using the TFL-TELO program (kindly provided by P Lansdorp of Terry Fox's laboratory) and calibrated using standard fluorescence beads.
Telomerase activity assay
Cell lysates were prepared from 5 × 105 cells per sample. Samples that were incubated at 85 °C for 10 min served as a negative control. Telomerase activity was measured by the TRAP assay using a TRAPEZE Telomerase Detection Kit (Millipore) according to the manufacturer's instructions.
Telomere CO-FISH
Cell samples from different days (OSKM/OSKMZ) were cultured as previously described41. The medium was changed daily. 5′-Bromo-2′-deoxyuridine (BrdU) was added for 18 h to allow BrdU incorporation for one cell cycle. Demecolcine solution (0.2 μg/ml, Sigma) was added for the final 5 h. Metaphase spreads were prepared. Slides were stained with 0.5 μg/ml Hoechst 33258 (Sigma), washed in 2× SSC, and then exposed to longwave radiation (e.g., 365 nm) in McIlvaine's buffer (pH 8.0) for 30 min. The BrdU-substituted DNA was digested with 3 U/μl Exonuclease III (Takara) for 10 min at room temperature. The leading strand telomeres were visualized with Cy3-conjugated DNA probes (Panagene). Chromosomes were counterstained with 1 μg/ml DAPI (Sigma). Three independent experiments were carried out for each sample.
G-SCE assay
Cell samples from different days (OSKM/OSKMZ) were cultured as previously described41. The medium was changed every day. BrdU was added for 18 h to allow BrdU incorporation for one cell cycle. Demecolcine solution (0.2 μg/ml, Sigma) was added for the final 5 h. Metaphase spreads were prepared. The slides were treated with 2 M HCl followed by 0.1 M Na2B4O7. BSA (1%, Sigma) was used to block nonspecific signal. The slides were incubated with anti-BrdU antibody (Sigma) at 4 °C overnight, followed by staining with the respective secondary antibodies conjugated to Alexa Fluor (Invitrogen). The cells were counterstained with DAPI (Sigma).
Western blotting
Cell samples from different days were extracted using cell lysis buffer containing PMSF (Beyotime). Insoluble material was pelleted by centrifugation at 13 200 rpm for 15 min at 4 °C. Protein concentrations were determined using the Bradford assay (Bio-Rad). The proteins were denatured, reduced, separated by SDS-PAGE, and transferred onto nitrocellulose membranes (PROTRAN; Whatman, Schleicher & Schuell). The membranes were blocked with 5% fat-free dry milk in TBS-Tween (TBST) for 60 min, incubated with primary antibodies overnight at 4 °C, washed three times with TBST for 10 min, incubated with peroxidase-conjugated secondary antibody in TBST with 5% fat-free dry milk for 90 min, and washed three times with TBST for 10 min. Immunoreactive proteins were detected using SuperSignal West Dura HRP Detection Kits (Pierce). Images were taken using an infrared imaging system (ImageQuant LAS 4000).
Immunofluorescence
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 1% BSA (Sigma). The cells were then stained with primary antibodies against γ-H2ax and Trf1, followed by staining with the respective secondary antibodies conjugated to Alexa Fluor (Invitrogen). The cells were counterstained with DAPI (Sigma).
Diploid and tetraploid blastocyst injection and embryo transfer
Diploid blastocysts were collected from the uterus of E3.5 superovulated ICR females mated with ICR males and maintained in KSOM medium with amino acids until iPS cell injection. For the blastocyst injection, cells were trypsinized, resuspended in mouse ES cell medium without LIF, and kept on ice. A flat tip microinjection pipette with an internal diameter of 15-18 μm was used for the injection. To generate mice by tetraploid embryo complementation, two-cell embryos were collected from the oviducts of ICR females and electrofused to produce one-cell tetraploid embryos that were then cultured in KSOM media. Approximately ten to fifteen iPS cells were injected into the tetraploid blastocyst cavity. The blastocysts were placed in KSOM with amino acids until embryo transfer. Ten to fifteen injected blastocysts were transferred to each uterine horn of 2.5-days-postcoitum pseudopregnant ICR females. Pregnant recipients with tetraploid embryos were subjected to caesarean section on day 18.5 of gestation.
Genotyping
The microsatellite markers used in SSLP were amplified using primer pair sequences obtained from the Mouse Genome Informatics website (The Jackson Laboratory, http://www.informatics.jax.org). DNA was extracted from the tail tips of the mice. Thirty-five cycles of PCR amplification were performed, and the products were separated on a 3% agarose gel before visualization. Allele sizes were approximated based on the known allele sizes of the various inbred strains. The primers used to detect exogenous factors are shown below.
Zscan4f-Retro-F: CTTTCGCCAATCATCCACTT, Zscan4f-Retro-R: CCTTGCAAAATGGCGTTACT
Oct4-Retro-F: GCTTGGATACACGCCGC, Oct4-Retro-R: TTCATGTCCTGGGACTCCTC
GAPDH-F: ACCACAGTCCATGCCATCAC, GAPDH-R: TCCACCACCCTGTTGCTGTA
Acknowledgments
We thank Drs S Dang and M Lei for critical and useful comments on this manuscript. We thank T Wei, Y Li and H Pang for technical assistance. This study was supported by the Ministry of Science and Technology of China (2009CB941101 to JL, 2009CB941000 to LL), the Chinese Academy of Sciences (XDA01010403 to JL), and the Shanghai Municipal Commission for Science and Technology (11ZR1443000 to XZ). JL gratefully acknowledges the support of SA-SIBS Scholarship Program.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Highly expressed factors in oocytes or early cleavage-stage embryos
Zscan4 greatly promotes iPS cell formation.
Characterization of OSKM and OSKMZ iPS cells.
References
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF. Stem cells: The dark side of induced pluripotency. Nature. 2011;471:46–47. doi: 10.1038/471046a. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Boue S, Izpisua Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming à la carte. Nat Rev Genet. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Zhao R, Doi A, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarig R, Rivlin N, Brosh R, et al. Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J Exp Med. 2010;207:2127–2140. doi: 10.1084/jem.20100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5:135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Hochedlinger K, Bell G, Jaenisch R. ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable. Proc Natl Acad Sci USA. 2006;103:933–938. doi: 10.1073/pnas.0510485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama S, Jakt ML, Suzuki M, et al. Equivalency of nuclear transfer-derived embryonic stem cells to those derived from fertilized mouse blastocysts. Stem Cells. 2006;24:2023–2033. doi: 10.1634/stemcells.2005-0537. [DOI] [PubMed] [Google Scholar]
- Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Bio. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Amano T, Nakatake Y, et al. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- Ikbal M, Atasoy M, Pirim I, Aliagaoglu C, Karatay S, Erdem T. The alteration of sister chromatid exchange frequencies in Behcet's disease with and without HLA-B51. J Eur Acad Dermatol Venereol. 2006;20:149–152. doi: 10.1111/j.1468-3083.2006.01386.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Erdmann N, Giannone RJ, Wu J, Gomez M, Liu Y. An increase in telomere sister chromatid exchange in murine embryonic stem cells possessing critically shortened telomeres. Proc Natl Acad Sci USA. 2005;102:10256–10260. doi: 10.1073/pnas.0504635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia N, Scholer HR. p53 connects tumorigenesis and reprogramming to pluripotency. J Exp Med. 2010;207:2045–2048. doi: 10.1084/jem.20101866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MG, Stagg CA, Falco G, et al. An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells. Gene Expr Patterns. 2008;8:181–198. doi: 10.1016/j.gep.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Yin Y, Ye X, et al. Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Res. 2012;22:757–768. doi: 10.1038/cr.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boeck G, Forsyth RG, Praet M, Hogendoorn PC. Telomere-associated proteins: cross-talk between telomere maintenance and telomere-lengthening mechanisms. J Pathol. 2009;217:327–344. doi: 10.1002/path.2500. [DOI] [PubMed] [Google Scholar]
- Jain D, Cooper JP. Telomeric strategies: means to an end. Annu Rev Genet. 2010;44:243–269. doi: 10.1146/annurev-genet-102108-134841. [DOI] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ding G, Lin J, et al. Different developmental potential of pluripotent stem cells generated by different reprogramming strategies. J Mol Cell Biol. 2011;3:197–199. doi: 10.1093/jmcb/mjr012. [DOI] [PubMed] [Google Scholar]
- Li J, Mombaerts P. Nuclear transfer-mediated rescue of the nuclear genome of nonviable mouse cells frozen without cryoprotectant. Biol Reprod. 2008;79:588–593. doi: 10.1095/biolreprod.108.069583. [DOI] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Loring J, et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci USA. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yuan D, Wei B, et al. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- Li W, Ding S. Small molecules that modulate embryonic stem cell fate and somatic cell reprogramming. Trends Pharmacol Sci. 2010;31:36–45. doi: 10.1016/j.tips.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Aasen T, Raya A, Barrero MJ, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biology. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Foreman R, Chevalier B, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darabi R, Arpke RW, Irion S, et al. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell. 2012;10:610–619. doi: 10.1016/j.stem.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Hansen NF, Zhao L, et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Bailey SM, Okuka M, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- Tanemura K, Ogura A, Cheong C, et al. Dynamic rearrangement of telomeres during spermatogenesis in mice. Dev Biol. 2005;281:196–207. doi: 10.1016/j.ydbio.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Huang J, Wang F, Okuka M, et al. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011;21:779–792. doi: 10.1038/cr.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl-Cachedenier I, Flores I, Klatt P, Blasco MA. Telomerase reverses epidermal hair follicle stem cell defects and loss of long-term survival associated with critically short telomeres. J Cell Biol. 2007;179:277–290. doi: 10.1083/jcb.200704141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Highly expressed factors in oocytes or early cleavage-stage embryos
Zscan4 greatly promotes iPS cell formation.
Characterization of OSKM and OSKMZ iPS cells.