Abstract
Human lung adenocarcinomas with activating mutations in EGFR (epidermal growth factor receptor) often respond to treatment with EGFRtyrosine kinase inhibitors(TKIs),butthe magnitude of tumour regression is variable and transient1,2. This heterogeneity in treatment response could result from genetic modifiers that regulate the degree to which tumour cells are dependent on mutant EGFR. Through a pooled RNA interference screen, we show that knockdown of FAS and several components of the NF-κB pathway specifically enhanced cell death induced by the EGFR TKI erlotinib in EGFR-mutant lung cancer cells. Activation of NF-κB through overexpression of c-FLIP or IKK (also known as CFLAR and IKBKB, respectively), or silencing of IκB (also known as NFKBIA), rescued EGFR-mutant lung cancer cells from EGFR TKI treatment. Genetic or pharmacologic inhibition of NF-κB enhanced erlotinib-induced apoptosis in erlotinib-sensitive and erlotinib-resistant EGFR-mutant lung cancer models. Increased expression of the NF-κB inhibitor IκB predicted for improved response and survival in EGFR-mutant lung cancer patients treated with EGFR TKI. These data identify NF-κB as a potential companion drug target, together with EGFR, in EGFR-mutant lung cancers and provide insight into the mechanisms by which tumour cells escape from oncogene dependence.
Despite marked clinical successes achieved with inhibitors of ‘driver’ kinases that promote tumour growth, responses are rarely complete and also vary in duration3,4. We hypothesized that the heterogeneity of treatment response may result from genetic modifiers that regulate the degree to which tumour cells are dependent upon the driver kinase and the response to TKI treatment.
Using lung cancer as a model to identify such modifiers, we conducted a screen for genes that, when silenced, enhance EGFR dependence in EGFR-mutant lung cancer cell lines. We intentionally selected H1650 cells for the primary screen because they are insensitive to EGFR TKIs despite expressing a mutant EGFR with an exon 19 deletion (ex19del) that predicts for erlotinib sensitivity in patients. H1650 cells harbour no known EGFR TKI resistance mechanisms other than functional PTEN loss, which does not fully account for their insensitivity5. To identify small hairpin RNAs (shRNAs) that might restore dependence on mutant EGFR in H1650 cells, we introduced a pooled, shRNA library targeting >2,000 ‘cancer-relevant’ genes6 (Supplementary Table 1) and treated these cells with vehicle or erlotinib, a standard EGFR TKI used in EGFR-mutant lung cancer patients. Hairpins targeting 36 genes reproducibly conferred erlotinib sensitivity in H1650 cells across three replicates (threshold 1.5-fold depletion in erlotinib/vehicle, P < 0.1; Supplementary Table 2).
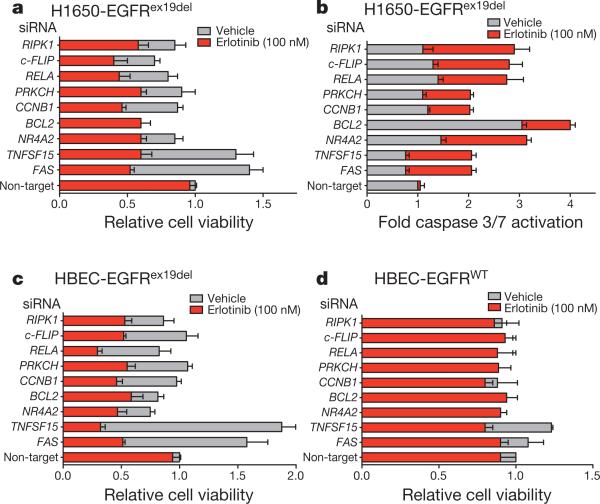
Among the primary screen hits, 18 of the targeted genes, including FAS, could be directly or indirectly linked to NF-κB signalling, which is known to have a role in survival signalling (Supplementary Table 2). Because recent data demonstrated that FAS and NF-κB signalling can promote tumour growth7–9, we proposed that FAS–NF-κB may rescue EGFR-mutant tumour cells from EGFR inhibition. First, we confirmed that shRNAs targeting six of the highest scoring NF-κB pathway-associated genes identified in the primary pooled screen effectively silenced expression of their targets in H1650 cells (Supplementary Fig. 1a). Then we validated their growth inhibitory effects in erlotinib-treated H1650 cells using independent siRNAs (Fig. 1a, Supplementary Fig. 1b). Importantly, this reduction in cell viability was associated with increased caspase 3/7 activity (Fig. 1b), indicating that knockdown of these genes promoted erlotinib-induced apoptosis. To address directly the role of the NF-κB pathway in EGFR TKI sensitivity, we knocked down the major NF-κB subunit RELA (not represented in the pooled library) and found that RELA knockdown also induced erlotinib sensitivity in H1650 cells using short term viability and apoptosis readouts as well as clonogenic assays (Fig. 1a, b and Supplementary Fig. 2). Furthermore, this sensitizing effect was specific to EGFR inhibition because stable RELA knockdown did not alter sensitivity to cisplatin, paclitaxel or ultraviolet treatment (Supplementary Fig. 2) or other TKI (imatinib) (Supplementary Fig. 3). Because c-FLIP and RIPK have been implicated as intermediate signalling proteins linking FAS to NF-κB10, we asked if these genes also regulate erlotinib sensitivity. Indeed, silencing of c-FLIP or RIPK induced erlotinib sensitivity in H1650 cells (Fig. 1a, b). The effects of the entire panel of siRNAs targeting nine different NF-κB pathway-associated genes were observed regardless of PTEN status (Supplementary Fig. 4a, b).
Figure 1. Mutant EGFR oncogene dependence requires downregulation of the FAS-NF-κB pathway.
a, Viability (CellTiter-Glo assay) of H1650 cells treated with vehicle or 100 nM erlotinib upon introduction of either a non-target siRNA pool or gene-specific siRNA pools targeting the genes. Relative cell viability is fold change in viability in erlotinib relative to vehicle non-target siRNA control (viability decrease >25% = validated). n = 3; mean + s.e.m. b, Caspase 3/7 activation (Caspase-Glo assay) in indicated cell lines treated as in a. c, d, Viability (CellTiter-Glo assay) of indicated isogenic HBEC cells treated as in a. n = 3; mean + s.e.m.
Next we asked whether the NF-κB pathway genes that scored in H1650 cells show similar effects in other EGFR mutant lung cancer cell lines. Whereas H1650 cells express EGFRex19del, 11-18 human lung cancer cells express EGFR(L858R) yet are also relatively insensitive to erlotinib without a resistance mechanism validated in patients11. Each of the siRNAs that scored in H1650 cells also scored in cell viability and survival assays in 11-18 cells (Supplementary Fig. 4c, d). Silencing of the same set of genes also enhanced the growth suppressive effects of erlotinib in HCC827 cells (expressing EGFRex19del) and in H3255 cells (expressing EGFR(L858R)), both of which are relatively more sensitive to EGFR TKIs (Supplementary Fig. 4e–h).
Because unknown alterations in human lung cancer cells could also influence erlotinib sensitivity, we used an isogenic system of EGFR-transduced human bronchial epithelial cells (HBEC) to test whether silencing of these genes cooperates with mutant EGFR to induce oncogene dependence. Consistent with prior data12, human bronchial epithelial cells (HBEC)-EGFR(L858R) and HBEC-EGFRex19del cells were not sensitive to erlotinib (100 nM). However, silencing of each of the nine genes studied in the lung cancer cell lines also induced erlotinib sensitivity in HBEC-EGFRex19del (Fig. 1c) and HBEC-EGFR(L858R) cells (Supplementary Fig. 5). Induction of erlotinib sensitivity seemed equivalent across both the exon19del and L858R EGFR genotypes. The erlotinib-sensitizing effect of silencing these genes was specific to mutant EGFR because no potentiating effect was seen in wild-type HBEC-EGFRWT cells (Fig. 1d).
Because FAS knockdown promoted erlotinib-induced apoptosis, we measured the activation state of three signalling pathways linked to cell survival (AKT, ERK, also known as MAPK1, and NF-κB) to determine which, if any, was associated with erlotinib-induced cell death. In erlotinib-sensitive HCC827 cells, erlotinib treatment alone led to reduced levels of phosphorylated pAKT, pERK and pRELA (a measure of NF-κB activity), regardless of FAS expression (Supplementary Fig. 6). But in erlotinib-resistant H1650 cells, these effects of erlotinib were only observed when FAS was silenced (Supplementary Fig. 6). In HBEC-EGFR(L858R) cells, erlotinib suppressed AKT and ERK activation regardless of FAS expression, but RELA phosphorylation was abolished only upon FAS silencing (Supplementary Fig. 6). Together these data implicate persistent NF-κB signalling in resistance to erlotinib-induced cell death.
NF-κB signalling was recently shown to be essential for KRAS-driven tumour growth13. Oncogenic KRAS and EGFR may drive tumour growth through a common signalling pathway. Because in our studies NF-κB knockdown alone was not lethal in EGFR-mutant lung cancer cell lines (HCC827, H3255), we compared the effect of silencing NF-κB in HBEC-KRAS12V cells versus HBEC-EGFR(L858R) cells directly. Knockdown of RELA alone impaired the growth of HBEC-KRAS12V but not HBEC-EGFR(L858R) cells (Supplementary Fig. 7), indicating that mutant EGFR does not function identically to mutant KRAS in this model.
To extend our findings to in vivo models, we used shRNAs targeting FAS, RELA or c-FLIP (distinct from the primary screen) to achieve stable knockdown (Supplementary Fig. 8) of the cognate protein in erlotinib-resistant H1650 cells. Silencing of each gene by stable shRNA enhanced erlotinib sensitivity, with a 1–2 log-fold decrease in IC50 (half maximal inhibitory concentration; Fig. 2a) and induction of apoptosis as measured by PARP cleavage (Fig. 2b). Similar results were observed in 11-18 and HBEC-EGFR(L858R) cells (Supplementary Fig. 9). We found that erlotinib treatment induced tumour regression and apoptosis only upon knockdown of FAS or RELA (Fig. 2c, d and Supplementary Fig. 10a, b) in H1650 human xenograft tumours.
Figure 2. Suppression of the FAS-NF-κB pathway enhances EGFR TKI response in EGFR-mutant cells and tumour models.
a, Erlotinib dose response in H1650 cells expressing a non-target control shRNA or a FAS (shRNA2), RELA (shRNA1), or c-FLIP(shRNA1) shRNA (5 μM erlotinib IC50 in parental H1650 cells). Cell viability was assayed as in Fig. 1. n = 3; mean + s.e.m. b, Immunoblots showing expression of indicated signalling proteins in H1650 cells generated in a. Data represent three independent experiments. NT, non-target; PARPcl, PARP cleaved. (The decrease in c-FLIP protein by shRNA was less complete than the decrease in c-FLIP mRNA level by siRNA; Supplementary Fig. 1). c, Effects of stable knockdown of FAS (shRNA3) or RELA (shRNA2) on erlotinib sensitivity in H1650 xenograft tumours, compared to non-target shRNA control H1650 tumours. Established tumours (>200 mm3,, n = 10 per treatment group) were randomized and treated for 7 days with 12.5 mg erlotinib per kg per day, n = 10. Data expressed as mean + s.e.m. d, Immunoblots showing expression of indicated proteins in representative H1650 tumour xenografts from c analysed at treatment day 7.
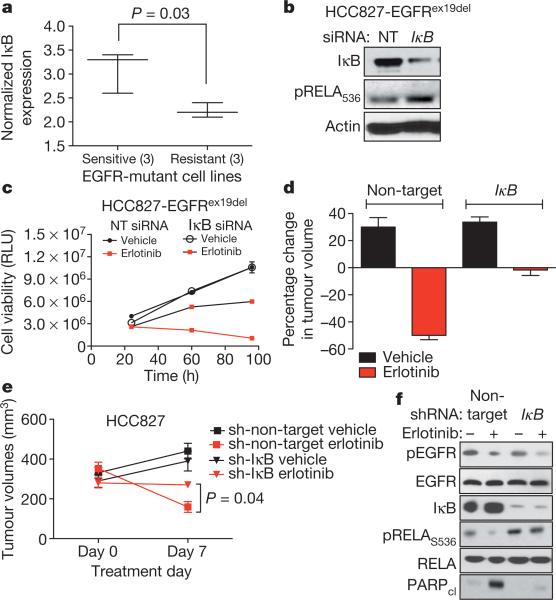
We next asked whether activation of NF-κB might be sufficient to confer erlotinib resistance in EGFR-mutant tumours. Canonical NF-κB signalling requires downregulation of the NF-κB inhibitor IκB14. Therefore we predicted that decreased IκB levels, leading to increased NF-κB signalling, might promote EGFR TKI resistance. First, we noted that IκB expression was lower in three erlotinib-resistant EGFR-mutant lung cancer cell lines that are resistant to erlotinib compared with three sensitive cell lines (Fig. 3a and Supplementary Fig. 11). Furthermore, we found that knockdown of IκB increased NF-κB phosphorylation (Fig. 3b) and conferred partial resistance to erlotinib in HCC827 cells in vitro (Fig. 3c) and in vivo (Fig. 3d–f). These effects were specific to EGFR inhibition because stable knockdown of IκB did not protect HCC827 cells from cisplatin, paclitaxel or ultraviolet treatment, as measured by clonogenic assays (Supplementary Fig. 12). c-FLIP has been implicated as an intermediate signalling molecule that activates NF-κB downstream of FAS10. Ectopic expression of c-FLIP induced persistent phosphorylation of RELA and resistance to erlotinib in HCC827 cells (Supplementary Fig. 13). Together these data suggest that NF-κB activation can rescue EGFR-mutant tumour cells from EGFR TKI.
Figure 3. NF-κB activation through IκB downregulation confers EGFR TKI resistance in EGFR-mutant lung cancer models.
a, Correlation of IκB expression with EGFR TKI sensitivity in EGFR-mutant lung cancer cells (IκB mRNA expression from Oncomine; sensitive <0.02 μM IC50: HCC827, H3255, HCC4006; resistant >1 μM IC50: H1650, H1975, H820). b, Immunoblots showing IκB and pRELA expression in lysates from HCC827 treated with non-targeting or IκB siRNA pools. c, Viability (CellTiter-Glo assay) of HCC827 cells treated with non-target siRNA pool or IκB siRNA pool and either vehicle or erlotinib (100 nM). RLU is relative luciferase units (n = 3; mean + s.e.m.). d, e, Effects of stable knockdown of IκB on erlotinib sensitivity in HCC827 tumour xenografts compared to non-target shRNA control HCC827 tumours. Tumours were established and treated as in Fig. 2c (n = 8 per treatment group, mean + s.e.m., 7-day treatment). f, Immunoblots showing expression of indicated proteins in representative HCC827 tumour xenografts analysed at treatment day 7.
Acquired resistance to erlotinib in patients can occur through second site, drug-resistant mutations in EGFR or through amplification of the MET kinase15,16. These same resistance mechanisms can also evolve in vitro with prolonged growth in EGFR TKI17,18. We therefore asked if NF-κB activation occurs naturally during the derivation of erlotinib-resistant HCC827 cells through prolonged and continuous EGFR TKI treatment. In parental HCC827 cells acute erlotinib treatment (100 nM, 24 h) inhibited EGFR, MET, AKT, ERK and RELA phosphorylation. Continuous erlotinib treatment resulted in the outgrowth of eight resistant HCC827 subclones (over >8 weeks) in which EGFR phosphorylation remained inhibited by erlotinib. Consistent with prior data18, we observed compensatory upregulation of phosphorylated MET in four erlotinib-resistant subclones. We also observed increased FAS expression in the absence of MET upregulation in four additional subclones. Increased RELA phosphorylation and decreased IκB expression were observed in three out of four sub-clones in which FAS was upregulated (Supplementary Fig. 14a). Knockdown of FAS in those subclones with FAS upregulation decreased pRELA and enhanced erlotinib sensitivity, but did not result in decreased pAKT or pERK (Supplementary Fig. 14b, c). Thus, FAS-mediated EGFR TKI resistance is distinct from MET-mediated EGFR TKI resistance, which occurs primarily through reactivation of pERK and pAKT in HCC827 cells18. Our results are consistent with recent data establishing both MEK-dependent19 and MEK-independent20 mechanisms of resistance of BRAF-mutant melanomas to the BRAF inhibitor PLX4032.
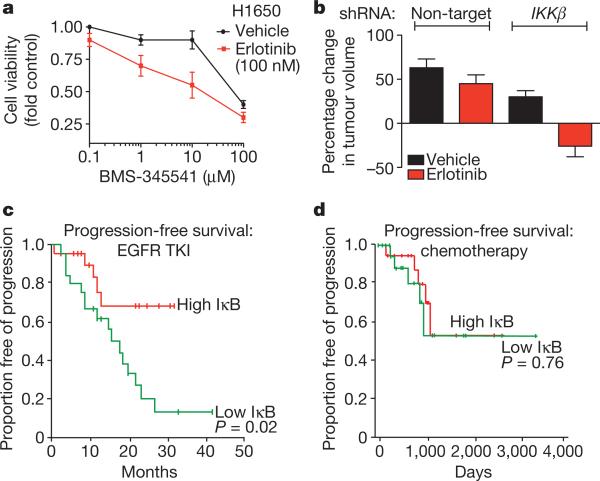
NF-κB pathway inhibition can be achieved through inhibition of IκB kinase (IKKβ, also known as IKBKB), the primary kinase that promotes IκB instability leading to NF-κB activation14. We therefore asked if pharmacologic or genetic inhibition of IKKβ enhances erlotinib sensitivity in our erlotinib-resistant lung cancer models. Treatment of H1650 (Fig. 4a) or HBEC-EGFR(L858R) cells (Supplementary Fig. 15a, b) with the IKKβ inhibitor BMS-345541 (ref. 21) inhibited RELA phosphorylation and restored erlotinib sensitivity. Because BMS-345541 has suboptimal pharmacokinetic properties we used an shRNA to knock down IKKβ in H1650 cells. The resulting H1650 cells, or those transduced with a non-target control shRNA, were then grown as xenograft tumours and treated with either vehicle or erlotinib. IKKβ knockdown resulted in increased levels of IκB and decreased NF-κB phosphorylation, as expected. Erlotinib treatment induced tumour regression and apoptosis only upon knockdown of IKKβ in this system (Fig. 4b and Supplementary Fig. 15c, d).
Figure 4. Rationale for combined NF-κB and EGFR inhibition in EGFR-mutant lung cancers.
a, Dose response in H1650 cells treated with BMS-345541 (IKK inhibitor) and additionally either vehicle or erlotinib (100 nM). Viability was measured as in Fig. 1 (n = 3, mean + s.e.m.). b, Effects of stable knockdown of IKKβ on erlotinib sensitivity in H1650 tumour xenografts compared to non-target shRNA control H1650 tumours. Established tumours (>200 mm3, n = 10 per treatment group) were randomized and treated for 7 days with 12.5 mg erlotinib per kg per day or vehicle. Data are expressed as in Fig. 2c (+ s.e.m.). c, d, Effects of IκkB expression on progression free survival in patients with EGFR-mutant lung cancers (c) treated with single agent EGFR TKI (n = 52) or (d) chemotherapy and surgery (n = 43). Clinical characteristics and responses were defined previously22. Median progression-free survival and overall survival for the entire EGFR TKI-treated cohort were 20 months (95% confidence interval, 13–26.9) and 33 months (95% confidence interval, 22.2–43.8), respectively.
To determine the clinical relevance of our findings, we examined the status of NF-κB activation in a cohort of 52 patients (part of a previously characterized cohort22) with EGFR mutant lung cancer who were treated with erlotinib and did not have evidence of the T790M mutation as a potential cause of erlotinib resistance. Because reduced IκB levels were associated with erlotinib resistance in cell lines and tumour models, we asked if IκB expression correlated with EGFR TKI response in patients with EGFR-mutant lung cancers. Low IκB expression (‘high-NF-κB’ activation state) was predictive of worse progression-free survival (Fig. 4c and Supplementary Table 4a) and decreased overall survival (Supplementary Fig. 16a and Supplementary Table 4b). This finding was specific to erlotinib because IκB expression did not predict outcome in a previouslycharacterized cohort23 of 43 EGFR-mutant lung cancer patients that were not treated with EGFR TKIs but instead with chemotherapy and surgery (Fig. 4d and Supplementary Fig. 16b). We also observed a significant positive correlation between FAS expression and RELA, c-FLIP and IκB levels in the erlotinib-treated patients (Supplementary Table 5). Together, our findings suggest that the extent to which EGFR-mutant tumour cells engage the NF-κB pathway may, in part, explain the non-uniform response to EGFR TKI treatment observed in EGFR-mutant lung cancer patients and provide rationale for testing an IKK inhibitor in combination with an EGFR TKI in EGFR-mutant lung cancer patients.
METHODS SUMMARY
Compounds and cell culture
H1650, 11-18, H3255, HCC827 cells were provided by W. Pao and were grown in RPMI supplemented with 10% FBS. H1650 PTEN-reconstituted cells have been described previously5. HBEC cell lines were provided by J. Minna and grown in K-SFM. Erlotinib was from LC Labs. BMS-354541 was from Sigma-Aldrich.
Antibodies and immunoblots
All antibodies were from Cell Signaling Technologies and used at 1:1,000 dilution according to manufacturer's instructions. Lysates from cell lines and xenograft tumours were generated using standard methods and RIPA buffer and assayed by immunoblots.
Pooled shRNA screen
H1650 cells were infected with 6,783 shRNAs targeting ~2,500 cancer-associated genes in a pooled format. Abundance of each hairpin within the total population over time was detected by hybridization to custom Agilent oligonucleotide microarrays with half-hairpin and barcode probes corresponding to each shRNA6. The abundance difference for the erlotinib-synthetic screen was determined as the average of the log2 signal from erlotinib treated cells at later time points (day 10 and day 21) minus the average of this log2 signal from vehicle treated cells at the later time points (day 10 and day 21). Significant hits were defined as those altered by 1.5 fold, P < 0.1.
RNA interference assays
Sequences for individual small interfering RNA (siRNA) or shRNA (from Sigma or Open Biosystems) are shown in Supplementary Table 3. For siRNA experiments, cells were treated in triplicate according to manufacturer's protocol. For shRNA experiments, cells were spin-infected with shRNA virus. Where indicated cells were treated with vehicle or erlotinib 24 h after RNAi treatment. Gene silencing was confirmed by mRNA isolation and gene-specific quantitative RT PCR and by immunoblots. Cell viability or survival was measured using CellTiter-Glo and Caspase-Glo, respectively, 24–96 h after drug treatment according to the manufacturer's instructions and normalized to cells with a non-target siRNA pool or shRNA hairpin.
Supplementary Material
Acknowledgements
We thank W. Pao, H. Varmus and members of the MSKCC Lung Cancer Oncogenome group for intellectual support. We thank J. Javier Sanchez for analysing human lung cancer gene expression data. We thank J. Wongvipat and E. Philips for technical assistance and W. Polkinghorn and members of the Sawyers Laboratory for critique of the manuscript. T.G.B. is supported by the MSKCC Clinical Scholars Fellowship funded by the Charles A. Dana Foundation, an ASCO YIA, and the Caine Halter Lung Cancer Research Fund/Uniting Against Lung Cancer Research Grant. C.L.S. and G.H. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Author Contributions T.G.B. and H.H. designed research, performed experiments, analysed data and co-wrote the paper. K.C., P.M., K.D., M.T. and C.C. provided reagents, performed experiments and analysed data. J.P. and V.A.M. analysed data. R.R., G.H. and C.L.S. designed research, analysed experiments and co-wrote the paper.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Haber DA, et al. Molecular targeted therapy of lung cancer: EGFR mutations and response to EGFR inhibitors. Cold Spring Harb. Symp. Quant. Biol. 2005;70:419–426. doi: 10.1101/sqb.2005.70.043. [DOI] [PubMed] [Google Scholar]
- 2.Miller VA, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J. Clin. Oncol. 2008;26:1472–1478. doi: 10.1200/JCO.2007.13.0062. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 4.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nature Rev. Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sos ML, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva JM, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peter ME, et al. The CD95 receptor: apoptosis revisited. Cell. 2007;129:447–450. doi: 10.1016/j.cell.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, et al. CD95 promotes tumour growth. Nature. 2010;465:492–496. doi: 10.1038/nature09075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Reilly LA, et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature. 2009;461:659–663. doi: 10.1038/nature08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green DR. Cancer: a wolf in wolf's clothing. Nature. 2010;465:433. doi: 10.1038/465433a. [DOI] [PubMed] [Google Scholar]
- 11.Gong Y, et al. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507). PLoS ONE. 2009;4:e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato M, et al. Multiple oncogenic changes (K-RASV12, p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 13.Meylan E, et al. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo JL, Kamata H, Karin M. IKK/NF-κB signaling: balancing life and death–a new approach to cancer therapy. J. Clin. Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bean J, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl Acad. Sci. USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelman JA, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J. Clin. Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Amiri KI, Burke JR, Schmid JA, Richmond A. BMS-345541 targets inhibitor of κB kinase and induces apoptosis in melanoma: involvement of nuclear factor κB and mitochondria pathways. Clin. Cancer Res. 2006;12:950–960. doi: 10.1158/1078-0432.CCR-05-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosell R, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 23.Chitale D, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28:2773–2783. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.