Abstract
Somatic reprogramming induced by defined transcription factors is a low efficiency process that is enhanced by p53 deficiency 1-5. To date, p21 is the only p53 target shown to contribute to p53 repression of iPSC (induced pluripotent stem cell) generation 1, 3, suggesting additional p53 targets may regulate this process. Here, we demonstrated that mir-34 microRNAs (miRNAs), particularly miR-34a, exhibit p53-dependent induction during reprogramming. mir-34a deficiency in mice significantly increased reprogramming efficiency and kinetics, with miR-34a and p21 cooperatively regulating somatic reprogramming downstream of p53. Unlike p53 deficiency, which enhances reprogramming at the expense of iPSC pluripotency, genetic ablation of mir-34a promoted iPSC generation without compromising self-renewal and differentiation. Suppression of reprogramming by miR-34a was due, at least in part, to repression of pluripotency genes, including Nanog, Sox2 and Mycn (N-Myc). This post-transcriptional gene repression by miR-34a also regulated iPSC differentiation kinetics. miR-34b and c similarly repressed reprogramming; and all three mir-34 miRNAs acted cooperatively in this process. Taken together, our findings identified mir-34 miRNAs as novel p53 targets that play an essential role in restraining somatic reprogramming.
Differentiated somatic cells can be induced to generate pluripotent stem cells that functionally resemble embryonic stem cells (ESCs) 6. This reprogramming process is rooted in the remarkable cellular plasticity retained during differentiation. The process can be triggered by exogenous expression of a set of defined ESC-specific transcription factors, Pou5f1 (Oct4), Sox2, Klf4, and c-Myc 6-9, which constitute the core regulatory circuits controlling pluripotency and self-renewal. Enforced expression of these reprogramming factors generates iPSCs with low efficiency and slow kinetics, suggesting the existence of cellular and molecular barriers to the process 10.
Recent studies have revealed considerable mechanistic overlap between somatic cell reprogramming and malignant transformation 11. Cellular mechanisms that enhance reprogramming, including cell proliferation and survival, evasion of DNA damage response, and cell immortalization, have long been known to promote tumorigenesis 3-5, 12. Several oncogenes and tumor suppressors also serve as essential regulators for reprogramming 6, 10, 11. Notably, the inactivation of p53, one of the most important tumor suppressors, significantly enhances iPSC generation 1-5, 13. As a tumor suppressor, p53's transcriptional regulation converges onto multiple target genes that collectively mediate its downstream effects, including cell cycle arrest, cellular senescence, apoptosis, DNA damage response, and genomic stability 14. Similarly, p53's role in repressing reprogramming also likely is mediated through multiple targets. To date, the cell-cycle regulator p21 is the only p53 target with a demonstrated role in repressing reprogramming. However, p21 deficiency only partially phenocopies that of p53 1, 3, 15, suggesting that p53 represses iPSC generation through as yet unidentified mechanism(s) and target(s).
Previous studies have identified the mir-34 miRNAs as bona fide p53 transcriptional targets, whose over-expression triggers cell cycle arrest or apoptosis in a cell type- and context-dependent manner 16-18. miRNAs, a large family of small non-coding RNAs, primarily repress gene expression post-transcriptionally, by pairing with partially complementary mRNA targets 19, 20. In response to p53 activation, induced mir-34 miRNAs can mediate p53 downstream effects by repressing specific targets, including cyclin D1, cyclin E2, Cdk4, Cdk6, Bcl2, and c-Met 21. Although p53 is primarily characterized for its role in transcriptional activation, it acts as a global gene regulator that both activates and represses gene expression 14. Direct transcriptional repression, together with indirect post-transcriptional repression through miRNAs such as mir-34, constitute two major mechanisms for p53-mediated gene repression.
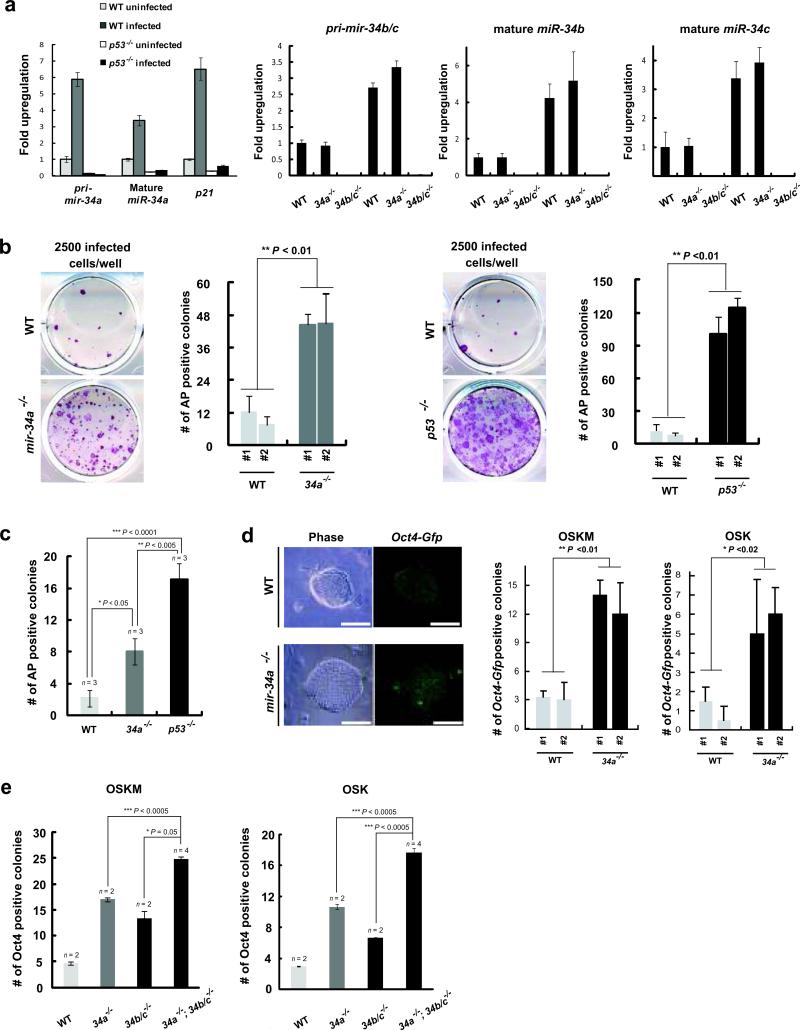
The mir-34 miRNAs belong to an evolutionarily conserved family, with three mammalian homologues, miR-34a, b and c, localized to two distinct genomic loci, mir-34a and mir-34b/c 16. All three mir-34 miRNAs were significantly induced when mouse embryonic fibroblast (MEF) reprogramming was triggered with Sox2, Oct4, and Klf4 in the presence or absence of c-Myc (Fig. 2a, data not shown). In both cases, mir-34 induction, like that of p21, was dependent on the activation of intact p53 (Fig. 2a). We therefore investigated whether mir-34 miRNAs are novel components of the reprogramming regulatory circuit downstream of p53. Among all mir-34 miRNAs, miR-34a exhibited the highest induction level during reprogramming; while miR-34b was the lowest. We therefore focused initially on the role of mir-34a in iPSC induction.
Fig. 2. Deficiency of miR-34 miRNAs increases reprogramming efficiency.
a. Four reprogramming factors triggered p53-dependent induction of miR-34 miRNAs. Three days after transduction, pri-mir-34a, mature miR-34a and p21 were measured in uninfected and four-factor induced WT and p53-/- MEFs. Induction of pri-mir-34a was dependent on the intact p53 response, and was comparable to that of p21. Induction of pri-mir-34b/c, mature miR-34b and c was determined in WT, mir-34a-/- and mir-34b/c-/- MEFs. Error bar, standard deviation, n=3. b. mir-34a deficiency significantly enhanced three-factor induced MEF reprogramming. 2500 three-factor infected WT, mir-34a-/- or p53-/- MEFs were plated to score reprogramming by AP-positive colonies with characteristic ESC morphology. A representative image and quantitative analysis is shown out of five independent experiments, comparing littermate-controlled WT and mir-34a-/- MEFs (left, **P < 0.01), as well as WT and p53-/- MEFs (right, **P < 0.01). Error bar, standard deviation, n=4. c. Single-sorted, four-factor infected MEFs were cultured at a density of one cell per well. Four weeks post-plating, AP-positive colonies with typical iPSC morphology were scored for WT, mir-34a-/- and p53-/- iPSCs. Four independent experiments confirmed this finding. *P < 0.05 for comparison between WT and mir-34a-/- MEFs. Error bar, standard error, n= experiments with independent MEF lines. d. mir-34a deficiency significantly enhanced MEF reprogramming as measured by Oct4-Gfp reporter expression. Three- or four-factor infected WT and mir-34a-/- MEFs that carry an Oct4-Gfp allele were sorted at the density of 2500 cells/well and 1000 cells/well, respectively. Reprogramming efficiency was quantified by GFP positive clones. Images of Oct4-Gfp positive iPSCs were shown on the left. A quantitative analysis for reprogramming efficiency triggered by four factor (left, **P<0.01) or three factor (right, *P<0.02) was shown. Scale bar, 100μm. Error bar, standard deviation, n=3. OSKM, Oct4, Sox2, Klf4 and Myc; OSK, Oct4, Sox2 and Klf4. e. miR-34a, b, and c cooperatively regulate somatic reprogramming. Deficiency in mir-34a or mir-34b/c alone significantly promoted somatic reprogramming, yet deficiency in all mir-34 miRNAs exhibited further increase. Two independent experiments confirmed this finding. Error bar, standard error, n = experiments with independent MEFs. All P-values were calculated based on two-tailed Student's t-test.
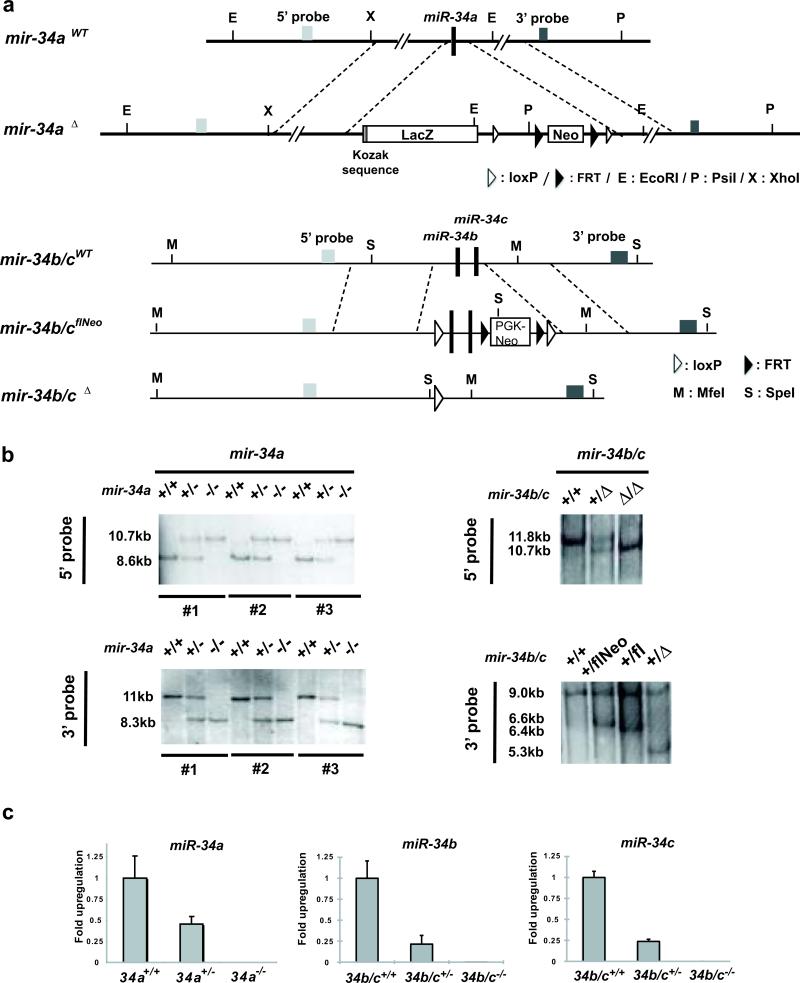
We generated mir-34a knockout mice using C57BL/6 ESCs (Fig. 1a), confirmed the germ line transmission of the targeted allele (Fig. 1b), and verified the genetic ablation of mir-34a by expression studies (Fig. 1c).mir-34a-/- mice were born at the expected Mendelian ratio, without obvious developmental or pathological abnormalities up to 12 months. However, when we induced the mir-34a-/- MEFs for reprogramming, we observed a significant increase in the reprogramming efficiency (Fig. 2b, 2d, S1a). Three-factor-infected mir-34a-/- MEFs exhibited a ~4.5-fold increase in alkaline phosphatase (AP) -positive colonies with typical iPSC morphology (Fig. 2b), while four-factor-infected mir-34a-/- MEFs yielded a ~4-fold increase (Fig. S1a). Furthermore, when we plated infected MEFs into 96-well plates at a density of one cell per well, mir-34a deficiency caused a similar increase in AP-positive colonies with typical iPSC morphology (Fig. 2c).
Fig. 1. Generation of mir-34a and mir-34b/c knockout MEFs.
A. Diagrams of endogenous mir-34a and mir-34b/c gene structure and the knockout construct. Using recombineering, we engineered the mir-34a targeting vector with a ~6kb homologous arm on both 5’ and 3’ ends, flanking a Kozak sequence, a lacZ cDNA and a FRT-neo-FRT cassette. The mir-34b/c targeting vector contains a ~6kb homologous arm at each end, with the mir-34b/c gene and a Neo selection cassette flanked by loxP sites. b. Validating the germline transmission of the mir-34a and mir-34b/c targeted allele using Southern analysis. Putative WT, mir-34a+/- and mir-34a-/- animals derived from three independently targeted ES line were analyzed by Southern blot using probes either 5’ or 3’ to the homologous arms (left). Similar validation was performed for WT, mir-34b/c+/- and mir-34b/c-/- animals (right). c. Confirming loss of mir-34 expression in mir-34a and mir-34b/c knockout MEFs. Littermate-controlled WT, mir-34a+/- and mir-34a-/- MEFs were analyzed by real-time PCR to quantify the expression of mir-34a. While WT MEFs showed robust miR-34a induction upon culture stress, no miR-34a expression was detected in mir-34a-/- MEFs. The miR-34a level in mir-34a+/- MEFs was approximately half that of WT MEFs. Similar validation was performed for mir-34b/c-/- MEFs. Error bar, standard deviation, n=3.
Using MEFs carrying an Oct4-Gfp knockin reporter allele22, we confirmed the effect of miR-34a in promoting reprogramming, scoring fully reprogrammed iPSCs based on endogenous Oct4 expression indicated by GFP. Consistently, a greater than 4-fold increase in reprogramming efficiency was observed in mir-34a-/-; Oct4-gfp/+ MEFs after three- or four-factor transduction (Fig. 2d). A significant increase in reprogramed Oct4-gfp positive colonies could also be achieved using a locked nucleic acid (LNA) inhibitor against miR-34a (Fig. S1b). Notably, mir-34a deficiency both enhanced the overall efficiency of iPSC generation and led to more rapid reprogramming kinetics. Small iPSC-like colonies first appeared 7 days post-infection in four-factor-transduced wild-type (WT) MEFs, but as early as post-infection day 5 in mir-34a-/- MEFs.
Since miR-34b and c share the miR-34a seed sequence and are similarly induced during reprogramming (Fig. 2a), they likely also regulate iPSC generation. We generated mir-34b/c knockout mice (Fig. 1a-1c). As with miR-34a deficiency, mir-34b/c knockout alone promoted somatic reprogramming, although to a lesser degree (Fig. 2e). Interestingly, MEFs deficient for all three mir-34 miRNAs exhibited an even greater increase in iPSC generation (Fig. 2e), suggesting a cooperative effect among these genes, although the exact molecular and cellular mechanisms underlying this cooperation still remained unclear. Since the mir-34a-/-; mir-34b/c-/- MEFs did not completely phenocopy p53-/- MEFs, additional mechanisms may act downstream of p53 to mediate the suppression of reprogramming.
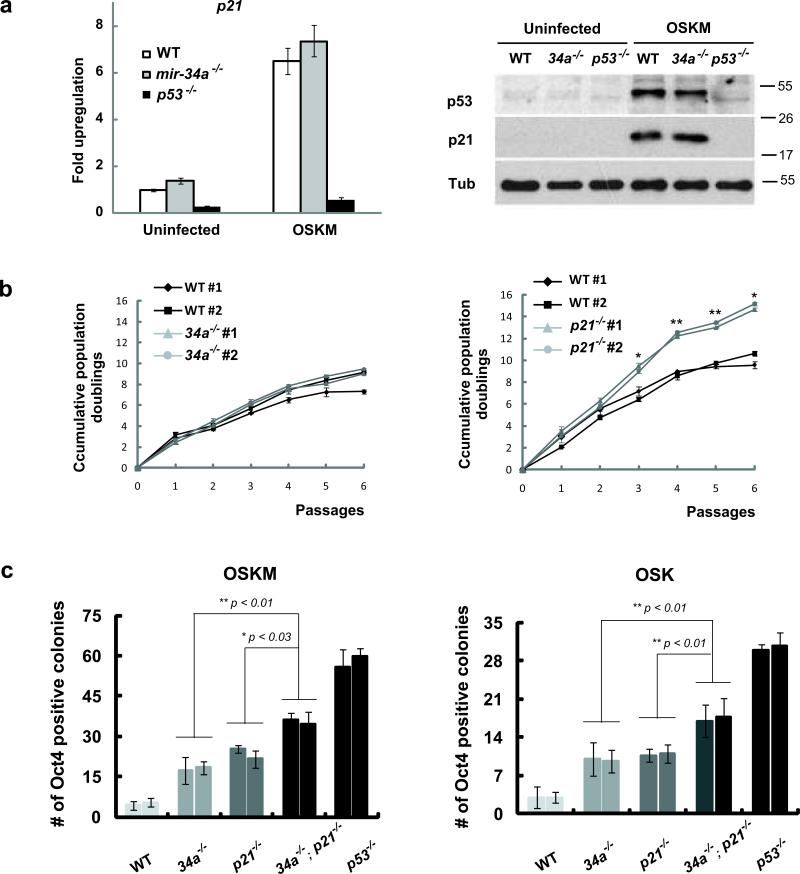
Previous studies have identified p21 as an important mediator of p53 suppression of reprogramming 1, 12. mir-34 and p21 both exhibited p53-dependent induction during reprogramming (Fig. 2a). p21 induction also was observed in mir-34a-/- MEFs (Fig. 3a). While MEFs deficient for mir-34a alone or p21 alone showed comparable increases in iPSC generation, MEFs deficient for both mir-34a and p21 exhibited a cooperative increase that recapitulated a significant fraction of the p53 effect (Fig. 3c). Given the functional similarities among the three mir-34 miRNAs, the cooperative effects among p21 and all mir-34 miRNAs could be even greater. Thus, the mir-34 miRNAs, together with p21, constitute important downstream effectors of p53 to mediate the repression of reprogramming.
Fig. 3. miR-34a and p21 cooperate to repress iPSCs generation.
a. p21 was induced in mir-34a-/- MEFs during somatic reprogramming. Three days after retroviral transduction of four reprogramming factors, both p21 mRNA (left) and p21 protein (right) exhibited a significant increase in WT and mir-34a-/- MEFs. This increase correlated well with the elevated level of p53 proteins (right). α-Tubulin (Tub) was used as a loading control. Error bar, standard deviation, n=3. b. p21-/- MEFs proliferate more rapidly than mir-34a-/- MEFs. Cumulative population doublings were measured for 6 consecutive passages in littermate-controlled WT and mir-34a-/- MEFs (left), and in WT and p21-/- MEFs (right). Compared to the WT counterparts, p21-/- MEFs exhibited an enhanced cell proliferation rate, while mir-34a-/- MEFs showed little differences. Error bar, standard deviation, n=3 for triplicate measurements at each time point. *P < 0.05; **P < 0.01 for comparisons between two lines of MEFs for each genotype. c. miR-34a and p21 cooperate to repress iPSCs generation. The reprogramming efficiency were compared among WT, mir-34a-/-, p21-/-, mir-34a-/-; p21-/- and p53-/- MEFs using either three (right) or four (left) reprogramming factors. Deficiency in mir-34a or p21 alone enhanced reprogramming efficiency to a comparable level. Deficiency in both mir-34a and p21 gave rise to an even greater reprogramming efficiency. Quantitative analyses of Oct4-positive colonies were carried out at 2 (four-factor induced reprogramming) or 3 (three-factor induced reprogramming) weeks post-plating using immunofluorescence analyses. Error bar, standard deviation, n=3. OSKM, Oct4, Sox2, Klf4 and Myc; OSK, Oct4, Sox2 and Klf4; MW, molecular weight. All P-values were calculated based on two-tailed Student's t-test.
p21 represses reprogramming efficiency primarily through its repression on cell proliferation12. Although miR-34a and p21 repress reprogramming efficiency similarly (Fig. 3c), their effects on cell proliferation were quite different. The increased cell proliferation in p21-/- MEFs was highly significant (Fig. 3b), yet within the time frame of our reprogramming experiments, mir-34a-/- MEFs exhibited little increase up to passage 6. We can not completely exclude the possibility that a mild increase in mir-34a-/- MEF proliferation contributed to the increased reprogramming. Yet unlike p21, whose effects on reprogramming are largely attributed to its inhibition of cell proliferation 12, miR-34a is likely to act mostly through a separate mechanism independent of cell proliferation. These two distinct mechanisms may underlie the cooperative regulation of reprogramming by miR-34a and p21.
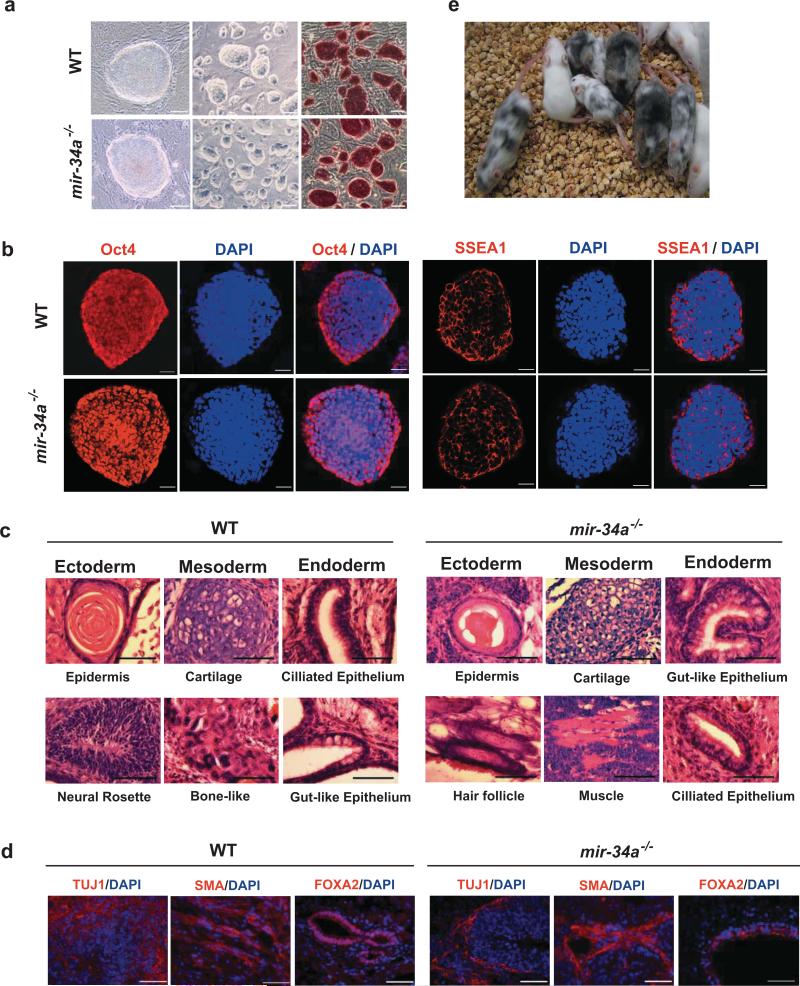
mir-34a-/- iPSCs resemble WT iPSCs and ESCs, exhibiting ESC-like morphology (Fig. 4a, S1c) and expressing key molecular markers for pluripotency. A high level of Oct4, Nanog, and SSEA1 were detected (Fig. 4b, S1d). mir-34a-/- iPSCs injected into immunocompromised nude mice yielded differentiated teratomas, containing terminally differentiated cell types from all three germ layers (Fig. 4c, 4d, S1e). Furthermore, three independent lines of four-factor-induced mir-34a-/- iPSCs all yielded healthy adult chimeric mice with a high percentage of iPSC contribution (Fig. 4e, S1g, Table S1). Taken together, mir-34a deficiency enhances the efficiency of iPSC generation without compromising self-renewal and pluripotency.
Fig. 4. mir-34a-/- iPSCs functionally resemble WT iPSCs.
a. iPSCs derived from both WT and mir-34a-/- MEFs exhibited ES-like morphology in culture, with robust AP expression. Scale bar, 20μm for the left panel, 100 μm for the middle and right panels. b. Both WT and mir-34a-/- iPSCs expressed pluripotency markers, including nucleus-localized Oct4 and membrane-localized SSEA1. Scale bar, 20μm; c, d. Wildtype and mir-34a-/- iPSCs both generated differentiated teratomas. Teratomas derived from four-factor induced WT (left) and mir-34a-/- (right) iPSCs were harvested from nude mice 4-6 weeks after subcutaneous injection. H&E staining(c), as well as immunofluorescence staining (d), revealed terminally differentiated cell types derived from all three germ layers. Scale bar in c, 25μm; in d, 50μm. e. Four-factor-induced mir-34a-/- iPSCs efficiently contribute to adult chimeric mice. We injected three independent lines of passage seven Oct4-Gfp/+, mir-34a-/- iPSCs into albino-C57BL/6/cBrd/cBrd/cr blastocysts. The iPSC contribution to adult chimeric mice was determined by coat color pigmentation.
Although p53 deficiency induced reprogramming more efficiently than mir-34a deficiency, the p53-/- iPSCs exhibited compromised self-renewal and differentiation capacity 1, 4. As also reported by the Yamanaka group 1, we have observed that four-factor-induced p53-/- iPSCs lost ESC-like morphology after 5-6 passages in culture, and failed to generate highly differentiated teratomas. In contrast, four-factor-induced mir-34a-/- iPSCs remained stable beyond passage 26, with no significant differences in self-renewal compared to WT iPSCs (Fig. S1f). Additionally, generation of healthy adult chimeras from p53-/- iPSCs was difficult. The percentage of p53-/- iPSC contribution was low, and the majority of such chimeras succumbed to tumorigenesis before 7 weeks 1. In contrast, mir-34a-/- iPSCs exhibited functional pluripotency: all three four-factor-induced mir-34a-/- iPSC lines tested gave rise to healthy adult chimeras with a high percentage of iPSC contribution (Fig. 4e, S1g, Table S1), which remained tumor-free for 6 months (to the time of manuscript preparation).
Consistent with these differences, we observed that four-factor-induced p53-/- iPSCs, but not mir-34a-/- or WT iPSCs, failed to silence retroviral transgenes, thus exhibiting lower levels of the corresponding endogenous genes (Fig. S3a-S3c). The exogenous expression of reprogramming factors, particularly c-Myc (Fig. S3e), may contribute to the impaired self-renewal and differentiation (Fig. S3d), and promote tumorigenesis 1. In contrast, pluripotency was established easily in mir-34a-/- iPSCs by the endogenous transcription factor circuitry, leaving the exogenous transgenes dispensable for the maintenance of pluripotency.
Similar to mir-34a-/- iPSCs, mir-34a-/-; mir-34b/c-/- iPSCs also exhibited robust self-renewal and differentiation capacity, with typical ESC morphology, key pluripotency markers, and the ability to generate differentiated teratomas (Fig. S2a-S2c). Yet their ability to generate chimera remains to be determined.
The increased reprogramming efficiency and compromised pluripotency of p53-/- MEFs may result from aberrant apoptosis and DNA damage response 4. However, mir-34a deficiency failed to protect cells from apoptosis or DNA damage response during reprogramming (Fig. S4a-S4c). Thus, the enhanced reprogramming efficiency observed in mir-34a-/- MEFs may reflect a mechanism largely independent of apoptosis and DNA damage response. These findings contrast to the ability of mir-34a overexpression to trigger apoptosis in specific tumor cells 17, 18. These mir-34a effects on apoptosis are likely cell-type-dependent, differing between primary fibroblasts 16 and specific cancer cell lines 17, 18. Cells deficient for mir-34a may exhibit apoptotic defects under conditions other than reprogramming. It is also possible that the lack of apoptotic protection in reprogramming mir-34a-/- MEFs reflects the functional redundancy of miR-34b and c.
Enhanced reprogramming in p53-deficient cells has also been attributed to cell immortalization and increased cell proliferation 1-5, 12, 15. While cell immortalization greatly promotes iPSC generation from p53-/- MEFs 5, increased cell proliferation is a key mechanism driving stochastic reprogramming of p53KD B cells 12. Although mir-34 overexpression induced growth arrest and cellular senescence in primary fibroblasts 16, no defects in senescence response were observed in mir-34a-/- MEFs during reprogramming (Fig. S4d and data not shown). As described above, we did not observe significant MEF proliferation difference caused by miR-34a deletion within the time frame of our reprogramming experiment. Presumably, miR-34a regulates reprogramming efficiency largely through a mechanism independent of cell proliferation and cell senescence.
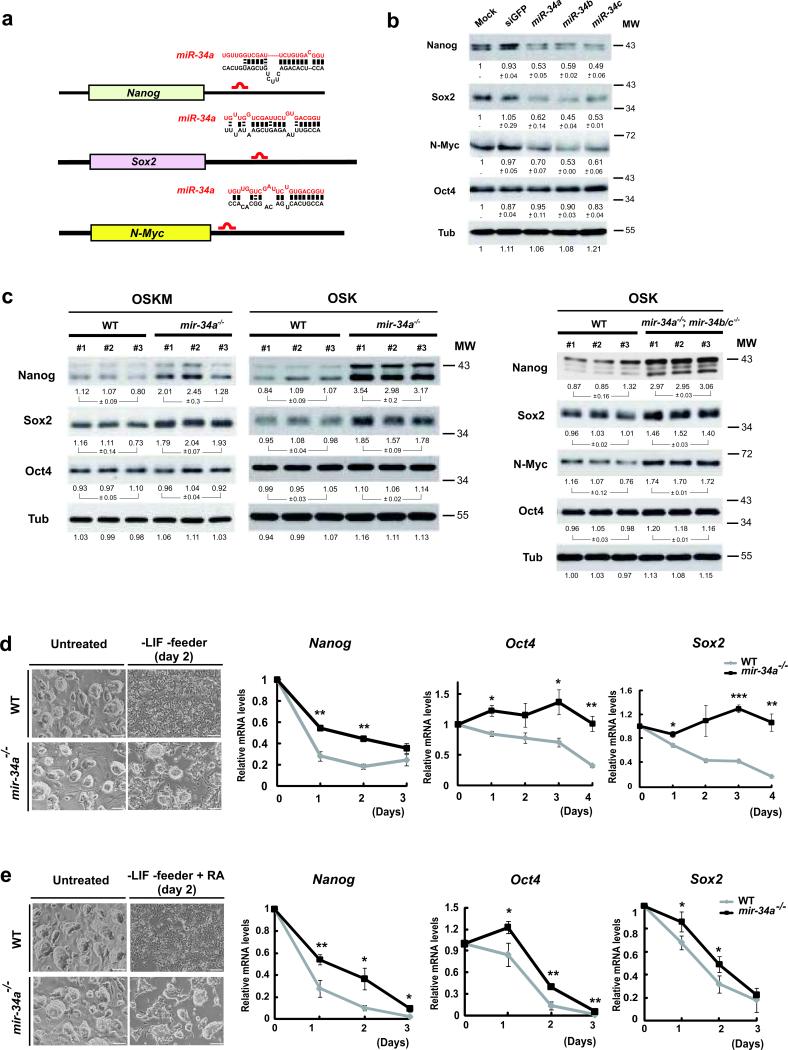
To better define the molecular mechanism of mir-34a in reprogramming, we initiated a search for its targets, specifically by examining genes that promotes the iPSC generation for possible miR-34a binding sites. RNA22 identified a number of such genes with predicted mir-34a sites 23 (Fig. S5d). Of all the genes tested, Nanog, Sox2, and Mycn (N-Myc) emerged as top candidates (Fig. 5a, S5a). All three exhibited mir-34a-dependent repression (Fig. 5b, S5c), and each had potent effects in promoting reprogramming 24, 25. ESCs over-expressing miR-34a, miR-34b, or miR-34c for 48 hours had decreased Nanog, Sox2 and N-Myc protein levels (Fig. 5b), but unaltered mRNA abundance (Fig. S5b). This reduction in Nanog, Sox2, and N-Myc protein levels was not due to ESC differentiation, because the level of Oct4, another pluripotency marker, remained unaltered at 48 hours post-transfection (Fig. 5b). Consistently, levels of Sox2 and Nanog proteins were elevated in mir-34a-/- iPSCs and mir-34a-/-; mir-34b/c-/- iPSCs compared to littermate- and passage-controlled WT iPSCs (Fig. 5c). We also observed increased N-Myc levels in mir-34a-/-; mir-34b/c-/- iPSCs, although this increase was moderate in mir-34a-/- iPSCs (Fig. 5c and data not shown). Real-time PCR analysis suggested effective silencing of the Sox2 transgene in mir-34a-/- and mir-34a-/-; mir-34b/c-/- iPSCs (Fig. S3a and data not shown); thus the increased Sox2 level was purely due to alterations in endogenous Sox2. Since Oct4 protein levels were largely unchanged among WT, mir-34a-/-, and mir-34a-/-; mir-34b/c-/- iPSCs, our data suggest that the increase in Sox2, Nanog, and N-Myc was specific for mir-34 deficiency, and not due to differences in iPSC pluripotency (Fig. 5c). Thus, when mir-34a-deficient cells undergo reprogramming, the post-transcriptional derepression of multiple pluripotency genes is likely to promote and accelerate the establishment of the endogenous regulatory circuitry for pluripotency, thereby enhancing reprogramming efficiency.
Fig. 5. mir-34a represses Nanog, Sox2 and N-Myc expression post-transcriptionally.
a. Schematic representation of the Nanog, Sox2 and N-Myc 3’UTR, and the predicted mir-34 binding sites. The mouse Nanog, Sox2 and N-Myc 3’UTR each contains one putative miR-34a binding site within their 3’UTRs. b. Enforced expression of mir-34a, b, and c in ESCs reduced the protein levels of Nanog, Sox2 and N-Myc, but not Oct4. Feeder-free ESCs were transfected with miRNA mimics for miR-34a, miR-34b and miR-34c, and a negative control, siGFP. At 48 hours post transfection, Western analysis indicated a significant reduction in the protein levels of Nanog, Sox2 and N-Myc, but not Oct4. The value of each band indicates the relative expression level normalized by the internal control, α-tubulin, averaged among two independent experiments, and presented as mean ± s.e.m. c. Derepression of Nanog, Sox2 and N-Myc was observed in mir-34 deficient iPSCs. A significant increase of Nanog and Sox2, but not Oct4, was observed in four factor induced mir-34a-/- iPSCs, when compared to passage matched, littermate controlled WT iPSCs. A similar comparison was performed for passage matched, three-factor induced WT and mir-34a -/-; mir-34b/c-/- double knockout iPSCs, where derepression of Nanog, Sox2 and N-Myc was observed. For this Western analysis, the quantitation of each band was performed by Quantity One software, and was normalized against its own internal tubulin control. The standard errors of three independent iPSC lines were shown for each genotype, n=3. d, e. mir-34a deficient iPSCs exhibited slower kinetics during differentiation. Wildtype and mir-34a-/- iPSCs were both triggered to differentiate by withdrawal of LIF in the presence (e) or absence (d) of RA treatment. The image of typical iPSC culture two days after each differentiation condition were shown on the top (d, e), and the quantitative analyses on the decline of Nanog, Sox2 and Oct4 transcripts in response to these differentiating conditions were shown on the bottom (d, e). Error bar, standard error, n=3. * P<0.05, ** P<0.01. Scale bar in d and e, 100 μm. MW, molecular weight.
N-Myc is a previously identified miR-34a target in neuroblastoma cell lines, whose downregulation is mediated through a miR-34a binding site within the 3’ untranslated region (UTR) 26, 27. To determine whether miR-34a directly targets Sox2 and Nanog, we constructed luciferase reporters that contained the WT 3’UTRs of these genes. Both Sox2 and Nanog 3’UTRs were repressed by exogenous expression of miR-34a in Dicer-deficient HCT116 cells and in WT ESCs (Fig. S5c and data not shown); mutation of the miR-34a binding sites of these reporters significantly compromised miR-34a-dependent regulation (Fig. S5c). These results indicate that miR-34a directly targets Nanog, Sox2, and N-Myc, thereby impeding iPSC generation. Interestingly, Nanog was previously identified as a direct target of p53-mediated transcriptional repression in multiple stem cell systems 28, 29. Thus, p53 mediates Nanog repression both by direct transcriptional silencing and by indirect post-transcriptional silencing through mir-34. Consistent with the role of miR-34a in negatively regulating multiple pluripotency genes, mir-34a-/- iPSCs exhibited delayed kinetics when triggered to differentiate by leukemia inhibitory factor (LIF) withdrawal, both in the presence or absence of retinoic acid (RA) (–LIF and –LIF+RA). Like ESCs, WT iPSCs differentiated quickly when subjected to the –LIF or –LIF+RA conditions, as evidenced by flattened cell morphology and rapid decline of Nanog, Oct4, and Sox2 expression (Fig. 5d, 5e). In contrast, mir-34a-/- iPSCs exhibited significant kinetic delay during differentiation, evidenced by the slower alterations in morphology and pluripotency gene expression (Fig. 5d, 5e). In mir-34a-/- iPSCs, the derepression of multiple pluripotency transcription factors may reinforce the regulatory circuitry to maintain self-renewal, causing less efficient silencing of the self-renewal program during differentiation. Conversely, the derepression of pluripotency genes in mir-34a-/- MEFs during reprogramming could promote and accelerate the establishment of the endogenous regulatory circuitry for pluripotency. Our results differ from a recent study where reduced miR-34a function did not impact ESC differentiation 30. This difference may reflect the limited efficacy of a nucleotide-based miR-34a inhibitor.
Current studies on gene regulation for pluripotency have focused primarily on transcription factor profiles. This emphasis may provide an incomplete picture, since post-transcriptional gene regulation by miRNAs could add robust and redundant controls to this process. The small size of miRNAs, combined with their imperfect target recognition, give them enormous capacity and versatility to regulate global gene expression 31. Along with transcription factors, miRNAs have emerged as essential gene regulators in self-renewal and differentiation of pluripotent stem cells 32, 33. ESCs with deficient global miRNA biogenesis exhibit proliferation and differentiation defects 34. In addition, RNA binding protein LIN28, which induces reprogramming by suppressing let-7 biogenesis 8, 35-37, has been identified as a major reprogramming factor in humans. Beside let-7 miRNAs, a number of miRNAs are specifically enriched or depleted in pluripotent stem cells and are demonstrated to regulate self-renewal and differentiation 30, 32, 34, 38-40. Here, we identify the miR-34 miRNAs as novel regulators that suppress reprogramming downstream of p53, providing direct evidence that modulation of miRNA abundance could constitute a critical step in establishing pluripotency. Since miRNA functions can be manipulated by oligonucleotide-based inhibitors or mimics, our findings suggest a paradigm for promoting reprogramming by manipulating specific miRNA functions.
Although p53 directly regulates hundreds of target genes 14, mir-34 miRNAs and p21 are the major downstream targets that cooperatively repress iPSC generation. p53 effects on reprogramming reflect changes in cell proliferation, immortalization, apoptosis, and DNA damage response 4, 5, 12. p21, a key downstream target of p53, represses cell proliferation. Interestingly, miR-34a deficiency alters MEF reprogramming not via cell proliferation but, at least in part, by posttranscriptional derepression of pluripotency genes. Thus, mir-34 miRNAs, together with other p53 targets, including p21, collectively mediate the suppression of somatic reprogramming via multiple critical cellular pathways.
Supplementary Material
Acknowledgements
We thank B. Zaghi, A Basila, A Perez, G Lai, M Foth, A Kemp, A Fresnoza, A Valeros, A Fritz, D Schichnes and H Noller for technical assistance and A Economides for stimulating discussions and helpful input. We also thank JM Halbleib, D Schaffer, and RM Harland for carefully reading the manuscripts, and I Lemischka, Q Li, M Hemann and B Vogelstein for their generosity to share reagents. In particular, we are grateful for the pathological expertise R Van Andel offered us. L.H. is a Searle Scholar, and is supported by a new faculty award by CIRM (RN2-00923-1), and a R01 (R01 CA139067) and a R00 grant (R00 CA126186) from the NCI. G. J. Hannon is a HHMI investigator, and is supported by a program project grant from the NCI. C.L. is supported by a Siebel postdoctoral fellowship.
Footnotes
Author Contributions
Y.C., C.L. and L.H. designed all experiments, and performed majority of the experiments shown in all figures and supplementary figures. J.H. and Y.Z. performed immunofluorescence analyses and teratoma analyses to characterize the pluripotency of the iPSCs. L.H., X.H. P.B. and G.J.H generated knockout constructs for mir-34a and mir-34b/c, and identified the correctly targeted ESC clones for mir-34a. N.O. and P.B. identified correctly targeted ESC clone for mir-34b/c, validated mir-34a and mir-34b/c targeting in mice by southern, and generated mir-34a-/-, mir-34b/c-/- and mir-34 triple knockout MEFs. S.K. and G.J.H. performed the blastocyst injection for mir-34a+/- and mir-34b/c+/- ESC clones. A.Z, S.K. and G.G.H. characterized the pluripotency of iPSCs using chimera assays.
Competing financial interests
The authors declare no competing financial interests.
Supplementary Information accompanies the paper
References
- 1.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marion RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamanaka S, Takahashi K. [Induction of pluripotent stem cells from mouse fibroblast cultures]. Tanpakushitsu Kakusan Koso. 2006;51:2346–2351. [PubMed] [Google Scholar]
- 7.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 9.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 10.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 15.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 18.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2009;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 22.Lengner CJ, et al. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci U S A. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei JS, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole KA, et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuijk EW, et al. PTEN and TRP53 independently suppress Nanog expression in spermatogonial stem cells. Stem Cells Dev. 2009;19:979–988. doi: 10.1089/scd.2009.0276. [DOI] [PubMed] [Google Scholar]
- 29.Lin T, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 30.Tarantino C, et al. miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. FASEB J. 2010;24:3255–3263. doi: 10.1096/fj.09-152207. [DOI] [PubMed] [Google Scholar]
- 31.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 32.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anokye-Danso F, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyoshi N, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.