Abstract
Ergosterol, the major sterol of fungal membranes, is essential for developmental growth and the main target of antifungals that are currently used to treat fatal fungal infections. Emergence of resistance to existing antifungals is a current problem and several secondary resistance mechanisms have been described in Aspergillus fumigatus clinical isolates. A full understanding of ergosterol biosynthetic control therefore appears to be essential for improvement of antifungal efficacy and to prevent antifungal resistance. An ergosterol biosynthesis pathway in A. fumigatus has been proposed with 14 sterol intermediates resulting in ergosterol and another secondary final compound C-24 ethyl sterol. Transcriptomic analysis of the A. fumigatus response to host-imposed stresses or antifungal agents is expanding our understanding of both sterol biosynthesis and the modes of action of antifungal drugs. Ultimately, the identification of new targets for novel drug design, or the study of combinatorial effects of targeting sterol biosynthesis together with other metabolic pathways, is warranted.
Keywords: Aspergillus, ergosterol biosynthesis, antifungal drugs, resistance mechanisms, transcriptome
ANTIFUNGALS TARGETING ERGOSTEROL AND STEROL BIOSYNTHESIS
Sterols are neutral lipids of eukaryotic cells, among which ergosterol is the main component of fungal membranes. Ergosterol is involved in numerous biological functions such as membrane fluidity, regulation, activity and distribution of integral membrane proteins, and control of the cell cycle (Bard et al., 1993; Gooday, 1995). The essential role of sterols in maintenance of cell membranes make ergosterol and its biosynthetic pathway essential for fungal growth, and a primary target for most, currently available, antifungal drugs to treat severe human fungal infections. Three main classes of antifungal drugs, namely polyenes, allylamines, and azoles, directly target ergosterol itself, or enzymatic steps of its biosynthetic pathway. The polyene amphotericin B (AMB) has represented, for more than 30 years, the standard antifungal therapy for invasive aspergillosis (IA). The AMB mode of action is still poorly understood. It is widely accepted that AMB kills yeast primarily via channel-mediated membrane permeabilization leading to fungal cell death (Ermishkin et al., 1976). However, recent research suggests that AMB kills yeast by simply binding ergosterol so that the channel formation represents a second complementary mechanism that further increases drug potency and the rate of cell death (Gray et al., 2012). The allylamine group of antimycotics interferes at the early-stage of ergosterol biosynthesis by inhibiting the enzyme squalene epoxidase (Erg1). Although squalene epoxidases from various origins have been investigated with respect to substrate requirements, cofactors, and inhibitors, no structural model is available; and the domains responsible for enzymatic activity and inhibitor interactions are not well understood (Ruckenstuhl et al., 2007). The cidal action of allylamines is closely associated with the development of high intracellular squalene concentrations, which are believed to interfere with fungal membrane function and cell wall synthesis (Ryder, 1992). Allylamines are not used to treat IA, however potential use in combination with azoles, polyenes, or echinocandins in the management of severe drug-resistant or refractory mycoses has been proposed (Krishnan-Natesan, 2009). Finally, the main category of antifungal agents used against Aspergillus is azole drugs, such as voriconazole (VCZ) and itraconazole, and a new generation of azoles that includes posaconazole, isavuconazole, and albaconazole. Azoles revolutionized medical mycology due to their broad spectrum and reduced toxicity compared to AMB. Azoles block the ergosterol biosynthesis pathway via inhibition of 14-α sterol demethylase (Cyp51/Erg11), a key enzyme that removes the methyl group at position C-14 of precursor sterols. Inhibition of ergosterol synthesis at this biochemical level results in toxic sterol accumulation and cell death (Ghannoum and Rice, 1999).
However, although the use of antifungal drugs has increased survival of patients with invasive fungal disease, the mortality rate associated with these infections remains extremely high. This situation becomes more complicated because emerging resistance to the existing antifungals is a current problem and secondary resistance mechanisms have been described in clinical isolates (Howard and Arendrup, 2011; Shapiro et al., 2011; van der Linden et al., 2011a). Full understanding of the ergosterol biosynthesis pathway in A. fumigatus is essential to new antifungal drug design and for improving the activity of existing ones. This review summarizes current knowledge about the enzymatic sequence and regulation of the biochemical reactions involved in ergosterol biosynthesis with a view to understanding the main mechanisms of antifungal resistance to drugs which target ergosterol or its biosynthesis.
Aspergillus fumigatus ERGOSTEROL BIOSYNTHESIS PATHWAY
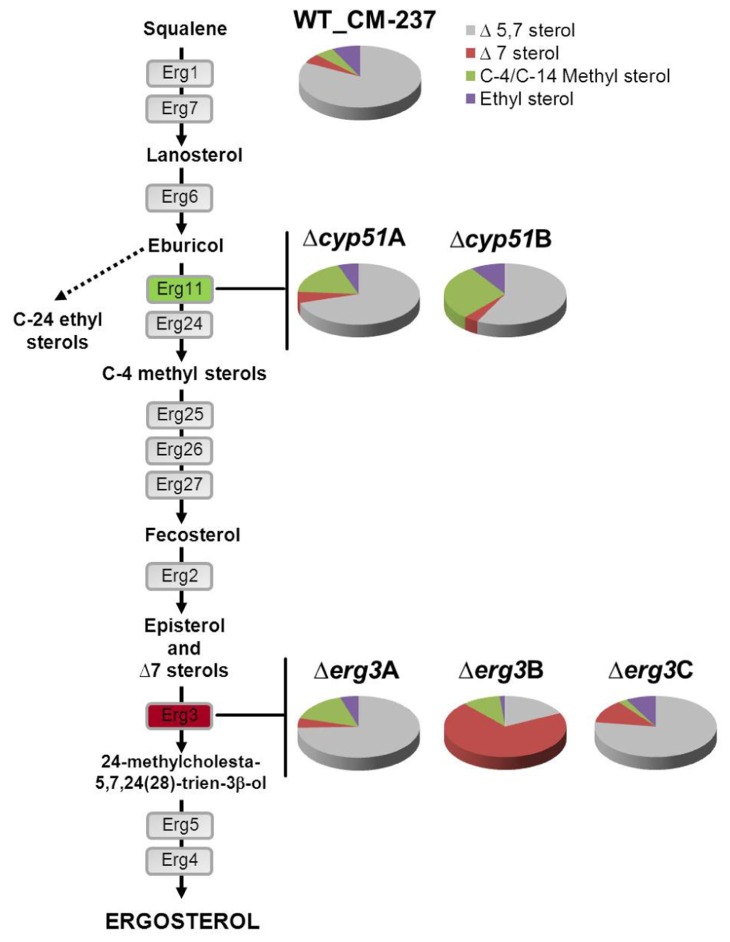
The biosynthesis of ergosterol involves about 20 enzymes and includes the synthesis of squalene from mevalonate (Ferreira et al., 2005; Alcazar-Fuoli et al., 2008). This route is well known in Saccharomyces cerevisiae which provides a model for studying this pathway in other eukaryotes (Fryberg et al., 1973). Depletion of ergosterol biosynthetic gene functions is lethal in S. cerevisiae, due to complete prevention of ergosterol production. Deviating from the prototypical pathway described in S. cerevisiae, other more complex biosynthetic pathways have been proposed in some fungi and also in plants, suggesting that the enzymatic pathways of ergosterol biosynthesis are specific to fungal taxa (Fryberg et al., 1973; Nes et al., 1989). In A. fumigatus, the study of this pathway at a genomic level, demonstrated the existence of multiple genes encoding key enzymes such as two distinct 14-α sterol demethylases (Cyp51A and Cyp51B; Mellado et al., 2001) and three C-5 sterol desaturases (Erg3; Alcazar-Fuoli et al., 2006). The classical ergosterol biosynthesis route was reconsidered in A. fumigatus based on the existence of multiple genes which putatively encode enzymes with the same or duplicated function in the pathway (Figure 1). Genetic and biochemical analysis of azole susceptible wild-type strains and a panel of clinical azole resistant strains identified a total of 14 sterols showing a similar sterol pattern composition, with ergosterol as the main sterol for all of them (Table 1). Each of the azole resistant strains had a different antifungal susceptibility pattern that correlated with different mutations in the cyp51A gene. The lack of differences between sterols highlights the fact that the resistance mechanism in those strains was target-dependent and not sterol-dependent, as will be discussed later. Instead, the sterol composition of single enzyme defective A. fumigatus mutants was rather different depending on the lacking enzyme (Alcazar-Fuoli et al., 2008). Mutants deleted in 14-α sterol demethylases (cyp51A and cyp51B) had less ergosterol than the parental strain and accumulated C-4, and C-14 methyl sterols; mainly eburicol (Figure 1). Accumulation of eburicol, together with a decrease in ergosterol, was much more prominent when the cyp51B gene was deleted compared to Δcyp51A strain. Downstream in the pathway, the synthesis of ergosterol involves the transformation of fecosterol, consisting of double-bond rearrangements in the steroid nucleus and in the side chain, isomerization of the double connection in the C-8 to the C-7 followed by the desaturation at C-5 and C-22, and the reduction of the C-24. The enzymatic sequence of these three last steps could differ between fungal taxa and growth conditions (Nes et al., 1989). Single gene deletion of each of the C-5 sterol desaturases (Erg3A, Erg3B, and Erg3C) revealed a different sterol profile in terms of total amount of ergosterol and sterol composition. Again this phenotype was different depending on the deleted enzyme. While the absence of the genes erg3A and erg3C did not significantly affect total ergosterol biosynthesis, the deletion of erg3B caused a dramatic ergosterol decrease (70%) and a vast accumulation of the Δ7 sterols (Figure 1). Although the sterol composition revealed differences according to the enzyme that was deleted, none of the single gene deletions, at 14-α sterol demethylase or C-5 sterol desaturase level, was lethal for A. fumigatus or influenced its virulence (Mellado et al., 2005; Alcazar-Fuoli et al., 2006). This suggests that although A. fumigatus might use one of the enzymes Cyp51 or Erg3 for normal growth, this fungal pathogen is able, including during infection, to adapt and compensate that lack of an enzyme function.
FIGURE 1.
Ergosterol biosynthetic pathway. Sterols of A. fumigatus wild-type strain (CM-237), and Δcyp51A, Δcyp51B (marked in green), Δerg3A, Δerg3B, and Δerg3C (marked in red) derived mutant strains. Sterols are clustered and represented in cake plots as the percentage of total sterols: Δ5 sterols (1, 2, 3, 4, 6, 8 in gray), C-4/C-14 methyl sterols (11, 12, 13, 14 in green), Δ7 sterols (5, 7, 9 in red), and ethyl sterols (10 in purple) according to the sterol identification in A. fumigatus (Table 1).
Table 1.
Relative amount of sterols (% of total sterols) in different A. fumigatus wild-type (CM-237), cyp51 deleted strains (Δcyp51A Δcyp51B), and azole resistance mutants strains with different cyp51A point mutations (CM-796:G54V; CM-3269:TRL98H; CM-2159: M220K).
| Sterol names | Strains | |||||
|---|---|---|---|---|---|---|
| CM-237* | Δcyp51A | Δcyp51B | CM-796* | CM-3269* | CM-2159* | |
| 1) 24-methylcholesta-5,7,9(11),22-tetraen-3β-ol | 1.09 | 3.69 | 2.74 | 0.85 | 1.18 | 0.92 |
| 2) 24-methylcholesta-5,8,22-trien-3β-ol | 0.91 | 0.87 | 0.77 | 0.90 | 0.80 | 0.85 |
| 3) 24-methylcholesta-5,7,9,22-tetraen-3β-ol | 0.71 | 5.69 | 4.57 | 0.00 | 0.00 | 0.00 |
| 4) 24-methylcholesta-5,7,22-trien-3β-ol (Ergosterol) | 78.12 | 59.48 | 48.82 | 84.08 | 83.70 | 88.44 |
| 5) 24-methylcholesta-7,22 (28)-dien-3β-ol | 2.62 | 1.84 | 1.18 | 1.51 | 1.67 | 1.09 |
| 6) 24-methylcholesta-5,7,22,24(28)-tetraen-3β-ol | 0.48 | 0.41 | 0.25 | 0.32 | 0.21 | 0.28 |
| 7) 24-methylcholesta-7,22,24(28)-trien-3β-ol | 0.66 | 1.44 | 0.71 | 0.82 | 0.51 | 0.20 |
| 8) 24-methylcholesta-5,7,24(28)-trien-3β-ol | 0.91 | 0.14 | 0.34 | 0.08 | 0.19 | 0.16 |
| 9) 24-methylcholesta-7,24(28)-dien-3β-ol (Episterol) | 1.51 | 2.55 | 2.01 | 3.59 | 1.84 | 1.08 |
| 10) 24-Ethylcholesta-5,7,22-trien-3β-ol | 7.48 | 5.48 | 9.26 | 2.43 | 6.46 | 5.10 |
| 11) 4,4,14-trimethylcholesta-8,24-dien-3β-ol (Lanosterol) | 1.06 | 1.39 | 2.87 | 3.63 | 0.29 | 0.18 |
| 12) 4α,24-dimethylcholesta-8,24(28)-dien-3β-ol | 0.99 | 2.03 | 0.98 | 0.73 | 0.00 | 0.82 |
| 13) 4,4,14,24-tetramethylcholesta-8,24(28)-dien-3β-ol (Eburicol) | 1.14 | 9.54 | 21.95 | 0.41 | 1.30 | 0.23 |
| 14) 4,4,24-trimethylcholesta-8,24(28)-dien-3β-ol | 2.04 | 5.44 | 3.56 | 0.65 | 0.62 | 0.65 |
Geometric mean of MICs to antifungal agents of A. fumigatus strains: CM-237: ITC (0.25 μg/ml), VRC (0.48 μg/ml), POS (0.06 μg/ml), FLC (1094.55 μg/ml), AMB (0.26 μg/ml), TRB (5.4 μg/ml); CM-796: ITC (16 μg/ml), VRC (1.19 μg/ml), POS (1.68 μg/ml), FLC (1094.55 μg/ml), AMB (0.15 μg/ml), TRB (2.38 μg/ml; CM-2159: ITC (16 μg/ml), VRC (1.19 μg/ml), POS (2 μg/ml), FLC (1094.55 μg/ml), AMB (0.3 μg/ml), TRB (3.17 μg/ml); CM-3269: ITC (16 μg/ml), VRC (4 μg/ml), POS (0.5 μg/ml), FLC (1094.55 μg/ml), AMB (0.2 μg/ml), TRB (2 μg/ml).
ANTIFUNGAL RESISTANCE MECHANISMS RELATED TO ERGOSTEROL OR ITS BIOSYNTHETIC PATHWAY
Three of the most widely used antifungal drugs, triazoles, polyenes, and allylamines, are aimed at ergosterol, and they are either fungicidal but toxic to the host (polyenes) or fungi static and more vulnerable to resistance (triazoles). Triazoles are inhibitors that target the ergosterol biosynthetic pathway by binding to the Cyp51 family of cytochrome P450s (14-α sterol demethylases) causing the depletion of ergosterol biosynthesis and the accumulation of lanosterol or eburicol (Kelly et al., 1995). A. fumigatus contains two different, but related Cyp51 proteins, encoded by cyp51A and cyp51B (Mellado et al., 2001) that are functional 14-α sterol demethylases (Martel et al., 2010; Warrilow et al., 2010). The lethality caused by double gene deletion has been shown, although neither gene is itself essential (Mellado et al., 2005; Hu et al., 2007). Moreover, the importance of A. fumigatus Cyp51 proteins in sterol biosynthesis and their significance in azole susceptibility have been extensively studied (Alcazar-Fuoli et al., 2008, 2011; Martel et al., 2010). Also, the in vitro and in vivo correlation of azole resistance has been widely documented; with a clear association of resistant A. fumigatus strain isolation and lack of patient response to therapy (van der Linden et al., 2011a). A. fumigatus azole resistance can arise either from modification of the Cyp51A target, or by its overexpression. Resistance acquired through exposure to azoles in the patientis correlated with cyp51A single mutations (Diaz-Guerra et al., 2003; Mann et al., 2003; Nascimento et al., 2003; Mellado et al., 2004; Howard et al., 2006; Albarrag et al., 2011), while resistance caused by cyp51A overexpression is specifically linked to the combination of a single amino acid substitution (a leucine to histidine change, L98H), together with the presence of two tandem copies of a 34-bp sequence in the promoter of the cyp51A gene. This latter type of resistance mechanism may have evolved in the environment through the exposure of the fungus to azole fungicides used in agriculture (Snelders et al., 2008). In isolation the tandem 34-bp promoter insertions cannot explain the observed azole cross resistant phenotype (Mellado et al., 2007), however recent work has demonstrated the essentiality of this 34-bp promoter sequence in maintaining the wild-type expression of cyp51A, and suggested the presence of other regulatory elements upstream of the tandem insertions (Paul et al., 2012). Further mechanisms of azole resistance include overexpression of drug transporters of the ABC- and/or MFS-type (Tobin et al., 1997; Slaven et al., 2002; Nascimento et al., 2003), however clinically observed resistance appears thus far to be limited to mutations which modify the Cyp51A target site. The sterol content of azole resistant A. fumigatus strains bearing different Cyp51A point mutations suggests that their azole resistant phenotypes are not the result of perturbations of the ergosterol biosynthetic pathway (Table 1). It is more likely that azole resistance in these variants results from a reduced affinity of drug binding to the Cyp51A enzyme. In this sense, 3D protein models of Cyp51A in combination with azoles have provided the basis to address how point mutations can affect azole drug resistance (Alcazar-Fuoli et al., 2011; Fraczek et al., 2011).
Regarding the polyene antifungals, the association between A. fumigatus susceptibility to AMB in vitro and clinical outcome is unclear. Secondary resistance to AMB is generally not observed, even in patients whose therapy has failed (Moosa et al., 2002), yet resistant mutants have been spontaneously induced in the laboratory (Manavathu et al., 1998) and resistance to AMB is well documented for many other Aspergillus species (Blum et al., 2008; van der Linden et al., 2011b; Hadrich et al., 2012). Acquired resistance to AMB has been most extensively evaluated in yeasts and is associated with mutations in the ERG3 gene, which is linked to qualitative and quantitative alterations of membrane lipids and an absence of ergosterol (Kelly et al., 1997; Morio et al., 2012). However, the deletion of three independent erg3-like genes in A. fumigatus did not affect susceptibility to AMB, despite marked alterations of sterol composition and a decrease in total ergosterol (Alcazar-Fuoli et al., 2006). Other studies have determined that neither ergosterol content, cell wall composition, or lipid peroxidation levels correlate with heightened A. terreus AMB resistance, only that a higher level of catalase production in A. terreus might contribute to AMB resistance since it promotes oxidative damage of fungal cell membranes through generation of reactive oxygen species (Blum et al., 2008).
Terbinafine belongs to the allylamine class of antifungals that inhibit squalene epoxidase (Erg1). Erg1 catalyzes the first oxygenation step in sterol biosynthesis and is suggested to be one of the rate-limiting enzymes in this pathway. Alterations in A. fumigatus erg1 gene dosage can promote terbinafine resistance (Liu et al., 2004), however, the first terbinafine resistance mechanism was described in A. nidulans and associated with the activity of salicylate 1-monooxygenase (salA), a well-characterized naphthalene-degrading enzyme, suggesting that resistance could follow degradation of the naphthalene ring contained in terbinafine (Graminha et al., 2004). Also, a point mutation, F391L, in the squalene epoxidase enzyme was found to confer a strong terbinafine resistance phenotype. The equivalent mutation was introduced into the homologous gene of A. fumigatus resulting in terbinafine resistance (Rocha et al., 2006). Since terbinafine is not currently used in the management of IA, the appearance of terbinafine-resistant strains in the clinical setting is unlikely, however, its potential use in combination with other antifungal drugs should be considered.
ALTERNATIVES TO IMPROVE ANTIFUNGAL EFFICACY AND TO MINIMIZE DRUG RESISTANCE
Global gene expression studies identified that ergosterol biosynthesis is likely to be highly sensitive to environmental perturbations. Among them, adaptation to the host environment when A. fumigatus initiates disease is particularly interesting. The transcriptome of A. fumigatus during early stages of infection in the neutropenic murine lung identified reduced expression of genes encoding ergosterol pathway functions (Erg11, Erg24, and Erg3) relative to laboratory cultures (McDonagh et al., 2008).
Although the target site of azole activity is well studied, the role of other proteins in the mode of action of these drugs in fungi is poorly understood. Recently, a critical role for SrbA-mediated regulation of ergosterol biosynthesis and triazole drug interactions in A. fumigatus has been reported (Blosser and Cramer, 2012). SrbA was identified by transcriptional profiling under hypoxia conditions as a regulator of ergosterol biosynthetic genes in response to low oxygen levels. The ΔsrbA mutant strain was attenuated in virulence and it was hypersusceptible to fluconazole and VCZ (Willger et al., 2008). Also, several genes encoding enzymes that require high levels of oxygen were found to be transcriptionally repressed in the absence of SrbA, including the enzymes Erg6, Erg11, Erg24, Erg25, and Erg3. The relationship between ergosterol regulation and hypoxia was also demonstrated by the analysis of the sterol profile of the ΔsrbA which showed a decrease in ergosterol levels and accumulation of C-4 methyl sterols, suggesting a blockage of C-4 demethylation. In addition, the apparent control of cyp51 transcript levels by SrbA suggests an additional target for drug development. A role for SrbA in the development of triazole resistance has also been suggested, since alterations in this transcription factor or its DNA-binding affinity could transcriptionally initiate changes that could alter target abundance and would lead to triazole resistance (Blosser and Cramer, 2012).
In a similar manner, analysis of sterol composition in iron-depleted and replete cultures revealed decreased ergosterol biosynthesis during iron starvation and the accumulation of two types of sterol intermediates C-4 and Δ7. The genome-wide expression response of A. fumigatus in a shift from iron-depleted to iron-replete conditions (10, 30, 60, 120, and 240 min compared with that from 0 min) showed that Erg24 and Erg25 functions were down-regulated for all of the time points, and in contrast Erg11 was up-regulated at 30 and 60 min of iron exposure (Schrettl et al., 2008). In conditions of iron limitation, A. fumigatus uses siderophore-assisted iron uptake, which in turn uses mevalonate as a precursor. The results described above, together with the characterization of functional siderophore deleted mutants, probed the link between fungal ergosterol and siderophore biosynthesis in A. fumigatus via the compound mevalonate. The latter would appear to feed both biochemical pathways (Yasmin et al., 2012). Also, in hypoxic conditions SrbA was found to activate siderophore-mediated iron uptake in response to hypoxia and iron starvation in part by transcriptional activation of another transcription factor, HapX (Blatzer et al., 2011).
Transcriptional regulation of ergosterol biosynthesis in A. fumigatus has also been studied under antifungal exposure in laboratory cultures. Microarray analysis indicated down-regulation of the erg6 gene under both AMB and VCZ exposure (da Silva Ferreira et al., 2006; Gautam et al., 2008). Erg6 has a role in the synthesis of secondary sterols (ethyl sterols), and its down-regulation suggests that A. fumigatus alters the synthesis of secondary sterols in response to AMB or VCZ. However, different results were found for the A. fumigatus Cyp51-encoding genes, which were both down-regulated under VCZ treatment and up-regulated with AMB. These results may indicate the existence of differential responses for overcoming the distinct stresses imposed by these drugs. In addition, genes encoding for Erg24, Erg25, and Erg3 were differentially expressed when A. fumigatus cells were cultured in the presence of VCZ (da Silva Ferreira et al., 2006).
Collectively, the above observations suggest that ergosterol biosynthesis is prone to perturbation by environmental conditions, and in a manner dependent upon Erg6, Erg11, Erg24, Erg25, and Erg3 functions, thereby highlighting these gene functions as alternative or synergic inhibitors of the pathway. An important finding in A. fumigatus was the identification of C-24 ethyl sterols. These sterols can also be final metabolites of the sterol pathway and they are predominantly found in higher plants, though absent in mammalian cells, which cannot alkylate the C-24 of sterols. In plants C-24 ethyl sterols have multiple roles to play in growth and development, however, few reports exist on the detection of 24-ethyl sterols in fungi, and their role in A. fumigatus remains unknown. C-24 alkylation is catalyzed by S-adenosyl-methionine-sterol-C-methyltransferases (SAMs). In S. cerevisiae the methylation is catalyzed by erg6 which converts zymosterol into fecosterol, and at least two methyltransferases are involved in consecutive methylation reactions leading to 24-ethyl sterols in higher plants (Bouvier-Nave et al., 1998). In A. fumigatus, only one Erg6 has been identified that could transform lanosterol to eburicol and may also be involved in the production of C-24 ethyl sterols. The sterol profile analysis of A. fumigatus strains showed that the synthesis of 24-Ethylcholesta-5,7,22-trien-3β-ol is sensitive to pathway perturbations. In a Δcyp51B mutant a lack of ergosterol is compensated by an increase of 24-Ethylcholesta-5,7,22-trien-3β-ol, probably because the activity of the pathway becomes diverted toward the synthesis of alkyl sterols. However, when erg3B is suppressed, ergosterol levels are decreased together with 24-ethylcholesta-5,7,22-trien-3β-ol, confirming that this enzyme would also be involved in the desaturation of C-5,7 alkyl sterols.
Genome wide studies are providing us with a very useful platform from which to dissect the linkages between cellular sterol biosynthesis and other cell functions or metabolic pathways. Such new findings might be further explored for novel drug development or for possible combinatorial therapeutic strategies to fight invasive fungal diseases. Among them, genes encoding enzymes involved in cellular stress, cell wall synthesis, and transport have been found to be differentially expressed under antifungal exposure or underhost imposed stresses. To conclude, A. fumigatus is able to synthesize ergosterol, even with suppression of several enzymes in the pathway. Biosynthetic control under different environmental circumstances is observed, demonstrating that Aspergillus has alternatives to overcome severe drawbacks. Therefore, the precise knowledge of this complex biosynthetic pathway could facilitate the future development of novel and more selective antifungal drugs in order to improve efficacy and to minimize drug resistance.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Emilia Mellado was supported by the European Science Foundation: Fuminomics 06-RNP-132 and the Research Projects from the Spanish Ministry of Science and Innovation: SAF2008-04143 and ERA-NET Pathogenomics (7th FP), BFU2008-04709-E/BMC. Laura Alcazar-Fuoli is funded by Spanish Fondo de Investigación Sanitaria with a Miguel Servet fellowship: MPY 991/12.
REFERENCES
- Albarrag A. M., Anderson M. J., Howard S. J., Robson G. D., Warn P. A., Sanglard D., et al. (2011). Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob. Agents Chemother. 55 5113–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar-Fuoli L., Cuesta I., Rodriguez-Tudela J. L., Cuenca-Estrella M., Sanglard D., Mellado E. (2011). Three-dimensional models of 14α-sterol demethylase (Cyp51A) from Aspergillus lentulus and Aspergillus fumigatus: an insight into differences in voriconazole interaction. Int. J. Antimicrob. Agents 38 426–434 [DOI] [PubMed] [Google Scholar]
- Alcazar-Fuoli L., Mellado E., Cuenca-Estrella M., Sanglard D. (2011). Probing the role of point mutations in the cyp51A gene from Aspergillus fumigatus in the model yeast Saccharomyces cerevisiae. Med. Mycol. 49 276–284 [DOI] [PubMed] [Google Scholar]
- Alcazar-Fuoli L., Mellado E., Garcia-Effron G., Buitrago M. J., Lopez J. F., Grimalt J. O., et al. (2006). Aspergillus fumigatus C-5 sterol desaturases Erg3A and Erg3B: role in sterol biosynthesis and antifungal drug susceptibility. Antimicrob. Agents Chemother. 50 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar-Fuoli L., Mellado E., Garcia-Effron G., Lopez J. F., Grimalt J. O., Cuenca-Estrella J. M., et al. (2008). Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 73 339–347 [DOI] [PubMed] [Google Scholar]
- Bard M., Lees N. D., Turi T., Craft D., Cofrin L., Barbuch R., et al. (1993). Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids 28 963–967 [DOI] [PubMed] [Google Scholar]
- Blatzer M., Barker B. M., Willger S. D., Beckmann N., Blosser S. J., Cornish E. J., et al. (2011). SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PLoS Genet. 7:e1002374 10.1371/journal.pgen.1002374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blosser S. J., Cramer R. A. (2012). SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A). Antimicrob. Agents Chemother. 56 248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G., Perkhofer S., Haas H., Schrettl M., Würzner R., Dierich M. P., et al. (2008). Potential basis for amphotericin B resistance in Aspergillus terreus. Antimicrob. Agents Chemother. 52 1553–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier-Nave P., Husselstein T., Benveniste P. (1998). Two families of sterol methyltransferases are involved in the first and the second methylation steps of plant sterol biosynthesis. Eur. J. Biochem. 256 88–96 [DOI] [PubMed] [Google Scholar]
- da Silva Ferreira M. E., Malavazi I., Savoldi M., Brakhage A. A., Goldman M. H., Kim H. S., et al. (2006). Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr. Genet. 50 32–44 [DOI] [PubMed] [Google Scholar]
- Diaz-Guerra T. M., Mellado E., Cuenca-Estrella M., Rodriguez-Tudela J. L. (2003). A point mutation in the 14alpha-Sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47 1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermishkin L. N., Kasumov K. M., Potzeluyev V. M. (1976). Single ionic channels induced in lipid bilayers by polyene antibiotics amphotericin B and nystatine. Nature 262 698–699 [DOI] [PubMed] [Google Scholar]
- Ferreira M. E., Colombo A. L., Paulsen I., Ren Q., Wortman J., Huang J., et al. (2005). The ergosterol biosynthesis pathway, transporter genes, and azole resistance in Aspergillus fumigatus. Med. Mycol. 43 S313–S319 [DOI] [PubMed] [Google Scholar]
- Fraczek M. G., Bromley M., Bowyer P. (2011). An improved model of the Aspergillus fumigatus CYP51A protein. Antimicrob. Agents Chemother. 55 2483–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryberg M., Oehlschlager A. C., Unrau A. M. (1973). Biosynthesis of ergosterol in yeast. Evidence for multiple pathways. J. Am. Chem. Soc. 95 5747–5577 [DOI] [PubMed] [Google Scholar]
- Gautam P., Shankar J., Madan T., Sirdeshmukh R., Sundaram C. S., Gade W. N., et al. (2008). Proteomic and transcriptomic analysis of Aspergillus fumigatus on exposure to amphotericin B. Antimicrob. Agents Chemother. 52 4220–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum M. A., Rice L. B. (1999). Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance Clin. Microbiol. Rev. 12 501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooday G. W. (1995). “Cell membrane,” in The Growing Fungus eds Gow N. A. R., Geoffrey M. G. (London: Chapman & Hall; ) 62–64 [Google Scholar]
- Graminha M. A., Rocha E. M., Prade R. A., Martinez-Rossi N. M. (2004). Terbinafine resistance mediated by salicylate 1-monooxygenase in Aspergillus nidulans. Antimicrob. Agents Chemother. 48 3530–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K. C., Palacios D. S., Dailey I., Endo M. M., Uno B. E., Wilcock B. C., et al. (2012). Amphotericin primarily kills yeast by simply binding ergosterol. Proc. Natl. Acad. Sci. U.S.A. 109 2234–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadrich I., Makni F., Neji S., Cheikhrouhou F., Bellaaj H., Elloumi M., et al. (2012). Amphotericin B in vitro resistance is associated with fatal Aspergillus flavus infection. Med. Mycol. 50 829–834 [DOI] [PubMed] [Google Scholar]
- Howard S. J., Arendrup M. C. (2011). Acquired antifungal drug resistance in Aspergillus fumigatus: epidemiology and detection. Med. Mycol. 49 S90–S95 [DOI] [PubMed] [Google Scholar]
- Howard S. J., Webster I., Moore C. B., Gardiner R. E., Park S., Perlin D. S., et al. (2006). Multi-azole resistance in Aspergillus fumigatus. Int. J. Antimicrob. Agents 28 450–453 [DOI] [PubMed] [Google Scholar]
- Hu W., Sillaots S., Lemieux S., Davison J., Kauffman S., Breton A., et al. (2007). Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 3:e24 10.1371/journal.ppat.0030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. L., Lamb D. C., Corran A. J., Baldwin B. C., Kelly D. E. (1995). Mode of action and resistance to azole antifungals associated with the formation of 14 alpha-methylergosta-8,24(28)-dien-3 beta,6 alpha-diol. Biochem. Biophys. Res. Commun. 207 910–915 [DOI] [PubMed] [Google Scholar]
- Kelly S. L., Lamb D. C., Kelly D. E., Manning N. J., Loeffler J., Hebart H., et al. (1997). Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Lett. 400 80–82 [DOI] [PubMed] [Google Scholar]
- Krishnan-Natesan S. (2009). Terbinafine: a pharmacological and clinical review. Expert Opin. Pharmacother. 10 2723–2733 [DOI] [PubMed] [Google Scholar]
- Liu W., May G. S., Lionakis M. S., Lewis R. E., Kontoyiannis D. P. (2004). Extra copies of the Aspergillus fumigatus squaleneepoxidase gene confer resistance to terbinafine: genetic approach to studying gene dose-dependent resistance to antifungals in A. fumigatus. Antimicrob. Agents Chemother. 48 2490–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathu E. K., Alangaden G. J., Chandrasekar P. H. (1998). In-vitro isolation and antifungal susceptibility of amphotericin B-resistant mutants of Aspergillus fumigatus. J. Antimicrob. Chemother. 41 615–619 [DOI] [PubMed] [Google Scholar]
- Mann P. A., Parmegiani R. M., Wei S. Q., Mendrick C. A., Li X., Loebenberg D., et al. (2003). Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14alpha-demethylase. Antimicrob. Agents Chemother. 47 577–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C. M., Parker J. E., Warrilow A. G., Rolley N. J., Kelly S. L., Kelly D. E. (2010). Complementation of a Saccharomyces cerevisiae ERG11/CYP51 (sterol 14α -demethylase) doxycycline-regulated mutant and screening of the azole sensitivity of Aspergillus fumigatus isoenzymes CYP51A and CYP51B. Antimicrob. Agents Chemother. 54 4920–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonagh A., Fedorova N. D., Crabtree J., Yu Y., Kim S., Chen D., et al. (2008). Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 4:e1000154 10.1371/journal.ppat.1000154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E., Diaz-Guerra T. M., Cuenca-Estrella M., Rodriguez-Tudela J. L. (2001). Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39 2431–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E., Garcia-Effron G., Alcazar-Fuoli L., Melchers W. J., Verweij P. E., Cuenca-Estrella M., et al. (2007). A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51 1897–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E., Garcia-Effron G., Alcazar-Fuoli L., Cuenca-Estrella M., Rodriguez-Tudela J. L. (2004). Substitutions at methionine 220 in the 14alpha-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48 2747–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E., Garcia-Effron G., Buitrago M. J., Alcazar-Fuoli L., Cuenca-Estrella M., Rodriguez-Tudela J. L. (2005). Targeted gene disruption of the 14-alpha sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49 2536–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosa M. Y., Alangaden G. J., Manavathu E., Chandrasekar P. H. (2002). Resistance to amphotericin B does not emerge during treatment for invasive Aspergillosis. J. Antimicrob. Chemother. 49 209–213 [DOI] [PubMed] [Google Scholar]
- Morio F., Pagniez F., Lacroix C., Miegeville M, Le Pape P. (2012). Amino acid substitutions in the Candida albicans sterol {Delta}5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. J. Antimicrob. Chemother. 67 2131–2138 [DOI] [PubMed] [Google Scholar]
- Nascimento A. M., Goldman G. H., Park S., Marras S. A., Delmas G., Oza U., et al. (2003). Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47 1719–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes W. D., Xu S. H., Haddon W. F. (1989). Evidence for similarities and differences in the biosynthesis of fungal sterols. Steroids 53 533–558 [DOI] [PubMed] [Google Scholar]
- Paul S., Klutts J. S., Moye-Rowley W. S. (2012). Analysis of promoter function in Aspergillus fumigatus. Eukaryot. Cell 11 1167–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. M., Gardiner R. E., Park S., Martinez-Rossi N. M., Perlin D. S. (2006). A Phe389Leu substitution in ergA confers terbinafine resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 50 2533–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckenstuhl C., Lang S., Poschenel A., Eidenberger A., Baral P. K., Kohút P., et al. (2007). Characterization of squaleneepoxidase of Saccharomyces cerevisiae by applying terbinafine-sensitive variants. Antimicrob. Agents Chemother. 51 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder N. S. (1992). Terbinafine: mode of action and properties of the squaleneepoxidase inhibition. Br. J. Dermatol. 126 S2–S7 [DOI] [PubMed] [Google Scholar]
- Schrettl M., Kim H. S., Eisendle M., Kragl C., Nierman W. C., Heinekamp T., et al. (2008). SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70 27–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R. S., Robbins N., Cowen L. E. (2011). Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 75 213–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E., van der Lee H. A., Kuijpers J., Rijs A. J., Varga J., Samson R. A., et al. (2008). Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5:e219 10.1371/journal.pmed.0050219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaven J. W., Anderson M. J., Sanglard D., Dixon G. K., Bille J., Roberts I. S., et al. (2002). Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet. Biol. 36 199–206 [DOI] [PubMed] [Google Scholar]
- Tobin M. B., Peery R. B., Skatrud P. L. (1997). Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene 200 11–23 [DOI] [PubMed] [Google Scholar]
- van der Linden J. W., Snelders E., Kampinga G. A., Rijnders B. J., Mattsson E., Debets-Ossenkopp Y. J., et al. (2011a). Clinical implications of azole resistance in Aspergillus fumigatus, The Netherlands, 2007–2009. Emerg. Infect. Dis. 17 1846–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden J. W., Warris A., Verweij P. E. (2011b). Aspergillus species intrinsically resistant to antifungal agents. Med. Mycol. 49(Suppl. 1) S82–S89 [DOI] [PubMed] [Google Scholar]
- Warrilow A. G., Melo N., Martel C. M., Parker J. E., Nes W. D., Kelly S. L., et al. (2010). Expression, purification, and characterization of Aspergillus fumigatus sterol 14-alpha demethylase (CYP51) isoenzymes A and B. Antimicrob. Agents Chemother. 54 4225–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willger S. D., Puttikamonkul S., Kim K. H., Burritt J. B., Grahl N., Metzler L. J., et al. (2008). A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 4:e1000200. 10.1371/journal.ppat.1000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin S., Alcazar-Fuoli L., Gründlinger M., Puempel T., Cairos T., Blatzer M., et al. (2012). Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proc. Natl. Acad. Sci. U.S.A. 109 E497–E504 [DOI] [PMC free article] [PubMed] [Google Scholar]